Background: The sialic acid N-glycolylneuraminic acid (Neu5Gc) shows conserved suppression of expression in vertebrate brains, suggesting brain-specific toxicity.
Results: α2–8-Linked Neu5Gc incorporated into the neural glycan polysialic acid (polySia) resists sialidase breakdown through conformational effects.
Conclusion: Neu5Gc in brain would prevent rapid turnover of surface polySia.
Significance: This mechanism potentially underlies the evolutionary suppression of Neu5Gc expression in vertebrate brains.
Keywords: Carbohydrate Chemistry, Carbohydrate Metabolism, Glycobiology, Glycosidases, Sialic Acid, N-Glycolylneuraminic Acid, Polysialic Acid, Sialidase
Abstract
The sialic acid (Sia) N-acetylneuraminic acid (Neu5Ac) and its hydroxylated derivative N-glycolylneuraminic acid (Neu5Gc) differ by one oxygen atom. CMP-Neu5Gc is synthesized from CMP-Neu5Ac, with Neu5Gc representing a highly variable fraction of total Sias in various tissues and among different species. The exception may be the brain, where Neu5Ac is abundant and Neu5Gc is reported to be rare. Here, we confirm this unusual pattern and its evolutionary conservation in additional samples from various species, concluding that brain Neu5Gc expression has been maintained at extremely low levels over hundreds of millions of years of vertebrate evolution. Most explanations for this pattern do not require maintaining neural Neu5Gc at such low levels. We hypothesized that resistance of α2–8-linked Neu5Gc to vertebrate sialidases is the detrimental effect requiring the relative absence of Neu5Gc from brain. This linkage is prominent in polysialic acid (polySia), a molecule with critical roles in vertebrate neural development. We show that Neu5Gc is incorporated into neural polySia and does not cause in vitro toxicity. Synthetic polymers of Neu5Ac and Neu5Gc showed that mammalian and bacterial sialidases are much less able to hydrolyze α2–8-linked Neu5Gc at the nonreducing terminus. Notably, this difference was not seen with acid-catalyzed hydrolysis of polySias. Molecular dynamics modeling indicates that differences in the three-dimensional conformation of terminal saccharides may partly explain reduced enzymatic activity. In keeping with this, polymers of N-propionylneuraminic acid are sensitive to sialidases. Resistance of Neu5Gc-containing polySia to sialidases provides a potential explanation for the rarity of Neu5Gc in the vertebrate brain.
Introduction
The sialic acid N-acetylneuraminic acid (Neu5Ac)7 and its derivative N-glycolylneuraminic acid (Neu5Gc) differ only by a single oxygen atom, are widely synthesized throughout most animals of the deuterostome lineage, and are commonly positioned as the terminal residues of the glycoconjugates that cover all cell surfaces (1, 2). The conversion of CMP-Neu5Ac to CMP-Neu5Gc is catalyzed by a single highly conserved enzyme, cytidine monophosphate N-acetylneuraminic acid hydroxylase (CMAH) (3–5). Decades of studies indicate that the Neu5Gc fraction of total sialic acids (Sias) is highly variable among tissues and between species (6–33). The notable exception appears to be the vertebrate brain. Although this tissue contains more total Sias than any other (34), Neu5Gc is reported to be present at only trace levels, if at all. Studies of mouse and pig tissues have accordingly found Cmah mRNA to be undetectable in brain (35, 36). As we confirm and expand in this study, no other tissue displays this unusual suppression of Neu5Gc expression across vertebrate taxa.
The evolutionarily conserved suppression of Neu5Gc expression in the vertebrate brain suggests that a brain-specific detrimental effect requires its consistent down-regulation. We have considered a number of possible mechanisms to explain this observation. There is no evidence to suggest that CMAH has any function beyond the conversion of CMP-Neu5Ac to CMP-Neu5Gc. The only known homologue of CMAH is N-acetylmuramic acid hydroxylase (namH), which also converts an N-acetyl group to an N-glycolyl group on muramic acid in the actinomycete bacteria (37). Also, Cmah−/− mice do not manifest any gross defects in brain development (38). It is therefore reasonable to assume that CMAH is responsible primarily for the synthesis of Neu5Gc and that any detrimental effect results from the presence of Neu5Gc and not from a secondary effect of CMAH.
Neu5Ac is widely prevalent on a variety of sialoglycoconjugates (SGCs) in vertebrate brain tissue. We considered the possibility that Neu5Gc may interfere with the synthesis, recognition, or function of one or more SGCs. However, although sialyltransferases or Sia-binding proteins may have a relative preference for Neu5Ac or Neu5Gc, they will generally accept either Sia (39). Even in the cases where there is selectivity, biological function is not likely to require such a remarkably low fraction of Neu5Gc to be maintained. Similarly, whereas neural lectins such as the myelin-associated glycoprotein do prefer Neu5Ac to Neu5Gc (40), a minority of Neu5Gc should not interfere with the recognition of a majority of Neu5Ac. Rather, based on the remarkably low amounts of Neu5Gc that are maintained in vertebrate brains, we presume that even a very small amount of Neu5Gc must cause toxicity. As a first step toward testing this hypothesis, we propose that the presence of Neu5Gc in SGCs in the vertebrate brain critically impairs their degradation. This model allows for even a very small fraction of Neu5Gc to exert substantial toxicity through widespread resistance of underlying glycans to breakdown.
Of course, any proposed mechanism underlying Neu5Gc toxicity must selectively impact brain tissue. As a first step toward testing our hypothesis that Neu5Gc impairs degradation of brain SGCs, we propose a candidate glycan for this mechanism, polysialic acid (polySia, sometimes called PSA). PolySia is a homopolymer of α2–8-linked Neu5Ac, most prevalent on the neural cell adhesion molecule (NCAM) (41, 42), and is also found in the capsules of certain neuroinvasive bacteria (43). Polysialylated NCAM is highly expressed through embryonic and early postnatal development and plays critical roles in neurite outgrowth and plasticity (44). Its absence in mice causes severe developmental phenotypes (45, 46), and its dysregulation has been implicated in neural disorders such as schizophrenia (46–50).
The cellular mechanisms responsible for polySia breakdown are not well understood. The molecule has been shown to undergo an intramolecular self-cleavage into short oligomers over many hours at mildly acidic pH (51), under conditions that would be found in the lysosome. However, the resulting oligomers are resistant to further intramolecular self-cleavage, and these would presumably require further digestion by the prevalent lysosomal mammalian sialidase NEU1. Notably, NEU1 has also been implicated in desialylation of cell surface SGCs (52, 53), another process that could be critical in the appropriate regulation of polySia.
It is well established that bacterial and viral sialidases have a relative preference for Neu5Ac over Neu5Gc in α2–3 and α2–6 linkages (54–56). Relatively little is known about the four mammalian sialidases NEU1–4 in this regard (57). Only NEU2 has been crystallized to date (58), although the other related sialidases have been homology-modeled to this structure (59). Unlike bacterial sialidases, human NEU2 has been demonstrated to have very similar activity on Neu5Ac and Neu5Gc in α2–3 or α2–6 linkage to an underlying galactose residue (60). To our knowledge, there are no reports examining the cleavage of α2–8-linked Neu5Gc by vertebrate exosialidases.
We have taken a biochemical approach to address the question of whether NEU1 and other vertebrate sialidases are able to digest α2–8-linked Neu5Gc. We compare here the chemical and enzymatic breakdown of polymers of α2–8-linked Neu5Ac and Neu5Gc under various conditions, showing that polymers containing Neu5Gc exhibit an unusual resistance to breakdown by sialidases, including NEU1. Using molecular dynamics modeling, we propose a structural mechanism by which Neu5Gc in an α2–8 linkage is less likely to be found in the conformation optimal for enzymatic cleavage. The proposed mechanism is supported by replacement of the glycolyl moiety with a propionyl group, which, while occupying a similar spatial arrangement to the glycolyl residue, negates the stability. Given the critical importance of developmental regulation of polySia in the central nervous system and the likely need for its rapid turnover under specific circumstances, this finding provides the first steps toward an explanation for the evolutionary conservation of the suppression of Neu5Gc synthesis in the vertebrate brain.
EXPERIMENTAL PROCEDURES
DMB-HPLC Analysis of Sialic Acids
Quantification of Sia content and type on acid-hydrolyzed samples of vertebrate tissues, polymers, and disaccharides was done using previously described methods of DMB derivatization at 50 °C for 2.5 h (61) followed by HPLC on a Phenomenex C18 column using an isocratic elution in 85% water, 7% methanol, and 8% acetonitrile. Other details are in an accompanying paper (see Ref. 62).
For samples in which heated DMB derivatization would cause unwanted breakdown of polySia in solution (partial enzymatic and acid-catalyzed breakdown reactions of polymers and disaccharides), an extended derivatization reaction was used, for 48 h at 4 °C, that allowed labeling of all reducing ends in solution without the destruction of polymers (63). To quantify the monomer following such reactions, the derivatization reaction was not quenched, and samples were run on a Phenomenex C18 column as described above. To observe polySias, reactions were quenched with the addition of 0.2 volume 1 m NaOH and analyzed on a Dionex DNAPac PA-100 column along a 2–35% gradient of 1 m NaNO3 in water (63).
Studies of Vertebrate Tissue Neu5Gc Content
Brain samples from mouse, rat, pig, and cow were obtained from Pel-Freez Biologicals, Rogers, AR; chimpanzee brain samples were from the Yerkes Primate Center, Atlanta, GA; dolphin liver and brain samples were obtained from the Southwest Fisheries Science Center, La Jolla, CA. Dolphin milk samples were obtained from the United States Navy marine mammal program, Point Loma. African and Asian elephant liver samples were obtained from the Zoological Society of San Diego. All samples were rinsed in PBS to remove blood, and 1 g of sample was homogenized using a Polytron in 2 m acetic acid directly or in methanol chloroform for separate analysis of glycolipids and glycoproteins. Monomeric sialic acids were then released from bound glycans by treatment with 2 m acetic acid for 3 h at 80 °C and quantified by DMB-HPLC.
Cell Culture and Flow Cytometry
SH-SY5Y cells (ATCC) were cultured in DMEM/F-12 (Invitrogen) with 10% human serum (Valley Biomedical Inc.) and supplemented with 2 mm Neu5Ac or Neu5Gc (Nacalai) for 1 week. Cells were lifted with 2 mm EDTA in PBS. Neu5Gc incorporation in the 2 mm Neu5Gc-supplemented cells was confirmed as 76.5% of total Sias by complete acid hydrolysis of cell pellets in 0.1 m trifluoroacetic acid (TFA) for 4 h at 80 °C followed by DMB-HPLC. Neu5Gc was not detected in 2 mm Neu5Ac-supplemented cells. One aliquot of suspended cells was treated with endo-NF (40 ng/μl) (64) in PBS for 45 min on ice. Cells were stained with 12E3 (10 μg/ml) followed by anti-mouse IgM secondary antibody. Concurrently, a separate aliquot was stained with inactive endo-NA-GFP (10 μg/ml) (65). Flow cytometry was conducted on a FACSCalibur. Cells were gated on a population of cells positive for polySia and analyzed with FlowJo software (TreeStar).
Preparation of Sialic Acid Dimers
A mixed polymer of Neu5Ac and Neu5Gc (1 mg) was hydrolyzed with 50 mm sodium acetate buffer, pH 4.8, at 50 °C for 20 h. The hydrolysate was subjected to DEAE-Sephadex A-25 anion-exchange chromatography (1 × 10 cm, pre-equilibrated in 10 mm Tris-HCl, pH 8.0). After washing the column with 3 volumes of 10 mm Tris-HCl, pH 8.0, oligo-Sias were eluted with a linear gradient of NaCl (0–0.4 m NaCl in 10 mm Tris-HCl, pH 8.0) (66). The di-Sia fraction was collected, desalted with Sephadex G-25 chromatography (1 × 10 cm, water), and lyophilized. Dried di-Sia was then dissolved in 200 μl of water. Neu5Acα2–8Neu5Ac, Neu5Acα2–8Neu5Gc, Neu5Gcα2–8Neu5Ac, and Neu5Gcα2–8Neu5Gc were separated and purified by preparative thin layer chromatography (tlc) as described previously (66). Each fraction was subjected to the Sephadex G-25 chromatography and lyophilized. The identity of each disaccharide was confirmed by hydrolysis in 0.1 m HCl for 1 h at 80 °C followed by DMB-HPLC.
Synthesis of Sialic Acid Polymers
De-N-acetylation of PolySia
PolySia was de-N-acetylated as described previously (67). N-Acetyl-polySia (Lipoxen, 500 mg, 1.6 mmol) was dissolved in a solution of 2 m sodium hydroxide (20 ml) with sodium borohydride (50 mg, 1.3 mmol). The solution was heated at reflux. To achieve over 90% deacetylation, the polymer was refluxed for 50 h, during which time a white precipitation was seen. The deacetylated polymer intermediate was dialyzed (dialysis tubing molecular mass cutoff of 3000 daltons) with a solution of ammonium carbonate (0.01 m) for 4 h and then water overnight. The solution was dried under vacuum to yield a white powder (197 mg, 45%). The polymer could now be reacetylated, glycolated, or propionylated as described below.
Ac100
Deacetylated polymer (98 mg, 0.04 mmol) was slurried in water (5 ml) with sodium bicarbonate (492 mg, 6 mmol). Acetic anhydride (612 μl, 6 mmol) was added, and the solution stirred vigorously for 30 min. The solution was filtered and then dialyzed as described above. The N-acetylation procedure was repeated on the isolated material. The solution was dried under vacuum to yield a white powder (99 mg, 89%). DMB-HPLC analysis of the acid-hydrolyzed polymer revealed one single peak, which correlated (qualitatively and quantitatively) with an Neu5Ac standard. In addition, ion-exchange polymer profiles of Ac100 were comparable with the polySia starting material. For 1H NMR (D2O, 500 MHz), the CH region of the polymer was broad, due to the polymeric nature of the molecule; characteristic peaks were seen in the spectra, which was in good agreement with a standard of the polySia starting material (ppm) as follows δ = 1.5 (m, 1H, CHH), 1.8 (s, 3H, OCH3), 2.6 (m, 1H, CHH), and 3.4–4.2 (CH region, 7H).
Gc100
Deacetylated polymer (70 mg, 0.03 mmol) was slurried in water (2 ml) with sodium bicarbonate (176 mg, 2 mmol). Acetoxyacetyl chloride (118 μl, 1.5 mmol) was added, and the reaction mixture was stirred vigorously for 30 min. An additional portion of sodium bicarbonate and then acetoxyacetyl chloride were added, and the reaction mixture was stirred for an additional 30 min. The solution was filtered and then dialyzed as described above. The product was dissolved in 0.1 m sodium hydroxide solution (5 ml) and heated at 50 °C for 30 min. The solution was then dialyzed as described above and then dried under vacuum to yield a white powder (74 mg, 76%). The white powder was analyzed for composition by DMB-HPLC. To ensure no free amine residues remained, the product was put through the acetoxy-acetylation procedure described above again, and the composition was analyzed again by DMB-HPLC. If no difference was seen in composition of Sia content, we then interpreted this as no free amines remaining. If a difference was seen, the procedure was repeated until the composition remained constant. For 1H NMR (D2O, 500 MHz), the characteristic peaks were seen (ppm): δ = 1.5 (m, 1H, CHH), 1.8 (s, 0.1H, OCH3), 2.6 (m, 1H, CHH), 3.4–4.2 (CH region, 7H), and 4.0 (s, 1.9H, COCH2OH).
Mixed Polymers, Gc40 and Gc60
Mixed polymers were made and analyzed using a similar procedure as described above for Gc100, using a mixture of acetic anhydride and acetoxyacetyl chloride. Gc40 was generated using a 1:1 premix of acetoxyacetyl chloride and acetic anhydride. Gc60 was generated using a 5:3 premix of acetoxyacetyl chloride and acetic anhydride, respectively.
Pr70
Polymers containing monomeric residues of N-propionylneuraminic acid (Neu5Pr) were generated in a similar way as described under the mixed polymer method. Pr70 was generated using a 4:1 premix of propionyl chloride and acetic anhydride, respectively. The composition was confirmed as described above, namely checking the composition after running the polymer through additional acetylation reactions.
Endo-NF Digests of Polymers
200 μg of polymers were subjected to digest with 40 ng of endo-NF (64) in 100 mm sodium phosphate buffer, pH 7.4, for 45 min at 37 °C. The reaction was terminated by the addition of an equal volume of ice-cold EtOH; samples were dried under vacuum and resuspended in water. Digestion to oligomers of 3–7 Sias in length was confirmed by extended DMB labeling of reducing Sias followed by HPLC on a DNAPac anion-exchange column (63, 68). To determine concentration of polymers, endo-NF-digested samples were then hydrolyzed in 0.1 m TFA at 80 °C for 4 h and quantified using DMB-HPLC.
Preparation of Inactive Endo-NA-GFP Probe
The pQE31-based construct for inactive endo-NA-eGFP (65) was transformed to the M15 [pREP4] (Qiagen) expression strain and expressed as a histidine-tagged fusion protein. The cells were grown in SOB medium (2% tryptone, 0.5% yeast extract, 8.5 mm NaCl, 10 mm MgCl2, 10 mm MgSO4) to A600 nm ∼0.4, and expression was induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside for 3 h at room temperature. After centrifugation at 3000 × g for 20 min, the pellet was stored at −80 °C. The cells were thawed on ice and resuspended in 50 mm sodium phosphate buffer, pH 7.4, 300 mm NaCl, 20 mm imidazole. After adding complete EDTA-free protease inhibitor mixture (Roche Applied Science) and lysozyme to a final concentration of 5 mg/ml, the cells were incubated for 30 min on ice and lysed by sonication. Cell wall debris was separated by centrifugation twice at 10,000 × g at 4 °C for 20 min. Nickel-nitrilotriacetic acid resin (Qiagen) equilibrated with the above phosphate buffer was added to the supernatant and stirred at 4 °C for 1 h. The protein-resin complex was washed five times as a batch with 8× (v/v) excess of the buffer and packed into a column. The packed column was washed with the buffer until A280 ≤0.01. Bound protein was eluted by adding imidazole to 100 mm concentration. The buffer was changed using Amicon Ultra filter devices (Millipore) to 50 mm sodium phosphate, pH 7.4, 300 mm NaCl.
Preparation of Sialidases
Murine NEU1
NEU1 sialidase was purified from mouse kidney tissue by affinity chromatography on a concanavalin A-Sepharose column followed by fast protein liquid chromatography gel filtration on Superose 6 column, as described previously (69).
Rat NEU1
COS-7 cells (2 × 106 cells) were transfected with rat NEU1 (rNEU1) plasmid or mock plasmid using the GeneJuice transfection reagent (Novagen) and incubated for 48 h. After washing, transfected cells were collected, sonically disrupted in PBS containing protease inhibitors (1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml of leupeptin, 1 μg/ml pepstatin A, and 1 mm EDTA) and centrifuged at 600 × g at 4 °C for 10 min to remove debris and nucleus. The supernatant was ultracentrifuged at 100,000 × g at 4 °C for 1 h. The precipitate was washed with cold PBS containing protease inhibitors and ultracentrifuged. The precipitate, representing the membrane fraction, was dissolved in PBS containing protease inhibitors, and then used as the rNEU1 enzyme fraction.
Murine NEU2 and NEU4
COS-7 cells cultured in 10-cm dishes in Eagle's minimal essential medium supplemented with 10% (v/v) fetal calf serum (Wisent) and 5% DMSO were transfected with pCTAP-Neu2 and pCTAP-Neu4 plasmids using Lipofectamine LTX (Invitrogen) as described in the manufacturer's protocol. 48 h post-transfection, cells were washed with PBS and harvested by scraping. Cell pellets from 10 dishes were resuspended in 2 ml of lysis buffer from InterPlay TAP purification kit (Stratagene) supplemented with 0.1% Nonidet P-40 and Sigma protease and phosphatase inhibitor mixture (P8340, 10 μl per ml of cell suspension). The homogenates were sonicated for 5 s to solubilize proteins. The suspension was then centrifuged at 13,000 × g for 30 min. The supernatant was first passed through 0.4 ml of avidin-agarose resin (Sigma A9207) and then affinity purification of TAP-tagged Neu2 and Neu4 was performed using streptavidin resin (Stratagene) according to the manufacturer's protocol. Purified enzymes were stabilized in 20% glycerol and stored at −20 °C until use.
AUS
Arthrobacter ureafaciens sialidase (AUS) was purchased from EY Laboratories.
Enzyme Digests of 4-Methylumbelliferyl Sialic Acids
4-Methylumbelliferyl (4MU) Neu5Ac was purchased from Nacalai (Japan). 4MU-Neu5Gc and 4MU-Kdn were from Dr. Kimio Furuhata (70). 25 nmol of 4MU-Sia was digested in 90 μl of 50 mm sodium acetate, pH 4.75 (NEU1), or PBS, pH 6.5 (AUS). 20-μl aliquots were quenched at each time point by the addition of 180 μl of stop buffer (0.1 m glycine, 25% EtOH, pH 10.7). Fluorescence was read in a 96-well plate reader at 365 nm excitation and 450 nm emission.
Rat NEU1 digests were conducted separately at 37 °C in 50 mm sodium acetate buffer, pH 4.5, 100 μm 4MU-Sia, 0.1% BSA, 0.1% Triton X-100 and enzyme fractions (10 μl) in a final volume of 0.1 ml. The reaction was terminated by addition of 1.0 ml of 0.25 m glycine-NaOH, pH 10.4, and the amount of released 4-MU was fluorometrically determined with FluoroMax-3 (365 nm excitation and 448 nm emission). The rat NEU1 activity was calculated based on the activity for membrane fraction of mock-transfected COS-7 cells and natural degradation of the substrate (negative control).
Enzyme Digests of PolySia
1 nmol (Sia equivalent) of endo-NF-digested polymers Ac100, Gc40, Gc60, Gc100, and Pr70 were further digested at 37 °C in 50 mm sodium acetate, pH 4.75 (NEU1 and NEU4), pH 5.5 (NEU2), or PBS, pH 6.5 (AUS). For all samples except Pr70, 250 pmol of Neu5Pr were added to each tube as an internal standard. All samples were run in parallel with a no-enzyme and enzyme-only control. Total acid hydrolysis in 0.1 m TFA at 80 °C for 4 h was performed in parallel. At each time point, aliquots were removed and flash-frozen in liquid nitrogen. All samples were diluted 5-fold in water and derivatized for 48 h at 4 °C using an extended DMB protocol that allows labeling of monomer but does not cause further breakdown of polySia. Analysis was then performed by HPLC on a Phenomenex C18 column, with fluorescence detection.
Total monomeric Neu5Ac, Neu5Gc, and Neu5Pr were quantified by integration of spectra compared with standards for each sample. Each run was normalized according to known concentrations of Neu5Pr (this was not done for the Pr70 sample). No-enzyme and enzyme-only control values were subtracted from the corresponding time point for each sample. Values at t = 0 were also subtracted from values for all time points. Total enzymatic breakdown of Neu5Ac, Neu5Gc (Gc40, Gc60, and Gc100), and Neu5Pr (Pr70 only) was determined as a fraction of total monomer calculated from TFA-hydrolyzed samples. To independently measure Neu5Ac and Neu5Gc breakdown within the Gc40 polymer, released monomer was calculated as a fraction of the total Neu5Ac or Neu5Gc content of the polymer.
Digests of 1 nmol of disaccharides of Neu5Ac and Neu5Gc were conducted and analyzed as described above. Reactions were conducted in triplicate. In parallel, disaccharides were hydrolyzed in 0.1 m HCl for 1 h at 80 °C to confirm total possible monomer release.
Analysis of Polymer Breakdown under Acidic Conditions
Analysis of Monosaccharide Release by DMB-HPLC
Ac100 or Gc100 polymers were dissolved in HCl-KCl, citrate phosphate, or phosphate-based buffers at pH of 1, 2, 4.5, and 7.5 at a concentration of 20 μg/ml (total volume = 500 μl). Samples were heated at 80 °C, and aliquots (40 μl) were taken at hourly intervals. The pH of samples was corrected to pH 1.0 on ice using TFA (13 m) prior to DMB derivatization. Samples were derivatized using the extended DMB reaction followed by HPLC analysis of monomer release.
Analysis of Glycosidic Linkage Hydrolysis at pH 4.5 and 37 °C Using a Reducing Sugar Assay
Ac100 or Gc100 polymers (0.4 mg) were dissolved in an acetate buffer (pH 4.5, 200 μl) and incubated at 37 °C. 20-μl samples were taken at regular time intervals over a 200-h time period. Before analysis, samples were stored at −20 °C. In parallel, complete hydrolysis was achieved by taking a sample of the polymer in acetate buffer and heating at 80 °C with 0.1 m H2SO4 for 1 h. Samples were analyzed using a reducing end assay variant of the Mopper-Grindler method (71, 72). Standards of monomeric Neu5Ac or Neu5Gc (1–20 nmol) were used to quantify the hydrolysis of the polymer linkage. A fractional value was obtained by comparing the molar breakdown to a completely hydrolyzed polymer sample.
Molecular Dynamics Modeling
Hexamers of α2–8-linked Neu5Ac, Neu5Gc, and Neu5Pr were developed using the GLYCAM06 structure library and parameter sets (73). Charges for Neu5Ac and Neu5Gc were obtained from this parameter set. Charges for the Neu5Pr residue were calculated from a quantum mechanically optimized Neu5Pr with a reducing O-methyl aglycone (HF/6–31++g**). This model was used to obtain the restrained electrostatic potential at HF/6–31++g**. Ensemble averaged charges for Neu5Pr were obtained from 100 equally spaced snapshots from a 10-ns molecular dynamics simulation using HF/6–31++g** and a restrained electrostatic potential weight of 0.01 (74), consistent with the formalism used in GLYCAM06 (73). Simulations were performed using the pmemd module of AMBER 11 (75). All simulations followed a standard protocol of minimization (5000 steps conjugate gradient and 5000 steps steepest descent), heating (50 ps from 5 to 300 K), equilibration (300 K temperature throughout 100 ps for ensemble averaged charge development and 10 ns for the hexasaccharides), and production (300 K throughout 10 ns for ensemble averaged charge and 0.5 μs for the hexasaccharides). The molecular dynamics simulations were performed using an nPT setup at 1 atm, employing a Berendsen-type (76) thermostat and barostat, with temperature and pressure coupling constants of 10 and 0.1 ps, respectively. Sodium ions were employed to neutralize the system charge, and a cubic box of transferable intermolecular potential 3 point waters (77) was placed with a minimum of 12 Å between the box edge and solute. Direct nonbonded interactions were truncated at 10 Å, and the Particle Mesh Ewald method (78) was used to treat long range electrostatics beyond this cutoff. Scaling factors for 1,4-interactions were not employed, and high frequency motions involving hydrogen atoms were restrained using the SHAKE algorithm (79). Initial conformations for all three homopolymers were symmetrized from the dominant conformer identified from a previous study by Yongye et al. (80) for di- and trisaccharides of similar systems. Conformations were extracted and analyzed every 1 ps throughout the molecular dynamics simulation. Glycosidic torsion angles ϕ (C1-C2-O8′-C8′), ψ (C2-O8′-C8′-H8′), ω8 (H8-C8-C7-H7), and ω7 (H7-C7-C6-H6) between the nonreducing terminal residues were used to check for convergence and identify the geometries and relative populations. NMR J-couplings for the entire hexasaccharide were consistent with experimental di- and trisaccharide values presented in Yongye et al. (80).
RESULTS
Relative Absence of Neu5Gc Is a Conserved Feature of Vertebrate Brain
Prior studies of sialic acids in the vertebrate brain have either failed to find Neu5Gc or reported it to be present only at very low levels (10, 26, 81–88). To confirm and extend these findings, we collected samples of brain tissue from chimpanzee, mouse, rat, cow, pig, and dolphin. Elephant liver and dolphin liver and milk were also analyzed. High performance liquid chromatography (HPLC) was used to characterize the Neu5Ac and Neu5Gc fractions of total Sias in tissues of these mammalian taxa. The combined data, including those from prior publications, are shown in Table 1. These data support previous observations that although Neu5Gc is widely and variably distributed in other tissues in various species, it is relatively low or absent from all vertebrate brains. This is corroborated by RT- quantitative PCR data of mouse tissues from our laboratory (data not shown), as well as published Cmah gene expression data from pig that demonstrate highly variable expression in most tissues, with the brain as the only tissue with markedly lowered levels (35). It is clear from these results that Neu5Gc and CMAH are otherwise highly variable in their expression, both between species and among tissues within an individual species. In striking contrast, Neu5Gc is expressed at very low levels in the brain of all species studied. This suppression of expression is highly conserved across vertebrates, including the frog.
TABLE 1.
Distribution of Neu5Gc in vertebrate tissues
The Neu5Gc fraction of total Sias in tissues was compared across vertebrates. Table combines data from the literature with that obtained from samples studied in our laboratory. Neu5Ac and Neu5Gc fractions of samples in our laboratory were determined by total acid hydrolysis of tissue lysate followed by DMB-HPLC. Conserved suppression of Neu5Gc in the brain is unusual among vertebrate tissues. The following symbols are used: ++, major fraction; +, minor fraction; −, absent; Trace, present at 0.8–3%; NR = not reported. Literature references are in the main text.
| Species | Serum | RBC | Submaxillary gland | Liver | Kidney | Milk | Brain |
|---|---|---|---|---|---|---|---|
| Human | − | − | NR | NR | − | − | − |
| Chimpanzee | NR | ++ | NR | + | + | +a | Traceb |
| Macaque | + | + | NR | NR | NR | + | − |
| Mouse | + | +b | − | ++ | NR | NR | Traceb |
| Rat | + | +b | + | + | + | NR | Traceb |
| Rabbit | Trace | + | NR | − | + | NR | − |
| Pig | NR | ++ | ++ | + | + | NR | Traceb |
| Cow | ++ | ++ | + | NR | ++ | Trace | Traceb |
| Sheep | + | ++ | Trace | + | + | ++ | Trace |
| Elephant African | NR | NR | NR | ++a | NR | + | NR |
| Elephant Asian | NR | NR | NR | ++a | NR | − | NR |
| Dolphin | NR | NR | NR | ++a | ++ | +a | Tracea |
| Horse | + | ++ | Trace | − | + | NR | Trace |
| Chicken | − | − | − | − | − | − | − |
| Xenopus | NR | NR | NR | NR | NR | NR | − |
a Data are from our laboratory.
b Published data were confirmed in our laboratory.
Neu5Gc Is Efficiently Incorporated into Polysialic Acids on Neural Cells
Polysialic acid is synthesized in the mammalian brain by two polysialyltransferases, ST8SiaII (STX) and ST8SiaIV (PST) (89–92). Many sialyltransferases studied to date have some preference but not outright specificity for CMP-Neu5Ac or CMP-Neu5Gc (39). With regard to ST8SiaII and -IV, the situation is less clear. Although they have been found to be variably active on a number of unnatural sialic acids (93–96), their activity on CMP-Neu5Gc has not been reported.
We have previously shown that cells in culture will take up free Neu5Gc from the culture medium by macropinocytosis and, furthermore, that they will incorporate Neu5Gc into endogenous SGCs (97). We exploited this finding to determine whether neural cells are capable of incorporating Neu5Gc into polySia. SH-SY5Y cells, a human neuroblastoma line that expresses both ST8SiaII and ST8SiaIV and synthesizes PSA-NCAM (98), were grown for 1 week in medium supplemented with 2 mm Neu5Ac or Neu5Gc. This extended time line was chosen to allow adequate turnover of the long surface polymers. After 1 week, surface polySia was measured using flow cytometry. Cells were stained with 12E3, an antibody against α2–8-linked Neu5Ac (99, 100). 12E3 showed reduced binding in cells supplemented with 2 mm Neu5Gc (Fig. 1), indicating either reduced synthesis of polySia or impaired binding of the antibody due to Neu5Gc incorporation.
FIGURE 1.
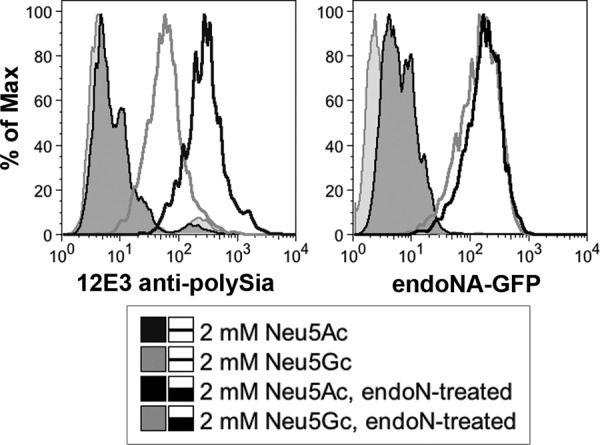
Neuroblastoma cells can incorporate Neu5Gc into endogenous PSA-NCAM. The human neuroblastoma cell line SH-SY5Y was cultured in the presence of 2 mm Neu5Ac or Neu5Gc for 1 week. An antibody to polymers of Neu5Ac (12E3, left panel) showed reduced staining in the Neu5Gc-supplemented cells. Cells were also stained with a fluorescent polySia-binding probe that does not demonstrate Neu5Ac or Neu5Gc specificity (inactive endo-NA-GFP, right panel). Here, staining was comparable between Neu5Ac- and Neu5Gc-supplemented cells. Endo-N treatment indicates negative control cells treated with endoneuraminidase from K1F.
To distinguish between these two scenarios, cells were further stained with inactive endo-NA-GFP (65). This molecule is a modified version of an endoneuraminidase from the bacteriophage PKIA. PK series endoneuraminidases such as this one have been shown to have homology to the endoneuraminidase from phage K1F (101), which has been shown to cleave polymers of both Neu5Ac and Neu5Gc (66, 102). In this inactivated form, endo-NA-GFP binds but does not cleave polySia. Unlike 12E3, inactive endo-NA-GFP demonstrated equal binding in both Neu5Ac- and Neu5Gc-supplemented cells.
Taken together, these data indicate that Neu5Gc can be incorporated into endogenous polySia in SH-SY5Y cells. Thus, the presence of Neu5Gc does not appear to impair the synthesis of polySia. If Neu5Gc were present in the vertebrate brain, it is therefore likely that it would be incorporated into PSA-NCAM on cell surfaces in vivo.
No Detectable Difference in Relative Rates of Release of α-Linked Neu5Ac and Neu5Gc by NEU1
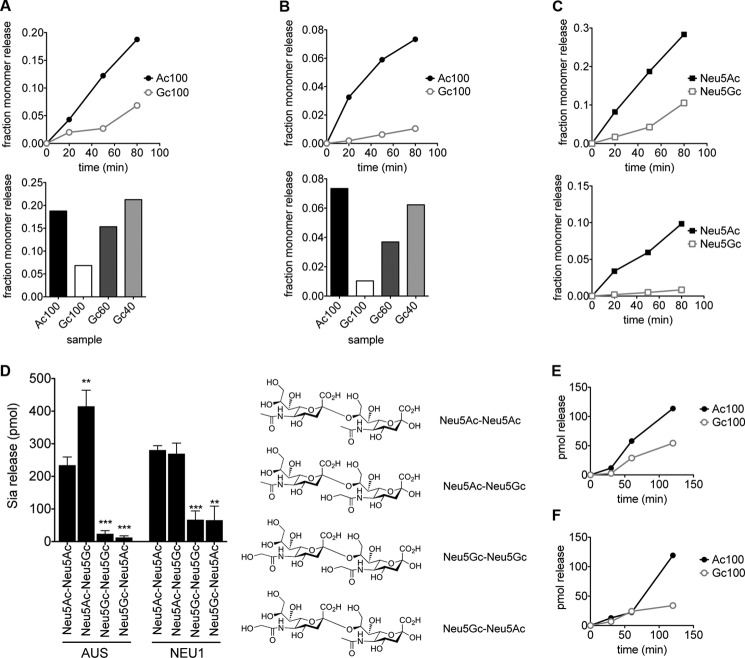
Many bacterial sialidases have been shown to exhibit a relative preference for α2–3- or 2–6-linked Neu5Ac over Neu5Gc (54, 55). To determine the intrinsic preference of sialidases for recognizing Neu5Ac or Neu5Gc independent of linkage, we assessed their ability to digest substrates of 4MU Neu5Ac, Neu5Gc, and the related nonulosonic acid, deaminoneuraminic acid (Kdn). Digests were performed either with the well characterized AUS or with preparations of murine NEU1 from liver and kidney. Both enzymes demonstrated very similar activity on substrates containing Neu5Ac and Neu5Gc (Fig. 2). Neither sialidase demonstrated appreciable activity on 4MU-Kdn. A similar preference was seen with a preparation of NEU1 from rat liver (data not shown).
FIGURE 2.
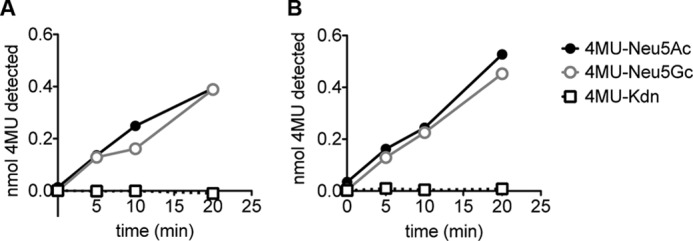
Sialidases exhibit no detectable preference for Neu5Ac or Neu5Gc. 1 nmol of substrates of Neu5Ac, Neu5Gc, and Kdn α-linked to the fluorescent molecule 4-methylumbelliferone were digested with a murine sialidase (NEU1, A) and a bacterial sialidase (AUS, B). Enzymatic breakdown at each time point was calculated by subtracting fluorescence in the absence of enzyme to fluorescence in enzyme digest samples.
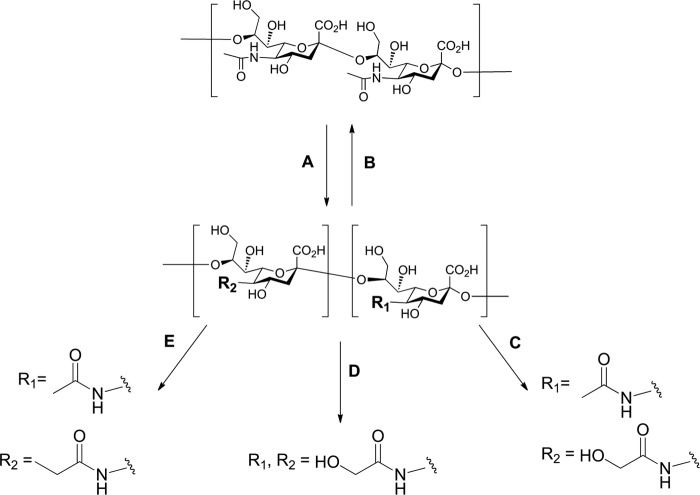
Synthesis and Characterization of α2–8-Linked Sialic Acid Polymers with Varying Ratios of Different N-Acyl Groups
Because natural sources of α2–8-linked sialic acid polymers containing Neu5Gc are rare, we adapted current synthetic methods (67, 103) to generate polymers containing varying levels of Neu5Gc and Neu5Ac (Fig. 3). Briefly, the N-acetyl groups of polySia were removed under strong basic conditions to reveal free amines. The molecule could then be re-N-acetylated, N-glycolylated, or a mixture of both obtained by reacting with acetic anhydride and/or acetoxyacetyl chloride in an aqueous solution of sodium bicarbonate. The final composition was determined using NMR spectroscopy and HPLC analysis. Fig. 4 shows the polymers used in this study as follows: 100% Neu5Ac (Ac100); 40% Neu5Gc and 60% Neu5Ac (Gc40); 60% Neu5Gc and 40% Neu5Ac (Gc60); 100% Neu5Gc (Gc100), and 70% Neu5Pr and 30% Neu5Ac (Pr70).
FIGURE 3.
Synthetic strategy to generate polymers of N-acetyl, N-glycolyl, or N-propionylneuraminic acid (Neu5Ac, Neu5Gc, and Neu5Pr, respectively; see also Table 2). A, from commercial colominic acid (bacterial N-acetyl polySia, Lipoxen), the de-acetylated form was accessed by refluxing in an aqueous solution of sodium hydroxide (2 m) with sodium borohydride. B, control polymer of Neu5Ac (Ac100) was made by reacetylation of the deacetylated polymer with acetic anhydride and an aqueous solution of sodium bicarbonate. C, mixed polymers of Neu5Ac and Neu5Gc (Gc60 and Gc40) were generated using a mixture of acetic anhydride and acetoxyacetyl chloride in an aqueous solution of sodium bicarbonate. D, polymer of Neu5Gc (Gc100) was generated using acetoxyacetyl chloride under the same mild basic conditions described above. E, Neu5Pr polymer was synthesized using propionyl chloride in the same way as described above (also see Fig. 8).
FIGURE 4.
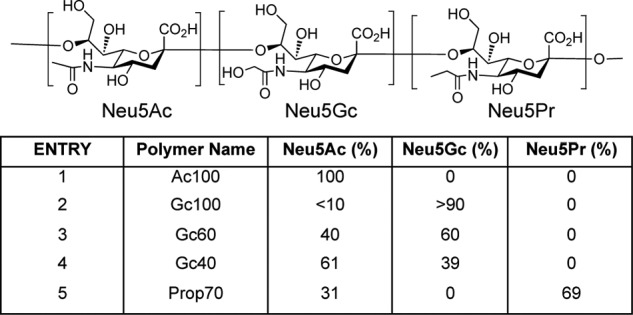
Polymers of Neu5Ac, Neu5Gc, and Neu5Pr used in these studies. Top panel indicates an oligomer of (from left to right) Neu5Ac, Neu5Gc, and Neu5Pr to depict the structure of the individual Sias. Bottom panel, the five polymer substrates used in these experiments and their respective percentages of each Sia, determined by DMB-HPLC and 1H NMR.
HPLC analysis on the DNAPac anion-exchange column showed that the process of synthesis caused some breakdown of long polymers, which presumably occurred during the base-catalyzed de-acetylation. To ensure consistency in length between synthesized polymers and control Ac100 polymers, we employed two methods. First, Ac100 was made in the same way as the Neu5Gc-containing polymers, i.e. from the de-acetylated polymer backbone (Fig. 3). Second, we treated all polymers with the enzymatically active phage endo-NF. This cleaves polySia into oligomers of 3–7 residues (endo-NF has previously been shown to digest polymers of Neu5Gc in salmonid eggs) (102). We confirmed that endo-NF digested all polymers of Neu5Ac and Neu5Gc by anion-exchange HPLC (size range, 4–7 residues, data not shown). We next tested the effect of varying levels of Neu5Gc within the polymer backbone on sialidase activity.
Major Difference in Relative Rates of Cleavage of Terminal Sialic Acids in α2–8-Linked Polymers of Neu5Ac and Neu5Gc by Various Sialidases
To determine the sensitivity of Neu5Ac and Neu5Gc to sialidase digestion in an α2–8 linkage, all Neu5Ac- and Neu5Gc-containing polymers (prepared as described above) were subjected to degradation by AUS or NEU1. Both enzymes hydrolyze the glycosidic linkage of the terminal residue, releasing a monomeric sialic acid residue. The released Neu5Ac and Neu5Gc monomer was detected by DMB-HPLC.
Although polymers of Neu5Ac are quite sensitive to breakdown by either AUS or NEU1, the polymers containing Neu5Gc exhibit significant resistance to both sialidases (Fig. 5A). This result was consistent across multiple experimental replicates. The same observation was made with a preparation of NEU1 from rat (data not shown). In mixed polymers, increasing the percentage composition of Neu5Gc within the polymer decreased sialidase sensitivity (Fig. 5B). Notably, even though the total Sia release from Gc40 is similar to that of Ac100, further analysis indicates that Neu5Ac, and not Neu5Gc, is the dominant product (Fig. 5C). This suggests that although the enzymes are still able to hydrolyze terminal Neu5Ac residues within the polymers, the presence of a Neu5Gc residue inhibits the hydrolysis rate.
FIGURE 5.
Relative rate of hydrolysis of α2–8-linked Sias by sialidases is markedly reduced when Neu5Gc residue is present at terminal positions. Polymer substrates (Fig. 4) and disaccharides of Neu5Ac and Neu5Gc were digested with NEU1 and AUS. Enzymatic breakdown was measured using DMB-HPLC. A, top panel, NEU1 demonstrates much greater activity on Ac100 than on Gc100. Bottom panel, total breakdown of mixed Neu5Ac and Neu5Gc polymers by NEU1 at 80 min shows increasing resistance with increasing Neu5Gc fraction. Fractions were calculated relative to total amount of monomer released by complete hydrolysis in 0.1 m TFA. Data shown are from a representative experiment. B, AUS activity on Neu5Ac and Neu5Gc polymers as in A. C, fractional release of monomeric Neu5Ac and Neu5Gc from the Gc40 polymer by NEU1 (top panel) and AUS (bottom panel) indicates preferential release of Neu5Ac from this polymer. Fractions are expressed relative to the total amount of the respective monomer (Neu5Ac or Neu5Gc) present in the sample. D, 1 nmol of α2–8-linked disaccharides of Neu5Ac and Neu5Gc (Neu5Ac-Neu5Ac, Neu5Ac-Neu5Gc, Neu5Gc-Neu5Ac, and Neu5Gc-Neu5Gc, nomenclature indicates nonreducing followed by reducing sugar) was digested by AUS and NEU1, and the released monomer was detected by DMB-HPLC. Background subtraction of PBS or acetate buffer controls as well as enzyme-only controls were applied to the values obtained. Neu5Gc at the nonreducing terminus conferred significant resistance to digestion by both AUS and NEU1. (n = 3. Error bars represent standard deviation. ***, p < 0.0005; **, p < 0.005, analyzed by t test relative to corresponding Neu5Ac-Neu5Ac sample.) E and F, as in A and B, polymers Ac100 and Gc100 were subjected to digestion by mammalian sialidases NEU2 (E) and NEU4 (F).
To further explore this idea, and determine the position of the inhibitory Neu5Gc moiety, we digested disaccharides of α2–8-linked Neu5Ac and Neu5Gc. These were Neu5Ac-Neu5Ac, Neu5Gc-Neu5Gc, Neu5Ac-Neu5Gc, and Neu5Gc-Neu5Ac (nomenclature indicates nonreducing Sia followed by reducing Sia). Both AUS and NEU1 were able to digest disaccharides with Neu5Ac at the nonreducing terminus; however, Neu5Gc showed significant resistance to breakdown (Fig. 5D). The underlying residue appeared to have no affect on the ability of the sialidases to break the glycosidic bond. AUS showed a slight preference for disaccharides with a terminal Neu5Ac and an underlying Neu5Gc, an effect that NEU1 did not exhibit. Taken together, these data indicate that the terminal Neu5Gc residue confers resistance of a polymer to sialidase digestion. Because we see no breakdown of the Neu5Gc-terminated disaccharides and the released monomer from the mixed polymers is predominantly Neu5Ac, this indicates that relatively low percentages of Neu5Gc within a polySia chain would dramatically inhibit enzymatic hydrolysis.
The other known vertebrate sialidases NEU2, NEU3, and NEU4 are far less prevalent than NEU1 in the brain (104). NEU3 has a strong preference for gangliosides rather than glycoproteins (105). We considered the possibility that the NEU2 or NEU4 may also be affected by the presence of Neu5Gc in polySia. Indeed, a recent study has implicated NEU4 as being involved in polySia degradation (106). Although this work demonstrated the ability of NEU4 to break down polySia, it did not show exclusivity of this enzyme in breaking down polySia in vivo. As NEU1 is present at much higher levels in brain tissue, it is likely that both enzymes are involved in polySia degradation.
To explore the impact of Neu5Gc on these other sialidases, the Ac100 and Gc100 substrates were digested with purified preparations of NEU2 and NEU4. These sialidases also exhibit a strong preference for Neu5Ac over Neu5Gc (Fig. 5, E and F). Notably, the difference in activity on the two polymers was greatest in NEU4 (Fig. 5F). Further studies are needed to determine whether NEU1 or NEU4 is most impacted by the presence of Neu5Gc on polySia in vivo.
α2–8-Linked Polymers of Neu5Gc Are More Sensitive to Acid Hydrolysis than α2–8-Linked Neu5Ac
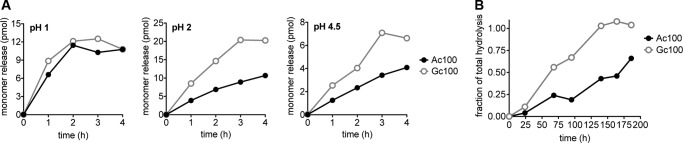
Our previous work demonstrated that polymers of α2–8-linked Neu5Ac undergo an intramolecular self-cleavage (hydrolysis of the glycosidic linkage) under mildly acidic conditions (51), similar to conditions found in the lysosome. To ask if polymers containing Neu5Gc were also more resistant to acidic degradation, we looked at the breakdown of Ac100 and Gc100 under various acidic pH conditions (Fig. 6). Using DMB-HPLC analysis, we were able to compare monomer release between Ac100 and Gc100 when heated at 80 °C and pH 1, 2, 4.5, and 7.5 (Fig. 6A).
FIGURE 6.
Neu5Gc in polySia increases relative susceptibility to acid-catalyzed breakdown. A, to determine differences in acid-catalyzed cleavage, Ac100 and Gc100 were incubated at 80 °C at pH of 1, 2, or 4.5. Monomer release was measured by DMB-HPLC. At pH of 2 and 4.5, Gc100 appears to break down faster. B, to determine breakdown of polymer chains to short oligomers, at lysosomal pH of 4.5 and physiological temperature of 37 °C, the fraction of linkages hydrolyzed was analyzed using the Moppler-Grindler reducing end assay. Fractions were calculated relative to complete hydrolysis of polymers in H2SO4. In keeping with A, Gc100 appeared to break down faster, with all linkages hydrolyzed by the end point.
Although there was no appreciable difference in breakdown at pH 1, at higher pH values of 2 and 4.5, it appeared that Gc100 was more sensitive to hydrolysis. At pH 7.5, no appreciable breakdown was seen in either polymer (data not shown). We also looked at the breakdown of Ac100 and Gc100 under conditions similar to that in the lysosome (37 °C, pH 4.5), this time analyzing the glycosidic linkages hydrolyzed over time using a reducing sugar assay (see under “Experimental Procedures”). As before, Gc100 was more sensitive to acid hydrolysis (Fig. 6B). We found that by 125 h, 100% of the glycosidic linkages in the Gc100 polymer had hydrolyzed, whereas ∼50% still remained in the Ac100 polymer. This may indicate that polymers of Neu5Ac are more likely to form protective lactone rings at acidic pH (107).
Regardless, taken together, these data suggest that spontaneous acid hydrolysis of polySia at lysosomal pH is not inhibited by the presence of Neu5Gc residues. Indeed, in striking contrast to enzymatic sensitivity, Gc100 polymers in fact appear to be more sensitive to acid-catalyzed hydrolysis. However, the time course of polySia breakdown by acid alone is extremely slow, over a period of several days. It is therefore unlikely that adequate breakdown of either polymer could occur in the absence of sialidase. Furthermore, this mechanism is not available at the neutral pH of the plasma membrane, and so it cannot impact surface turnover of polySia.
Molecular Modeling Shows Differences in the Three-dimensional Shape of α2–8-Linked Neu5Gc, Neu5Ac, and Neu5Pr
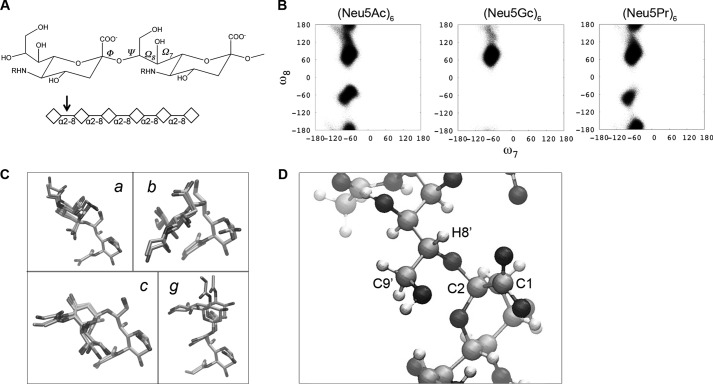
To investigate further the mechanism by which Neu5Gc residues inhibit sialidase activity, we modeled α2–8-linked hexamers of Neu5Ac (Ac100) and Neu5Gc (Gc100). Although sialidase is active on fragments as small as disaccharides, the use of a hexamer model was selected to introduce the three-dimensional structural effects of the polymer chain on the enzymatically cleaved nonreducing terminal residue pair. Many studies have indicated that poly-Neu5Ac has a helical conformation in solution; although the number of residues per turn appears to vary under different conditions, a recent crystallization of an endoneuraminidase estimated this number to be around 3.5 (108–110). However, studies of three-dimensional conformation have thus far been limited to di- and trisaccharides (80) and have never been performed on poly-Neu5Gc.
Hexamers of Neu5Ac and Neu5Gc were analyzed based on the torsion angles (ϕ, ψ, ω8, and ω7) around the bonds of the final glycosidic linkage (Fig. 7A). With the exception of the ω7 rotamer, which did not transition away from −60°, all other torsions frequently transitioned between states, suggesting a converged simulation by 0.5 μs (data not shown). Examining the glycosidic linkage between the terminal residues revealed the apparent absence of one conformer in Neu5Gc hexamers, indicating a constriction within the ω8 conformational space for Neu5Gc relative to Neu5Ac (Fig. 7B).
FIGURE 7.
Molecular dynamics modeling demonstrates conformational similarities and differences between hexamers of Neu5Ac, Neu5Gc, and Neu5Pr. Hexamers were modeled and analyzed for conformational differences. A, location of ψ, ϕ, ω7, and ω8 bond torsion angles across a glycosidic linkage. The arrow indicates the position of the terminal glycosidic linkage of the hexamer whose data is shown in Table 2. B, plots for the nonreducing terminal glycosidic linkage torsion angles (°) for ω7 and ω8, demonstrating the absence of one state (b, Table 2) in Neu5Gc hexamers. C, three-dimensional structures of the two terminal disaccharides in the most populated rotamers (state a, Table 2) and those with significant differences between Neu5Ac, Neu5Pr, and Neu5Gc (states b, c, and g, Table 2). Each of the sialic acid models is shown overlaid. D, molecular model indicating a conformation close to the likely transition state for lactonization. The molecular modeling predicts this conformation (and therefore the transition state of lactonization) is not easily accessible by the poly-Neu5Gc molecules.
Grouping the conformation states based on the torsion angles (ϕ, ψ, ω8, and ω7) produced seven states between the hexamer models (Table 2 and Fig. 7C). The most populated state, centered near (−60°, 0°, 80°, and −60°) (entry a, Table 2), was common to both of the hexamers and was observed for about 40% of each simulation, indicating that the overall structure of the polymers is largely similar. However, one of the states centered near −75°, 15°, −60°, and −68° (entry b, Table 2) was identified as absent in Neu5Gc; this state represented 24% of the simulation time for Neu5Ac. Another rotamer, centered at −90°, 160°, −170°, and −50° (entry g, Table 2) was observed to be exclusive to Neu5Gc but only for 6% of the simulation.
TABLE 2.
States and populations from 0.5-μs molecular dynamics simulations of Sia hexamers
Bond torsion angles across the glycosidic linkage at the nonreducing terminus of hexamers were calculated for Neu5Ac, Neu5Gc, and Neu5Pr as indicated in Fig. 7B. Seven total populations were grouped and state centers calculated, indicating average bond angles for Neu5Ac, Neu5Gc, and Neu5Pr within each state. Percentages of Neu5Ac, Neu5Gc, and Neu5Pr hexamers are indicated for each conformer.
| State ID | State centers ϕ, ψ, ω8, ω7 | Neu5Ac population | Neu5Gc population | Neu5Pr population |
|---|---|---|---|---|
| ° | % | % | % | |
| a | −62, 1, 81, −64 | 42 | 43 | 37 |
| −60, 5, 77, −66 | ||||
| −64, 1, 80, −63 | ||||
| b | −76, 16, −58, −60 | 24 | 0 | 9 |
| – | ||||
| −77, 16, −68, −75 | ||||
| c | −76, −53, 74, −64 | 8 | 24 | 12 |
| −92, −54, 67, −66 | ||||
| −90, −56, 71, −67 | ||||
| d | −50, 63, −178, −60 | 7 | 2 | 7 |
| −48, 56, 173, −65 | ||||
| −44, 57, −179, −54 | ||||
| e | −164, −15, 77, −57 | 7 | 10 | 9 |
| −166, −16, 76, −59 | ||||
| −164, −18, 80, −56 | ||||
| f | −52, 49, 81, −61 | 6 | 8 | 6 |
| −54, 46, 78, −63 | ||||
| −53, 48, 80, −61 | ||||
| g | – | 0 | 6 | 13 |
| −85, 159, −169, −49 | ||||
| −89, 163, −170, −53 | ||||
The accessibility of the b rotamer in hexameric Neu5Ac may represent a partial explanation for its increased susceptibility to enzymatic breakdown. This conformer may be accommodated by the catalytic site better than some of the other states, reducing the entropic penalty to the recognition of Neu5Ac. Alternatively, the b state may allow access to a transition state required for enzymatic activity.
The differences observed between Neu5Gc and Neu5Ac could be the result of increased steric repulsion from a larger substituent or may be an effect of the polar hydroxyl on the three-dimensional structure. To test this, we additionally modeled a hexamer of N-propionylneuraminic acid (Neu5Pr); the propionyl group is of a similar size as the glycolyl but does not contain the polar group (Fig. 3B and Table 2). Again, the same major conformation (a) was observed for about 40% of the simulation. The b state absent in Neu5Gc was observed in Neu5Pr, as in Neu5Ac, but for a shorter percentage of the simulation (9%), suggesting that the hydroxyl in Neu5Gc may in fact inhibit access to this conformation. However, the minor conformer g, centered at −90°,160°, −50°, and −170° and found at 6% in Neu5Gc, was also observed in Neu5Pr for 13% of the simulation. Although this could indicate a role for the length of the N-substituent in affecting the conformation, it could also suggest that convergence had not yet been achieved after 0.5 μs.
It is important to note that catalytic domain properties of the enzyme itself will also play a critical role in sialidase recognition. Homology modeling of NEU1 performed by Magesh et al. (59) revealed a hydrophobic pocket where the N-acetyl, N-glycolyl, or N-propionyl group is expected to reside. However, their homology model for NEU4 shows a more polar pocket, which may account for slight differences in the enzymatic activities of NEU4 and NEU1. Of course, the hydrophobicity of this pocket may not be the only factor at play as the hydroxyl group in the glycolyl moiety may also play a role in perturbing the orientation of the enzymatic machinery. The overall reduction in activity on Neu5Gc by sialidases may therefore be a result of multiple features impacting the enzyme-substrate interaction.
We further considered these models in the context of the enhanced sensitivity of Neu5Gc polymers to acid hydrolysis. It has been previously shown that at low pH, the polymers of Sia will form lactones that stabilize the molecule to acid breakdown (107). Modeling of a poly-Neu5Ac lactone showed that ϕ and ψ angles of −45° and −50° and 56° and 63°, respectively, would be required within the transition state of lactonization (Fig. 7D). Interestingly, molecular modeling data predicted that this transition state would fall close to conformation b and d (Table 2). As b cannot be achieved in polymers of Neu5Gc, the transition state for lactonization is likely not easily accessible. Therefore, a propensity toward protective lactonization may explain the relative stability of poly-Neu5Ac to acid-catalyzed self-cleavage.
Replacement of N-Glycolyl Groups with N-Propionyl Groups Restores Susceptibility to Sialidase Cleavage
The addition of an oxygen atom in Neu5Gc from Neu5Ac may conceivably have an array of effects, including steric hindrance of enzymatic binding, increased solvation of the polymer, or altered interaction with the sialidase binding pocket. To address these possibilities, polymers containing Neu5Pr and Neu5Ac residues were synthesized as discussed previously (Figs. 3E and 4, entry 5). We used a polymer with an average composition of 70% Neu5Pr and 30% Neu5Ac (Pr70). This is similar in composition ratios to the mixed polymer Gc60, which we previously showed had an inhibitory effect on sialidase activity (Fig. 5, A and B). In striking contrast to Gc60, Pr70 polymers show susceptibility to breakdown by the sialidases NEU1 and AUS (Fig. 8). In both cases, it appears that the presence of N-propionyl actually increased sensitivity to sialidase hydrolysis. This indicates that the resistance of Neu5Gc to NEU1 and AUS is not mediated by steric effects. Furthermore, as the b rotamer was found in equal fractions in the modeling of both the Neu5Ac and Neu5Pr hexamers (Table 2), this may indicate that conformational restriction is not a critical determinant of sialidase activity. This finding lends support to the idea that sialidase activity on α2–8-linked polymers may depend strongly on the effects of substrate binding and solvation, as the aliphatic pocket present at the binding site of NEU1 and AUS favors the longer propionyl group of Neu5Pr over the shorter acetyl group of Neu5Ac.
FIGURE 8.
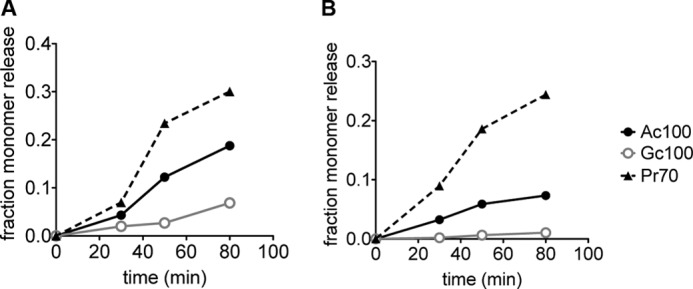
Replacement of the glycolyl hydroxyl group with a methyl group restores sialidase activity. To determine whether the addition of a glycolyl group in Neu5Gc confers resistance by steric effects, polymers of 70% Neu5Pr (Pr70) were digested with NEU1 (A) and AUS (B) as described in Fig. 5. For comparison, we have included enzymatic activity on Ac100 (black line) and Gc100 (gray line). In both cases Pr70 shows enhanced sensitivity to sialidase hydrolysis.
DISCUSSION
It is well established that sialidases exhibit relative preferences for the type of sialic acid or linkage that they are able to digest. However, most studies to date have focused on α2–3- or 2–6-linked Sias, and the differences found in vertebrate sialidases have been small (56). Here, we demonstrate a remarkable intrinsic stability of terminal α2–8-linked Neu5Gc residues to enzymatic digestion. This stability extends beyond simple enzymatic selectivity for the terminal Sia to a molecular stability in the presence of a terminal glycolyl moiety in combination with an α2–8-linkage. Molecular dynamics modeling suggests that conformational differences of α2–8 Neu5Gc, as well as a potentially decreased interaction of the glycolyl moiety with a sialidase binding pocket, may contribute to this finding. The significant inhibition of enzymatic activity by Neu5Gc-terminated disaccharides and the relatively low release of Neu5Gc in mixed polymers indicate that even a low percentage of Neu5Gc in polySia would dramatically inhibit its enzymatic hydrolysis by exosialidases. Because there are no endosialidases in vertebrates, this finding is expected to affect tissues expressing α2–8 polySia. Although further work is clearly needed to demonstrate a clear link, this finding provides a potential mechanism by which even small amounts of Neu5Gc may exert a detrimental effect in the vertebrate brain. We speculate that the resistance of α2–8 linked Neu5Gc to sialidase breakdown may therefore underlie the relative absence of Neu5Gc from all vertebrate brains studied to date.
PolySia is a strong candidate for this neural specific effect, being highly and widely expressed in neural development and playing critical roles in growth and plasticity. It is expressed during development in some extraneural tissues, but in much lower amounts (111), and so would not be expected to show major toxicity in other Neu5Gc-containing tissues. Furthermore, the α2–8 linkage that affords this resistance is enriched in brain, not only in polySia and on gangliosides but also on short di- and oligosialosyl SGC epitopes that are much rarer in other tissues (112). The large number of Sias present in each polymer must be broken down one by one by mammalian sialidases, which as exoglycosidases can only cleave a terminal sialic acid. This allows a very low fraction of Neu5Gc to render an entire chain relatively resistant to breakdown, due to the presence of a single terminal Neu5Gc residue.
We propose a model in which Neu5Gc can be incorporated into polySia but then renders the molecule relatively indigestible by vertebrate sialidases. The inability to rapidly desialylate NCAM may then result in widespread inappropriate polySia distribution in the brain. We show here that Neu5Gc can be incorporated into endogenous polySia by polysialyltransferases. Such polymers containing Neu5Gc are relatively resistant to breakdown by NEU1, NEU2, and NEU4. Previous work has indicated that polymers of Neu5Ac do undergo cleavage by both extramolecular and intramolecular protons at mild acid pH (51); however, it is a slow process that occurs over days via short oligomer intermediates, without much breakdown to the monomer. Although we find that polymers of Neu5Gc are actually relatively more susceptible to acid-catalyzed breakdown, it still occurs over a very long time course. In embryonic development, when PSA-NCAM is highly enriched in brain, lysosomal acid hydrolysis alone may be insufficient to degrade the large quantities of polySia present. Furthermore, this process would additionally be ineffective for the rapid desialylation of NCAM that may be necessary at the cell surface.
The enzymatic resistance of Neu5Gc-containing polySia may of course have effects in tissues outside the vertebrate brain. Although polySia is relatively rare in vertebrate tissues outside the developing brain, it is found in the capsule of certain bacteria such as Escherichia coli K1 and Neisseria meningitidis (43). However, no bacterium has yet been shown to be capable of Neu5Gc synthesis, and thus these capsules contain only Neu5Ac. There is a single report of Neu5Gc present in α2–8-linked polySia is in the eggs of salmonid fish (113). Although we are not aware of any organism that digests these glycoproteins, we can hypothesize that eggs that contain Neu5Gc may be more resistant to attack by sialidase-producing pathogens in the wild.
Further studies of Neu5Gc and polySia in vivo will of course be necessary to determine whether the observed enzymatic resistance is, in fact, sufficient to exert the proposed detrimental effect on vertebrate brain development. Regardless, this finding is an unusual biochemical consequence of exchanging Neu5Ac for Neu5Gc in a biological system, showing how a single oxygen atom can have dramatic consequences for the biology of a tissue-specific polysaccharide.
Acknowledgments
We thank Prof. Rita Gerardy-Schahn for the generous gift of endo-NF used in several of our experiments, and Prof. Kimio Furuhata, who kindly provided the 4MU-Neu5Gc and 4MU-Kdn. We also thank Dr. Anne Bergfeld for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM32373 and R01CA38701 (to A. V.) and GM094919 (EUREKA) (to R. J. W.).
- Neu5Ac
- N-acetylneuraminic acid
- CMAH
- CMP-Neu5Ac hydroxylase
- DMB
- 1,2-diamino-4,5-methylenedioxybenzene
- Neu5Gc
- N-glycolylneuraminic acid
- polySia
- polysialic acid
- 4MU
- 4-methylumbelliferyl
- AUS
- A. ureafaciens sialidase
- Sia
- sialic acid
- SGC
- sialoglycoconjugate
- Kdn
- deaminoneuraminic acid
- NCAM
- neural cell adhesion molecule
- endo
- endoneuraminidase
- PSA
- polysialic acid
- Neu5Pr
- N-propionylneuraminic acid.
REFERENCES
- 1. Schauer R. (1978) Characterization of sialic acids. Methods Enzymol. 50, 64–89 [DOI] [PubMed] [Google Scholar]
- 2. Varki A., Schauer R. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds) pp. 199–218, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 3. Schauer R., Schoop H. J., Faillard H. (1968) On biosynthesis of the glycolyl groups of N-glycolylneuraminic acid. Oxidative conversion of N-acetyl groups to glycolyl groups. Hoppe-Seyler's Z. Physiol. Chem. 349, 645–652 [PubMed] [Google Scholar]
- 4. Shaw L., Schauer R. (1988) The biosynthesis of N-glycoloylneuraminic acid occurs by hydroxylation of the CMP-glycoside of N-acetylneuraminic acid. Biol. Chem. Hoppe-Seyler 369, 477–486 [DOI] [PubMed] [Google Scholar]
- 5. Kawano T., Kozutsumi Y., Takematsu H., Kawasaki T., Suzuki A. (1993) Regulation of biosynthesis of N-glycolylneuraminic acid-containing glycoconjugates. Characterization of factors required for NADH-dependent cytidine 5′-monophosphate-N-acetylneuraminic acid hydroxylation. Glycoconj. J. 10, 109–115 [DOI] [PubMed] [Google Scholar]
- 6. Martensson E., Raal A., Svennerholm L. (1958) Sialic acids in blood serum. Biochim. Biophys. Acta 30, 124–129 [DOI] [PubMed] [Google Scholar]
- 7. Gottschalk A. (1960) The Chemistry and Biology of Sialic Acids and Related Substances, pp. 30–35, Cambridge University Press, London [Google Scholar]
- 8. Warren L., Spicer S. S. (1961) Biochemical and histochemical identification of sialic acid containing mucins of rodent vagina and salivary glands. J. Histochem. Cytochem. 9, 400–408 [DOI] [PubMed] [Google Scholar]
- 9. Yu R. K., Ledeen R. W. (1972) Gangliosides of human, bovine, and rabbit plasma. J. Lipid Res. 13, 680–686 [PubMed] [Google Scholar]
- 10. Ando S., Chang N. C., Yu R. K. (1978) High performance thin layer chromatography and densitometric determination of brain ganglioside compositions of several species. Anal. Biochem. 89, 437–450 [DOI] [PubMed] [Google Scholar]
- 11. Iwamori M., Nagai Y. (1978) GM3 ganglioside in various tissues of rabbit. Tissue-specific distribution of N-glycolylneuraminic acid-containing GM31. J. Biochem. 84, 1609–1615 [DOI] [PubMed] [Google Scholar]
- 12. Iwamori M., Nagai Y. (1981) Comparative study on ganglioside compositions of various rabbit tissues. Tissue specificity in ganglioside molecular species of rabbit thymus. Biochim. Biophys. Acta 665, 214–220 [DOI] [PubMed] [Google Scholar]
- 13. Corfield A., Schauer R. (1982) in Sialic Acids: Chemistry, Metabolism, and Function (Schauer R., ed) pp. 5–50, Springer-Verlag, New York [Google Scholar]
- 14. Hashimoto Y., Otsuka H., Yamakawa T. (1982) The occurrence of GM4 and GM2 in erythrocytes from inbred strains of mice. J. Biochem. 91, 1039–1046 [DOI] [PubMed] [Google Scholar]
- 15. Murayama J., Tomita M., Hamada A. (1982) Glycophorins of bovine erythrocyte membranes. Isolation and preliminary characterization of the major component. J. Biochem. 91, 1829–1836 [DOI] [PubMed] [Google Scholar]
- 16. Hashimoto Y., Suzuki A., Yamakawa T., Miyashita N., Moriwaki K. (1983) Expression of GM1 and GD1a in mouse liver is linked to the H-2 complex on chromosome 17. J. Biochem. 94, 2043–2048 [DOI] [PubMed] [Google Scholar]
- 17. Nagai K., Tadano-Aritomi K., Kawaguchi K., Ishizuka I. (1985) Acidic glycolipids from dolphin kidney. J. Biochem. 98, 545–559 [DOI] [PubMed] [Google Scholar]
- 18. Dahiya R., Brasitus T. A. (1988) Dexamethasone-induced alterations in the glycosphingolipids of rat kidney. Lipids 23, 863–868 [DOI] [PubMed] [Google Scholar]
- 19. Furukawa K., Chait B. T., Lloyd K. O. (1988) Identification of N-glycolylneuraminic acid-containing gangliosides of cat and sheep erythrocytes. 252Cf fission fragment ionization mass spectrometry in the analysis of glycosphingolipids. J. Biol. Chem. 263, 14939–14947 [PubMed] [Google Scholar]
- 20. Koizumi N., Hara A., Uemura K., Taketomi T. (1988) Glycosphingolipids in sheep liver, kidney, and various blood cells. Jpn. J. Exp. Med. 58, 21–31 [PubMed] [Google Scholar]
- 21. Sanai Y., Yamasaki M., Nagai Y. (1988) Monoclonal antibody directed to a Hanganutziu-Deicher active ganglioside, GM2 (NeuGc). Biochim. Biophys. Acta 958, 368–374 [DOI] [PubMed] [Google Scholar]
- 22. Lepers A., Shaw L., Schneckenburger P., Cacan R., Verbert A., Schauer R. (1990) A study on the regulation of N-glycoloylneuraminic acid biosynthesis and utilization in rat and mouse liver. Eur. J. Biochem. 193, 715–723 [DOI] [PubMed] [Google Scholar]
- 23. Wang D. Q., Fukui Y., Ito T., Nakajima K., Kato S., Naiki M., Kurimura T., Wakamiya N. (1990) Heterogeneity of Hanganutziu-Deicher antigen glycoproteins in different species of animal sera. Nippon Juigaku Zasshi 52, 567–572 [DOI] [PubMed] [Google Scholar]
- 24. Menzeleev R. F., Smirnova G. P., Chekareva N. V., Zvonkova E. N., Krasnopol'skii IuM, Shvets V. I. (1993) Ganglioside GM3 from horse erythrocytes. Structure and effect on cell proliferation. Bioorg. Khim. 19, 817–824 [PubMed] [Google Scholar]
- 25. Muchmore E. A., Diaz S., Varki A. (1998) A structural difference between the cell surfaces of humans and the great apes. Am. J. Phys. Anthropol. 107, 187–198 [DOI] [PubMed] [Google Scholar]
- 26. Rizzo A. M., Berra B., Rossi F., Guerra A., Gornati R., Bernardini G., Taki T., Kasama T., Mauri L., Sonnino S. (2002) Structure of the main ganglioside from the brain of Xenopus laevis. Glycoconj. J. 19, 53–57 [DOI] [PubMed] [Google Scholar]
- 27. Martin M. J., Rayner J. C., Gagneux P., Barnwell J. W., Varki A. (2005) Evolution of human-chimpanzee differences in malaria susceptibility. Relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. U.S.A. 102, 12819–12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osthoff G., Dickens L., Urashima T., Bonnet S. L., Uemura Y., van der Westhuizen J. H. (2008) Structural characterization of oligosaccharides in the milk of an African elephant (Loxodonta africana africana). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150, 74–84 [DOI] [PubMed] [Google Scholar]
- 29. Uemura Y., Asakuma S., Yon L., Saito T., Fukuda K., Arai I., Urashima T. (2006) Structural determination of the oligosaccharides in the milk of an Asian elephant (Elephas maximus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 145, 468–478 [DOI] [PubMed] [Google Scholar]
- 30. Zeleny R., Kolarich D., Strasser R., Altmann F. (2006) Sialic acid concentrations in plants are in the range of inadvertent contamination. Planta 224, 222–227 [DOI] [PubMed] [Google Scholar]
- 31. Spichtig V., Michaud J., Austin S. (2010) Determination of sialic acids in milks and milk-based products. Anal. Biochem. 405, 28–40 [DOI] [PubMed] [Google Scholar]
- 32. Cabezas J. A. (2011) El ácido siálico N-glicolilneuraminico. Su relación con la biodiversidad y con procesos inmunitarios e infecciosos. An. R. Acad. Nac. Farm 77, 72–86 [Google Scholar]
- 33. Tao N., Wu S., Kim J., An H. J., Hinde K., Power M. L., Gagneux P., German J. B., Lebrilla C. B. (2011) Evolutionary glycomics. Characterization of milk oligosaccharides in primates. J. Proteome Res. 10, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang B. (2009) Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29, 177–222 [DOI] [PubMed] [Google Scholar]
- 35. Kawano T., Koyama S., Takematsu H., Kozutsumi Y., Kawasaki H., Kawashima S., Kawasaki T., Suzuki A. (1995) Molecular cloning of cytidine monophospho-N-acetylneuraminic acid hydroxylase. Regulation of species- and tissue-specific expression of N-glycolylneuraminic acid. J. Biol. Chem. 270, 16458–16463 [DOI] [PubMed] [Google Scholar]
- 36. Song K. H., Kang Y. J., Jin U. H., Park Y. I., Kim S. M., Seong H. H., Hwang S., Yang B. S., Im G. S., Min K. S., Kim J. H., Chang Y. C., Kim N. H., Lee Y. C., Kim C. H. (2010) Cloning and functional characterization of pig CMP-N-acetylneuraminic acid hydroxylase for the synthesis of N-glycolylneuraminic acid as the xenoantigenic determinant in pig-human xenotransplantation. Biochem. J. 427, 179–188 [DOI] [PubMed] [Google Scholar]
- 37. Raymond J. B., Mahapatra S., Crick D. C., Pavelka M. S. (2005) Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280, 326–333 [DOI] [PubMed] [Google Scholar]
- 38. Hedlund M., Tangvoranuntakul P., Takematsu H., Long J. M., Housley G. D., Kozutsumi Y., Suzuki A., Wynshaw-Boris A., Ryan A. F., Gallo R. L., Varki N., Varki A. (2007) N-Glycolylneuraminic acid deficiency in mice. Implications for human biology and evolution. Mol. Cell. Biol. 27, 4340–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Higa H. H., Paulson J. C. (1985) Sialylation of glycoprotein oligosaccharides with N-acetyl-, N-glycolyl-, and N,O-diacetylneuraminic acids. J. Biol. Chem. 260, 8838–8849 [PubMed] [Google Scholar]
- 40. Vyas A. A., Schnaar R. L. (2001) Brain gangliosides. Functional ligands for myelin stability and the control of nerve regeneration. Biochimie 83, 677–682 [DOI] [PubMed] [Google Scholar]
- 41. Finne J., Finne U., Deagostini-Bazin H., Goridis C. (1983) Occurrence of α2–8-linked polysialosyl units in a neural cell adhesion molecule. Biochem. Biophys. Res. Commun. 112, 482–487 [DOI] [PubMed] [Google Scholar]
- 42. Cunningham B. A., Hoffman S., Rutishauser U., Hemperly J. J., Edelman G. M. (1983) Molecular topography of the neural cell adhesion molecule N-CAM. Surface orientation and location of sialic acid-rich and binding regions. Proc. Natl. Acad. Sci. U.S.A. 80, 3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Troy F. (1979) The chemistry and biosynthesis of selected bacterial capsular polymers. Annu. Rev. Microbiol. 33, 519–560 [DOI] [PubMed] [Google Scholar]
- 44. Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 45. Eckhardt M., Bukalo O., Chazal G., Wang L., Goridis C., Schachner M., Gerardy-Schahn R., Cremer H., Dityatev A. (2000) Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. J. Neurosci. 20, 5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinhold B., Seidenfaden R., Röckle I., Mühlenhoff M., Schertzinger F., Conzelmann S., Marth J. D., Gerardy-Schahn R., Hildebrandt H. (2005) Genetic ablation of polysialic acid causes severe neurodevelopmental defects rescued by deletion of the neural cell adhesion molecule. J. Biol. Chem. 280, 42971–42977 [DOI] [PubMed] [Google Scholar]
- 47. Barbeau D., Liang J. J., Robitalille Y., Quirion R., Srivastava L. K. (1995) Decreased expression of the embryonic form of the neural cell adhesion molecule in schizophrenic brains. Proc. Natl. Acad. Sci. U.S.A. 92, 2785–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arai M., Yamada K., Toyota T., Obata N., Haga S., Yoshida Y., Nakamura K., Minabe Y., Ujike H., Sora I., Ikeda K., Mori N., Yoshikawa T., Itokawa M. (2006) Association between polymorphisms in the promoter region of the sialyltransferase 8B (SIAT8B) gene and schizophrenia. Biol. Psychiatry 59, 652–659 [DOI] [PubMed] [Google Scholar]
- 49. Hildebrandt H., Mühlenhoff M., Oltmann-Norden I., Röckle I., Burkhardt H., Weinhold B., Gerardy-Schahn R. (2009) Imbalance of neural cell adhesion molecule and polysialyltransferase alleles causes defective brain connectivity. Brain 132, 2831–2838 [DOI] [PubMed] [Google Scholar]
- 50. Isomura R., Kitajima K., Sato C. (2011) Structural and functional impairments of polysialic acid by a mutated polysialyltransferase found in schizophrenia. J. Biol. Chem. 286, 21535–21545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Manzi A. E., Higa H. H., Diaz S., Varki A. (1994) Intramolecular self-cleavage of polysialic acid. J. Biol. Chem. 269, 23617–23624 [PubMed] [Google Scholar]
- 52. Katoh S., Miyagi T., Taniguchi H., Matsubara Y., Kadota J., Tominaga A., Kincade P. W., Matsukura S., Kohno S. (1999) Cutting edge. An inducible sialidase regulates the hyaluronic acid binding ability of CD44-bearing human monocytes. J. Immunol. 162, 5058–5061 [PubMed] [Google Scholar]
- 53. Pshezhetsky A. V., Hinek A. (2011) Where catabolism meets signaling. Neuraminidase 1 as a modulator of cell receptors. Glycoconj. J. 28, 441–452 [DOI] [PubMed] [Google Scholar]
- 54. Corfield A. P., Veh R. W., Wember M., Michalski J. C., Schauer R. (1981) The release of N-acetyl- and N-glycolloyl-neuraminic acid from soluble complex carbohydrates and erythrocytes by bacterial, viral and mammalian sialidases. Biochem. J. 197, 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Corfield A. P., Higa H., Paulson J. C., Schauer R. (1983) The specificity of viral and bacterial sialidases for α(2–3)- and α(2–6)-linked sialic acids in glycoproteins. Biochim. Biophys. Acta 744, 121–126 [DOI] [PubMed] [Google Scholar]
- 56. Cao H., Li Y., Lau K., Muthana S., Yu H., Cheng J., Chokhawala H. A., Sugiarto G., Zhang L., Chen X. (2009) Sialidase substrate specificity studies using chemoenzymatically synthesized sialosides containing C5-modified sialic acids. Org. Biomol. Chem. 7, 5137–5145 [DOI] [PubMed] [Google Scholar]
- 57. Monti E., Bonten E., D'Azzo A., Bresciani R., Venerando B., Borsani G., Schauer R., Tettamanti G. (2010) Sialidases in vertebrates. a family of enzymes tailored for several cell functions. Adv. Carbohydr. Chem. Biochem. 64, 403–479 [DOI] [PubMed] [Google Scholar]
- 58. Chavas L. M., Tringali C., Fusi P., Venerando B., Tettamanti G., Kato R., Monti E., Wakatsuki S. (2005) Crystal structure of the human cytosolic sialidase Neu2. Evidence for the dynamic nature of substrate recognition. J. Biol. Chem. 280, 469–475 [DOI] [PubMed] [Google Scholar]
- 59. Magesh S., Savita V., Moriya S., Suzuki T., Miyagi T., Ishida H., Kiso M. (2009) Human sialidase inhibitors. Design, synthesis, and biological evaluation of 4-acetamido-5-acylamido-2-fluorobenzoic acids. Bioorg. Med. Chem. 17, 4595–4603 [DOI] [PubMed] [Google Scholar]
- 60. Li Y., Cao H., Yu H., Chen Y., Lau K., Qu J., Thon V., Sugiarto G., Chen X. (2011) Identifying selective inhibitors against the human cytosolic sialidase NEU2 by substrate specificity studies. Mol. Biosyst. 7, 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Spiro R. G., Yasumoto Y., Bhoyroo V. (1996) Characterization of a rat liver Golgi sulfotransferase responsible for the 6-O-sulfation of N-acetylglucosamine residues in β-linkage to mannose. Role in assembly of sialyl-galactosyl-N-acetylglucosamine 6-sulfate sequence of N-linked oligosaccharides. Biochem. J. 319, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Banda K., Gregg C. J., Chow R., Varki N., Varki A. (2012) Metabolism of vertebrate amino sugars with N-glycolyl groups. Mechanisms underlying gastrointestinal incorporation of the non-human sialic acid xeno-autoantigen, N-glycolylneuraminic acid. J. Biol. Chem. 287, 28852–28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Inoue S., Lin S. L., Lee Y. C., Inoue Y. (2001) An ultrasensitive chemical method for polysialic acid analysis. Glycobiology 11, 759–767 [DOI] [PubMed] [Google Scholar]
- 64. Mühlenhoff M., Stummeyer K., Grove M., Sauerborn M., Gerardy-Schahn R. (2003) Proteolytic processing and oligomerization of bacteriophage-derived endosialidases. J. Biol. Chem. 278, 12634–12644 [DOI] [PubMed] [Google Scholar]
- 65. Jokilammi A., Ollikka P., Korja M., Jakobsson E., Loimaranta V., Haataja S., Hirvonen H., Finne J. (2004) Construction of antibody mimics from a noncatalytic enzyme-detection of polysialic acid. J. Immunol. Methods 295, 149–160 [DOI] [PubMed] [Google Scholar]
- 66. Sato C., Kitajima K., Tazawa I., Inoue Y., Inoue S., Troy F. A. (1993) Structural diversity in the α2→8-linked polysialic acid chains in salmonid fish egg glycoproteins. Occurrence of poly(Neu5Ac), poly(Neu5Gc), poly(Neu5Ac, Neu5Gc), poly(KDN), and their partially acetylated forms. J. Biol. Chem. 268, 23675–23684 [PubMed] [Google Scholar]
- 67. Roy R., Pon R. A. (1990) Efficient synthesis of α(2–8)-linked N-acetyl and N-glycolylneuraminic acid disaccharides from colominic acid. Glycoconj. J. 7, 3–12 [Google Scholar]
- 68. Sato C., Inoue S., Matsuda T., Kitajima K. (1999) Fluorescent-assisted detection of oligosialyl units in glycoconjugates. Anal. Biochem. 266, 102–109 [DOI] [PubMed] [Google Scholar]
- 69. Pshezhetsky A. V., Potier M. (1996) Association of N-acetylgalactosamine-6-sulfate sulfatase with the multienzyme lysosomal complex of β-galactosidase, cathepsin A, and neuraminidase. Possible implication for intralysosomal catabolism of keratan sulfate. J. Biol. Chem. 271, 28359–28365 [DOI] [PubMed] [Google Scholar]
- 70. Yamakawa N., Sato C., Miyata S., Maehashi E., Toriyama M., Sato N., Furuhata K., Kitajima K. (2007) Development of sensitive chemical and immunochemical methods for detecting sulfated sialic acids and their application to glycoconjugates from sea urchin sperm and eggs. Biochimie 89, 1396–1408 [DOI] [PubMed] [Google Scholar]
- 71. Waffenschmidt S., Jaenicke L. (1987) Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal. Biochem. 165, 337–340 [DOI] [PubMed] [Google Scholar]
- 72. Mopper K., Gindler E. M. (1973) A new noncorrosive dye reagent for automatic sugar chromatography. Anal. Biochem. 56, 440–442 [DOI] [PubMed] [Google Scholar]
- 73. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J. (2008) GLYCAM06. A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Basma M., Sundara S., Calgan D., Vernali T., Woods R. J. (2001) Solvated ensemble averaging in the calculation of partial atomic charges. J. Comput. Chem. 22, 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Case D. A., Darden T. A., Cheatham I. T., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., Kolossváry I., Wong K. F., Paesani F., Vanicek J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M. J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., Kollman P. A. (2010) AMBER, Version 11, pp. 1–300 University of California, San Francisco [Google Scholar]
- 76. Berendsen H. J., Postma J. P., Vangunsteren W. F., Dinola A., Haak J. R. (1984) Molecular dynamics with coupling to and external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 77. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 78. Darden T., York D., Pederson L. (1993) Particle mesh Ewald-an. NlogN method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 79. Ryckaert J. P., Ciccotti G., Berendsen H. J. (1977) Numerical integration of the Cartesian equations of motion of a system with constraints. Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 [Google Scholar]
- 80. Yongye A. B., Gonzalez-Outeiriño J., Glushka J., Schultheis V., Woods R. J. (2008) The conformational properties of methyl α-(2,8)-di/trisialosides and their N-acyl analogues. Implications for anti-Neisseria meningitidis B vaccine design. Biochemistry 47, 12493–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yu R. K., Ledeen R. W. (1970) Gas-liquid chromatographic assay of lipid-bound sialic acids. Measurement of gangliosides in brain of several species. J. Lipid Res. 11, 506–516 [PubMed] [Google Scholar]
- 82. Ghidoni R., Sonnino S., Tettamanti G., Wiegandt H., Zambotti V. (1976) On the structure of two new gangliosides from beef brain. J. Neurochem. 27, 511–515 [DOI] [PubMed] [Google Scholar]
- 83. Iwamori M., Nagai Y. (1978) A new chromatographic approach to the resolution of individual gangliosides. Ganglioside mapping. Biochim. Biophys. Acta 528, 257–267 [PubMed] [Google Scholar]
- 84. Chigorno V., Sonnino S., Ghidoni R., Tettamanti G. (1982) Densitometric quantification of brain gangliosides separated by two-dimensional thin layer chromatography. Neurochem. Int. 4, 397–404 [DOI] [PubMed] [Google Scholar]
- 85. Nakao T., Kon K., Ando S., Hirabayashi Y. (1991) A NeuGc-containing trisialoganglioside of bovine brain. Biochim. Biophys. Acta 1086, 305–309 [DOI] [PubMed] [Google Scholar]
- 86. Terabayashi T., Ogawa T., Kawanishi Y. (1992) A comparative study on ceramide composition of cetacean brain gangliosides. Comp. Biochem. Physiol. B 103, 721–726 [DOI] [PubMed] [Google Scholar]
- 87. Casellato R., Brocca P., Li S. C., Li Y. T., Sonnino S. (1995) Isolation and structural characterization of N-acetyl- and N-glycolylneuraminic acid-containing GalNAc-GD1a isomers, IV4GalNAcIV3Neu5AcII3Neu5GcGgOse4Cer and IV4GalNAcIV3Neu5GcII3Neu5AcGgOse4Cer, from bovine brain. Eur. J. Biochem. 234, 786–793 [DOI] [PubMed] [Google Scholar]
- 88. Mikami T., Kashiwagi M., Tsuchihashi K., Daino T., Akino T., Gasa S. (1998) Further characterization of equine brain gangliosides: the presence of GM3 having N-glycolyl neuraminic acid in the central nervous system. J. Biochem. 123, 487–491 [DOI] [PubMed] [Google Scholar]
- 89. McCoy R. D., Vimr E. R., Troy F. A. (1985) CMP-NeuNAc:poly-α-2,8-sialosyl sialyltransferase and the biosynthesis of polysialosyl units in neural cell adhesion molecules. J. Biol. Chem. 260, 12695–12699 [PubMed] [Google Scholar]
- 90. Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Molecular characterization of eukaryotic polysialyltransferase-1. Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 91. Nakayama J., Fukuda M. N., Fredette B., Ranscht B., Fukuda M. (1995) Expression cloning of a human polysialyltransferase that forms the polysialylated neural cell adhesion molecule present in embryonic brain. Proc. Natl. Acad. Sci. U.S.A. 92, 7031–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kojima N., Yoshida Y., Tsuji S. (1995) A developmentally regulated member of the sialyltransferase family (ST8Sia II, STX) is a polysialic acid synthase. FEBS Lett. 373, 119–122 [DOI] [PubMed] [Google Scholar]
- 93. Horstkorte R., Mühlenhoff M., Reutter W., Nöhring S., Zimmermann-Kordmann M., Gerardy-Schahn R. (2004) Selective inhibition of polysialyltransferase ST8SiaII by unnatural sialic acids. Exp. Cell Res. 298, 268–274 [DOI] [PubMed] [Google Scholar]
- 94. Charter N. W., Mahal L. K., Koshland D. E., Jr., Bertozzi C. R. (2000) Biosynthetic incorporation of unnatural sialic acids into polysialic acid on neural cells. Glycobiology 10, 1049–1056 [DOI] [PubMed] [Google Scholar]
- 95. Mahal L. K., Charter N. W., Angata K., Fukuda M., Koshland D. E., Jr., Bertozzi C. R. (2001) A small molecule modulator of poly-α2,8-sialic acid expression on cultured neurons and tumor cells. Science 294, 380–381 [DOI] [PubMed] [Google Scholar]
- 96. Bork K., Gagiannis D., Orthmann A., Weidemann W., Kontou M., Reutter W., Horstkorte R. (2007) Experimental approaches to interfere with the polysialylation of the neural cell adhesion molecule in vitro and in vivo. J. Neurochem. 103, 65–71 [DOI] [PubMed] [Google Scholar]
- 97. Bardor M., Nguyen D. H., Diaz S., Varki A. (2005) Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. J. Biol. Chem. 280, 4228–4237 [DOI] [PubMed] [Google Scholar]
- 98. Hildebrandt H., Becker C., Glüer S., Rösner H., Gerardy-Schahn R., Rahmann H. (1998) Polysialic acid on the neural cell adhesion molecule correlates with expression of polysialyltransferases and promotes neuroblastoma cell growth. Cancer Res. 58, 779–784 [PubMed] [Google Scholar]
- 99. Seki T., Arai Y. (1991) Expression of highly polysialylated NCAM in the neocortex and piriform cortex of the developing and the adult rat. Anat. Embryol. 184, 395–401 [DOI] [PubMed] [Google Scholar]
- 100. Sato C., Kitajima K., Inoue S., Seki T., Troy F. A., 2nd, Inoue Y. (1995) Characterization of the antigenic specificity of four different anti-(α2→8-linked polysialic acid) antibodies using lipid-conjugated oligo/polysialic acids. J. Biol. Chem. 270, 18923–18928 [DOI] [PubMed] [Google Scholar]
- 101. Petter J. G., Vimr E. R. (1993) Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J. Bacteriol. 175, 4354–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kitajima K., Inoue S., Inoue Y., Troy F. A. (1988) Use of a bacteriophage-derived endo-N-acetylneuraminidase and an equine antipolysialyl antibody to characterize the polysialyl residues in salmonid fish egg polysialoglycoproteins. Substrate and immunospecificity studies. J. Biol. Chem. 263, 18269–18276 [PubMed] [Google Scholar]
- 103. Pearce O. M., Varki A. (2010) Chemo-enzymatic synthesis of the carbohydrate antigen N-glycolylneuraminic acid from glucose. Carbohydr. Res. 345, 1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shiozaki K., Koseki K., Yamaguchi K., Shiozaki M., Narimatsu H., Miyagi T. (2009) Developmental change of sialidase neu4 expression in murine brain and its involvement in the regulation of neuronal cell differentiation. J. Biol. Chem. 284, 21157–21164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Miyagi T., Wada T., Iwamatsu A., Hata K., Yoshikawa Y., Tokuyama S., Sawada M. (1999) Molecular cloning and characterization of a plasma membrane-associated sialidase specific for gangliosides. J. Biol. Chem. 274, 5004–5011 [DOI] [PubMed] [Google Scholar]
- 106. Takahashi K., Mitoma J., Hosono M., Shiozaki K., Sato C., Yamaguchi K., Kitajima K., Higashi H., Nitta K., Shima H., Miyagi T. (2012) Sialidase NEU4 hydrolyzes polysialic acids of neural cell adhesion molecules and negatively regulates neurite formation by hippocampal neurons. J. Biol. Chem. 287, 14816–14826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang Y., Lee Y. C. (1999) Acid-catalyzed lactonization of α2,8-linked oligo/polysialic acids studied by high performance anion-exchange chromatography. J. Biol. Chem. 274, 6183–6189 [DOI] [PubMed] [Google Scholar]
- 108. Schulz E. C., Schwarzer D., Frank M., Stummeyer K., Mühlenhoff M., Dickmanns A., Gerardy-Schahn R., Ficner R. (2010) Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein endo-NF. J. Mol. Biol. 397, 341–351 [DOI] [PubMed] [Google Scholar]
- 109. Yamasaki R., Bacon B. (1991) Three-dimensional structural analysis of the group B polysaccharide of Neisseria meningitidis 6275 by two-dimensional NMR. The polysaccharide is suggested to exist in helical conformations in solution. Biochemistry 30, 851–857 [DOI] [PubMed] [Google Scholar]
- 110. Brisson J. R., Baumann H., Imberty A., Pérez S., Jennings H. J. (1992) Helical epitope of the group B meningococcal α (2–8)-linked sialic acid polysaccharide. Biochemistry 31, 4996–5004 [DOI] [PubMed] [Google Scholar]
- 111. Finne J., Bitter-Suermann D., Goridis C., Finne U. (1987) An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J. Immunol. 138, 4402–4407 [PubMed] [Google Scholar]
- 112. Sato C., Fukuoka H., Ohta K., Matsuda T., Koshino R., Kobayashi K., Troy F. A., 2nd, Kitajima K. (2000) Frequent occurrence of pre-existing α2→8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins. Prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and polySia antibodies specific for defined chain lengths. J. Biol. Chem. 275, 15422–15431 [DOI] [PubMed] [Google Scholar]
- 113. Inoue S., Iwasaki M. (1980) Characterization of a new type of glycoprotein saccharides containing polysialosyl sequence. Biochem. Biophys. Res. Commun. 93, 162–165 [DOI] [PubMed] [Google Scholar]