Background: The possible role of ML-IAP in regulating MAPK signaling and cell migration is examined.
Results: ML-IAP directly binds to C-RAF and targets it for proteasomal degradation. Loss of ML-IAP leads to an increase in MAPK activity and cell migration. ML-IAP interacts directly with XIAP.
Conclusion: ML-IAP regulates C-RAF stability and cell migration.
Significance: There is a novel role of ML-IAP in regulating cell migration and MAPK signaling. IAPs exist in heteromeric complexes and regulate C-RAF stability.
Keywords: Cell Migration, MAP Kinases (MAPKs), Molecular Chaperone, RAF, Ubiquitination, XIAP, Chaperone, IAP-IAP, ML-IAP
Abstract
Inhibitor of apoptosis (IAPs) proteins are characterized by the presence of evolutionarily conserved baculoviral inhibitor of apoptosis repeat (BIR) domains, predominantly known for their role in inhibiting caspases and, thereby, apoptosis. We have shown previously that multi-BIR domain-containing IAPs, cellular IAPs, and X-linked IAP can control tumor cell migration by directly regulating the protein stability of C-RAF kinase. Here, we extend our observations to a single BIR domain containing IAP family member melanoma-IAP (ML-IAP). We show that ML-IAP can directly bind to C-RAF and that ML-IAP depletion leads to an increase in C-RAF protein levels, MAPK activation, and cell migration in melanoma cells. Thus, our results unveil a thus far unknown role for ML-IAP in controlling C-RAF stability and cell migration.
Introduction
Melanoma IAP (ML-IAP/Livin)2 is a member of the IAP family highly expressed in tumors (1). IAPs are a conserved class of proteins primarily characterized by the presence of one to three baculoviral IAP repeat (BIR) domains, which are protein-protein interaction motifs (2). BIR domains are zinc-coordinated folds stabilized by the presence of conserved cysteine and histidine residues, and these domains are required for the interaction of IAPs with various proteins including caspases (2). There are eight known mammalian IAPs: cellular IAPs (cIAP1 and cIAP2), XIAP, NAIP (Neuronal Apoptosis Inhibitory Protein), BRUCE (BIR repeat containing ubiquitin-conjugating enzyme), Survivin, ILP2, and ML-IAP. Five of these IAPs (cIAP1, cIAP2, XIAP, NAIP, ILP2, and ML-IAP) also possess a RING (really interesting new gene) domain that has two coordinated zinc atoms linked to seven cysteines and three histidines (3). RING domains act as E3 ubiquitin ligases and catalyze the conjugation of ubiquitin moieties to the substrates contributing to cellular signaling or substrate protein degradation via the proteasomes or lysosomes (4). These domains are also shown to be responsible for heteromerization between the IAPs and also for their cross-regulation (5). Some IAPs also possess an ubiquitin binding UBA (ubiquitin-associated) domain (6, 7).
The anti-apoptotic function of these IAPs makes them a potential target for cancer chemotherapy. Several of these IAPs have been linked to tumor cell survival (8), resistance to chemotherapy, and radiotherapy and tumor metastases (9–11). cIAP1 is recognized as an oncogene in hepatocellular carcinoma (12). ML-IAP is a single BIR domain containing IAP detected to be highly expressed in malignant melanoma and was therefore named melanoma IAP (1). Since then, ML-IAP has been found to be significantly increased in various other tumors like colon cancer (13), bladder cancer (14), renal cell carcinoma (15, 16), non-small cell lung carcinoma (17), leukemia (18), neuroblastoma (19), glioma (20), and so forth and, thus, have been associated with prognostic significance in several solid and liquid tumors. Further, ML-IAP is located in chromosome 20q13, a region frequently amplified in many tumors. ML-IAP exerts its anti-apoptotic function via both the BIR and RING domains. Apart from inhibiting caspases 3 and 7 in vitro and caspase 9 in vivo, it also promotes ubiquitin-proteasome-mediated degradation of Smac/DIABLO (Second mitochondria-derived activator of caspases/direct IAP binding protein with low pI), an important antagonist of IAPs (21, 22). Smac, on the other hand, has also been shown to antagonize ML-IAP by direct binding (23). Further, ML-IAP has been shown to be transcriptionally regulated by microphthalmia-associated transcription factor in melanoma cells (24) and β-catenin/TCF complex in non-small cell lung carcinoma (25) and MycN in neuroblastoma (26). Apart from being associated with apoptosis inhibition, ML-IAP has been shown to inhibit cell proliferation and induce cell cycle arrest in G0/G1 phase (27, 28).
C-RAF is a central member of the classical MAPK pathway and the first effector of RAS to be discovered (29, 30). C-RAF phosphorylates and activates MEK1/2, leading to further activation of ERK1/2, forming the prototype of a three-tier MAPK cascade to transmit signals from outside the cells to the nucleus (31, 32). This cascade regulates various cellular processes, including proliferation, migration, differentiation, and survival, and deregulation of the components of this pathway is often associated with cancers (33). Previous observations from our group have revealed an unexpected role of IAPs in MAP kinase signaling pathway by directly controlling the stability of C-RAF kinase (34). We have shown that cIAP1, cIAP2, and XIAP directly bind to C-RAF kinase to promote its polyubiquitination and proteasomal degradation. Also, down-regulation of IAPs either by the siRNA or IAP antagonist compound leads to an enhancement in MAPK-dependent cell migration in cancer cells (34, 35). Here, we extend our studies and show that ML-IAP also binds to C-RAF directly and that silencing of ML-IAP expression leads to stabilization of C-RAF protein in melanoma cells. This, in turn, leads to MAPK-dependent cell migration, revealing a novel role for ML-IAP. We characterize the mode of interaction between ML-IAP and C-RAF and further present evidence that IAPs interact with each other, thus emphasizing a role for IAP-IAP heteromeric complexes in regulating C-RAF stability.
EXPERIMENTAL PROCEDURES
Antibodies and Constructs
The following antibodies have been employed in this study: anti-ML-IAP mouse monoclonal antibody (R&D Systems), a second anti-ML-IAP mouse monoclonal antibody (a gift from Genentech), anti-C-RAF rabbit polyclonal C-12 antibody (Santa Cruz Biotechnology), anti-C-RAF-p-Ser-338 rabbit monoclonal antibody (Cell Signaling Technology), anti-C-RAF-p-Ser-259 rabbit polyclonal antibody (Cell Signaling Technology), anti-C-RAF-p-Ser-621 mouse monoclonal antibody (Santa Cruz Biotechnology), anti-β-actin rabbit polyclonal antibody (Sigma), anti-phospho-ERK1/2 rabbit polyclonal antibody (Thr-202/Tyr-204) (Cell Signaling Technology), anti-XIAP mouse monoclonal antibody (BD Biosciences), anti-cIAP1 goat antibody (R&D Systems), anti-HA probe (12CA5) mouse monoclonal antibody (Santa Cruz Biotechnology), normal rabbit IgG (Santa Cruz Biotechnology), and normal mouse IgG (Santa Cruz Biotechnology).
The following expression constructs have been employed in the study: pcDNA3 FLAG-ML-IAP WT, pcDNA3-FLAG ML-IAP D120A, pcDNA3-FLAG ML-IAP D138A, pcDNA3 FLAG-ML-IAP C124A, pcDNA3 FLAG-ML-IAPΔRING, pcDNA3 HA-C-RAF WT, pcDNA3 HA-C-RAF DD, and pcDNA3 EV. The various point mutations in the ML-IAP BIR domains were generated with a site-directed mutagenesis kit (Stratagene) following the instructions of the manufacturer.
Cell Culture and siRNA Knockdown
MEL-HO and Colo829 cells (a gift from Richard Marais) were cultured in RPMI 1640 medium supplemented with 10% FCS (Invitrogen) and 0.2% penicillin (100 units/ml)/streptomycin (100 μg/ml) (Invitrogen) at 37 °C in 5% CO2. 293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FCS (Invitrogen) and 0.2% penicillin (100 units/ml)/streptomycin (100 μg/ml) (Invitrogen) at 37 °C in 5% CO2. Sbcl2 cells (a kind gift from Richard Marais, Institute for Cancer Research, UK) were cultured in MCDB153 (Sigma)/L15 medium (Invitrogen, v/v:4/1) supplemented with CaCl2 (2 mmol/liter), insulin (5 μg/ml, Sigma), and 2% FCS. The cells were treated with U0126 (Calbiochem, catalog no. 662005) at a final concentration of 10 μm when needed.
To silence ML-IAP, XIAP, cIAP1, and C-RAF expression by RNA interference, ∼75,000 cells/well were seeded in a 12-well plate at least 20 h prior to transfection. Scrambled control siRNA as well as siRNAs directed against various genes were transfected using the Hiperfect (Qiagen), DharmaFECT® Duo (Dharmacon), or Lipofectamine RNAimax (Invitrogen) transfection kits. The cells were normally lysed at 48 h post-transfection. Unless otherwise mentioned, we transfected siRNAs at a final concentration of 60 nm. The following siRNAs were employed in this study: ML-IAP siRNA-1, target sequence TTGGATGCTTCTGAATAGAAA (Qiagen); ML-IAP siRNA-2, target sequence ATGGCTTAACTGTACCTGTTT (Qiagen); XIAP 3′UTR siRNA, target sequence CTGACTGATCTAATTGTATTA (Qiagen); cIAP1 siRNA-1, catalog no. J-004390-12-0010 (Dharmacon on target plus siRNA); cIAP1 siRNA-2, target sequence CTAGGAGACAGTCCTATTCAA (Qiagen); cIAP2 siRNA-1, target sequence AATTGGGAACCGAAGGATAAT (Qiagen); C-RAF 3′UTR siRNA, target sequence GTGGATGTTGATGGTAGTACA (Qiagen); and control siRNA, catalog no D-001210-01-20 (Dharmacon).
Transwell Migration Experiments
MEL-HO cells were transfected with siRNAs for 36 h and then seeded on to 8-μm transwell migration chambers (Corning, catalog no. 3422). The cells were stimulated with serum-free RPMI or RPMI with 10% FCS and added to the lower chamber. The cells were left to migrate for 12–14 h. Cells on the upper part of the membrane were scraped using a cotton swab, and the migrated cells were fixed in 3.7% (v/v) paraformaldehyde and stained with 0.4% crystal violet in 10% ethanol. The experiment was performed in triplicates for all conditions described. From every transwell, several images were taken under a phase contrast microscope at 10× magnification, and eight random fields per condition were considered for quantification. Analyses were performed using two special algorithms as described before (34). The results of the analysis are depicted as graphs. Student's t test was performed to check for the significance (*, p < 0.05; **, p < 0.01; ***, p < 0.005).
Reporter Gene Assays
SRE luciferase reporter assay was performed using two different expression systems. Firstly, using the 293T overexpression system, where the ELK1-GAL4 transactivator plasmid and reporter gene plasmid with a synthetic GAL4 promoter, GAL4-luciferase (a kind gift from Walter Kolch), were transiently transfected with C-RAF 340D/341D with or without ML-IAP. The luciferase activity was monitored using the dual luciferase reporter assay system (Promega, Mannheim, Germany) in accordance with the protocol of the manufacturer. Secondly, MEL-HO cells were transduced with the Cignal Lenti Pathway Reporter lentiviral particles using the Cignal Lenti SRE reporter (luc) kit (Qiagen, catalog no. CLS-010L). Following viral infection, the cells were cultured under puromycin selection to generate a homogenous population of transduced cells. The SRE luciferase assay to monitor the ERK1/2 activation was performed using the dual luciferase reporter assay system (Promega) in accordance with the protocol of the manufacturer. Briefly, transduced and selected MEL-HO cells were seeded on a 12-well plate and transfected with ML-IAP siRNAs using Lipofectamine RNAimax. The cells were harvested in lysis buffer 48 h after transfection, luciferase assays were performed in triplicates, and the activity of firefly luciferase was used as a reporter for detecting the activation of the MAPK pathway.
SDS-PAGE and Western Blotting
For SDS-PAGE, cells were lysed in 4× Laemmli buffer and boiled at 100 °C for 5 min before loading onto the polyacrylamide gels. After separation, the proteins were transferred to nitrocellulose membranes. For immunoblot analysis, membranes were blocked with 5% low fat milk in phosphate-buffered saline for 1 h and then incubated with various primary antibodies diluted in blocking buffer or in TBST (50 mm TRIS, 150 mm NaCl, and 0.05% Tween 20). Antigen antibody complexes were detected by incubating with horseradish peroxidase-coupled secondary antibodies followed by enhanced chemiluminescence (Amersham Biosciences). Quantification of Western blot analyses was performed by densitometry (ImageJ software, National Institutes of Health).
Immunoprecipitation
To immunoprecipitate endogenous proteins, cells were seeded on 10-cm dishes and, if required, transfected after 24 h and then lysed 48 h post-transfection. The cells were lysed with lysis buffer (50 mm Tris-HCl (pH 7.5), 250 mm NaCl, 1% Triton X-100, 1 mm NaVO3, 25 mm NaF, 1.5 mm MgCl2, 1 mm PMSF, β-mercaptoethanol (1:1000 dilution, Applichem), protease inhibitor mixture (1:100 dilution, Calbiochem), 10% glycerol) for 30 min on ice and sonicated twice for 10 s. Lysates were cleared by centrifugation for 15 min at 14,000 rpm. Endogenous ML-IAP or C-RAF proteins were then immunoprecipitated with respective antibodies for 15 h at 4 °C. The antigen-antibody complexes were precipitated by Sepharose-coupled protein A/G beads (Roche). The beads were then washed three times with the lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting. For immunoprecipitation of coexpressed proteins in 293T cells, we transfected 293T cells with various plasmids. The cells were lysed at 48 h post-transfection, and proteins were immunoprecipitated as mentioned above. Whenever needed, the cells were treated with proteasome inhibitor MG132 (Calbiochem) at a final concentration of 10 μm for 6 h before lysis. Controls immunoprecipitations were performed with IgG isotype control antibodies (Santa Cruz Biotechnology). Immunoprecipitation of purified proteins was done using the lysis buffer mentioned above. Briefly, the proteins were added to the lysis buffer and incubated with the XIAP antibody for 15 h at 4 °C, and then the complexes were processed as described above.
Expression of GST-tagged proteins and Their Purification
GST-XIAP- or GST-encoding plasmids were transformed into BL21-CodonPlus-competent cells, and transformation and protein purification was performed following standard protocols. Briefly, the transformed cells were grown in a liquid culture at 37 °C overnight with chloramphenicol and ampicillin with constant shaking. Part of this overnight culture was then added to fresh Luria-Bertani medium (Applichem) and incubated at 37 °C until it reached A600 0.6. Bacteria transformed with GST were then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C, whereas bacteria transformed with GST-XIAP were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 16 °C. The cells were pelleted and lysed in GST lysis buffer (50 mm Hepes (pH 7.5), 150 mm NaCl, 1 mm EDTA, 5% glycerol, 0.1% Nonidet P-40), 1 mm DTT, and 1 mm protease inhibitor mixture (Calbiochem). The GST proteins were bound to glutathione-Sepharose beads (GE Healthcare). ML-IAP and cIAP1 proteins were obtained from R&D Systems (catalog nos. 787-LV and 818-IA, respectively), and His-C-RAF was purified from SF9 cells in the laboratory of Ulf Rapp.
GST Pull-downs
GSH as well as GST beads were washed with GST pull-down buffer (20 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm DTT, 1% Nonidet P-40), and the GST-tagged proteins were added to the respective Eppendorf tubes with 3% BSA and incubated for 1 h at 4 °C. The beads were washed with washing buffer (20 mm HEPES (pH 7.5), 250 mm NaCl, 1 mm DTT, 1% Nonidet P-40) three times and then the untagged protein was added. The mix was incubated at 4 °C for 2 h. The beads were washed three times with wash buffer. The buffer was then removed completely using an insulin syringe. The samples were then prepared for SDS-PAGE.
Electronic Manipulation of Images
In some cases, the whole images were subjected to contrast or brightness adjustments. No other manipulations were performed unless stated otherwise.
RESULTS
ML-IAP Directly Interacts with C-RAF via Its BIR Domain
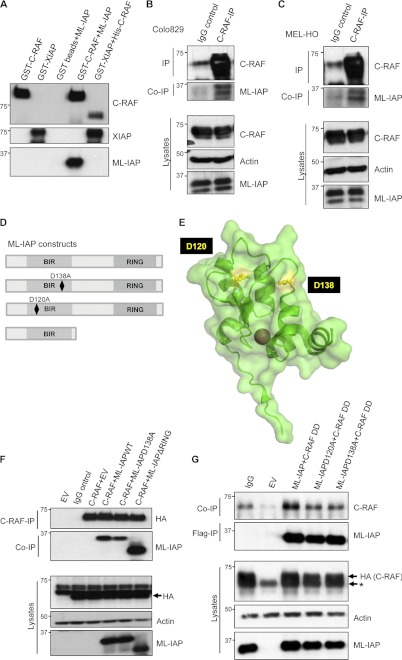
We have shown previously that C-RAF kinase can directly bind to multi-BIR domain-containing IAPs, cIAP1, cIAP2, and XIAP in a BIR domain-dependent manner (34). To further corroborate these observations and to further characterize the interaction, we generated several mutants of XIAP, including single BIR domains tagged to GST. Consistent with our previous studies, GST pull-down experiments with these mutants suggested a strong interaction between C-RAF and type II BIR domains (BIR2 and BIR3 domains) of XIAP. A XIAP mutant lacking the BIR1 domain binds more efficiently to C-RAF as compared with single BIR domains, and a XIAP mutant lacking BIR domains failed to interact with C-RAF under these settings (supplemental Figs. S1, A and B). We extended these studies to ML-IAP, a single BIR domain containing IAP. The ML-IAP BIR domain resembles the BIR2 domain of XIAP (1). To investigate whether ML-IAP can bind directly to C-RAF, we employed GST pull-down assays using purified proteins. These experiments revealed that ML-IAP could be precipitated by GST-tagged C-RAF and not by GST bead control (Fig. 1A). The interaction between His-C-RAF and GST-XIAP was used as a positive control. We tested a panel of tumor cell lines for endogenous expression of ML-IAP and found ML-IAP in detectable levels in only two melanoma cell lines, MEL-HO and Colo829, with two different ML-IAP antibodies. Therefore, we resorted to these two cell lines for our subsequent experiments. We tested for endogenous interaction between C-RAF and ML-IAP in these cells lines and could observe coprecipitating ML-IAP in C-RAF immunoprecipitates (Figs. 1, B and C). To further ascertain the residues responsible for driving the interaction between ML-IAP and C-RAF in vivo, we resorted to coimmunoprecipitation experiments in 293T cells. Various mutants of ML-IAP were generated, as shown in Fig. 1D. ML-IAPD120A and D138A are well characterized ML-IAP surface mutants lacking the capability to bind to IAP antagonist Smac (Fig. 1E) (23). HA-tagged C-RAF was expressed in 293T cells with the FLAG-tagged ML-IAP wild type and two other mutants, ML-IAPD138A and one lacking the RING domain (ML-IAPΔRING) (Fig. 1F). As expected, wild-type ML-IAP coprecipitated with C-RAF and, interestingly, the ML-IAPD138A mutant exhibited a reduced binding to C-RAF. Surprisingly, we detected a stronger interaction between C-RAF and ML-IAPΔRING as compared with ML-IAP, suggesting that the RING domain could possibly limit this interaction. As activated C-RAF exhibited a stronger interaction with XIAP (34), we explored whether a constitutively active C-RAF interacts with ML-IAP and whether this interaction can be disrupted by the mutations in the Smac binding domain of ML-IAP. A similar 293T overexpression experiment was performed with ML-IAP and its mutants to check for their interaction with a constitutively active C-RAF mutant, Y340DY341D (referred to here as C-RAF DD). These experiments revealed similar results, implying that mutations of residues Asp-120 and Asp-138 in the BIR domain of ML-IAP can largely impede C-RAF/ML-IAP interaction, although they are not sufficient to completely abolish this interaction (Fig. 1G).
FIGURE 1.
ML-IAP directly interacts with C-RAF via its BIR domain. A, ML-IAP was precipitated with GST-bound C-RAF, and the interaction was visualized by Western blot analysis. Interaction of GST-bound XIAP with C-RAF was used as a positive control for the experiment. B, ML-IAP interacts with C-RAF at endogenous levels in MEL-HO and Colo829 cells (C). Endogenous C-RAF was immunoprecipitated (IP), and the presence of coprecipitating (co-IP) ML-IAP was checked by immunoblot analyses. D, pictorial representation of ML-IAP WT and mutants used for overexpression experiments. E, representations of ML-IAP surface amino acid residues Asp-120 and Asp-138 adapted from the crystal structure of ML-IAP (PDB code 3F7G) generated using PyMOL software. F, Western blot analysis of HA-tagged C-RAF WT and FLAG-tagged ML-IAP WT and mutants expressed in 293T cells and immunoprecipitated using HA antibody. Arrow indicates the HA-tagged C-RAF. G, HA-tagged C-RAF Y340D Y341D (C-RAF DD) and FLAG-tagged ML-IAP WT and mutants were overexpressed in 293T cells, and ML-IAP was immunoprecipitated using FLAG antibody. Coprecipitated C-RAF was visualized by Western blotting. The asterisk represents residual endogenous C-RAF observed because of stripping and reprobing of blots after detection with C-RAF antibody. EV, empty vector control.
Loss of ML-IAP Leads to Stabilization in C-RAF Protein Levels
Mutations in NRAS and B-RAF are common in melanomas, and the V600E mutation in B-RAF enhances its kinase activity more than 500-fold. Although NRAS mutant melanomas are dependent on C-RAF kinase for the activation of the MEK1/2 and ERK1/2 kinases, B-RAF V600E mutant melanomas are completely dependent on B-RAF for their downstream signaling (36). To test whether IAPs influence C-RAF kinase in melanomas, we have screened various cell lines with mutations in NRAS or B-RAF V600E. We depleted IAPs in various cell lines, including Sbcl2 (NRAS*), A375, Colo829 (both expressing B-RAF V600E), and MEL-HO (Myc-amplified). As expected, C-RAF levels are increased in all these cases when individual IAPs are depleted with validated siRNAs (supplemental Fig. S2, A–D).
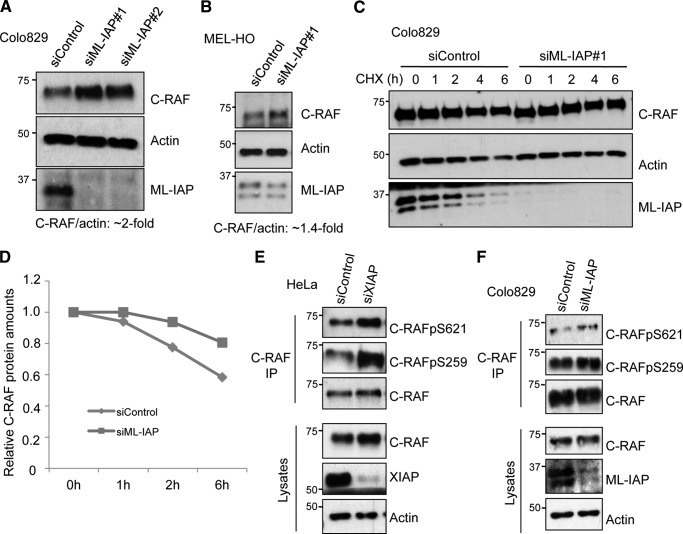
ML-IAP was then depleted in Colo829 and MEL-HO cells with two different siRNAs, and any alterations in the protein levels of C-RAF were tested by immunoblot analyses. We detected an increase (1.4–2-fold) in the protein level of C-RAF upon knockdown of ML-IAP in both cell lines (Fig. 2, A and B). We then tested whether the detected increase in the level of C-RAF protein is a result of increased protein stability. We performed cycloheximide chase experiments, which revealed that knockdown of ML-IAP in Colo829 as well as MEL-HO cells led to increased C-RAF protein stability (Fig. 2, C and D, and supplemental Fig. S3, A and B). These data confirm that ML-IAP regulates C-RAF protein stability in melanoma-derived cell lines.
FIGURE 2.
Loss of IAPs lead to stabilization in C-RAF protein levels. siRNA-mediated knockdown of ML-IAP in Colo829 cells (A) as well as in MEL-HO cells (B) leads to an increase in C-RAF levels, which was monitored by Western blot analysis. The levels of C-RAF were quantified using ImageJ software as detailed under “Experimental Procedures.” C, cycloheximide chase in Colo829 cells with knockdown of ML-IAP shows an increase in the C-RAF protein levels at all time points as compared with control cells using Western blotting. D, quantification of relative C-RAF protein levels in C by ImageJ software. HeLa cells (E) and Colo829 cells (F) were transfected with control and XIAP siRNAs, and the changes in C-RAF phosphorylation status were observed by Western blotting in the C-RAF immunoprecipitates (IP).
It has been shown previously that C-RAF kinase is stabilized in the cytosol by binding to 14-3-3, which is facilitated by phosphorylation at two residues, Ser-259 and Ser-621 (37). Ser-621 is an autophosphorylation site of C-RAF, preventing its proteasomal degradation, and Ser-259 has been identified as an inhibitory phosphorylation site (38, 39). Because binding of IAPs to C-RAF affects its stability, we wanted to test the effect of IAP knockdown on the phosphorylation status of Ser-621. We have shown previously that knockdown of X-/c-IAPs in HeLa cells leads to an increase in both the cytosolic and membrane pools of C-RAF with a consistent increase in the phosphorylation of both Ser-338 and Ser-259 (34). We knocked down XIAP in HeLa cells and ML-IAP in Colo829 cells and checked for the phosphorylation status of Ser-621 in the immunoprecipitates of endogenous C-RAF from control and knockdown cells by Western blotting (Fig. 2, E and F). We found that phosphorylation at Ser-621 and at Ser-259 in C-RAF is increased in IAP-depleted cells.
ML-IAP Is Required for Chaperone-mediated C-RAF Ubiquitination and Proteasomal Degradation
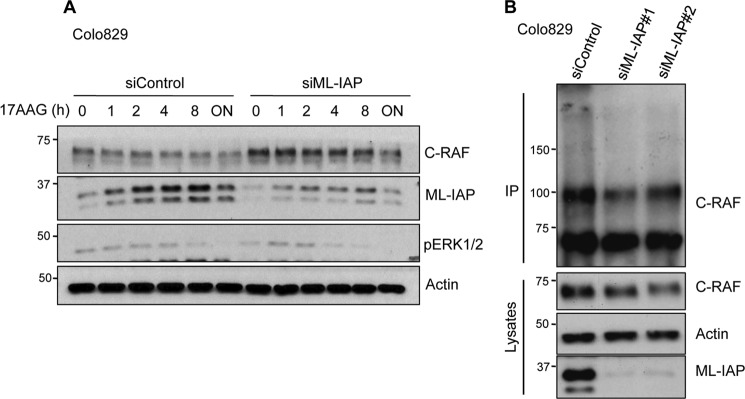
As C-RAF is a chaperone client protein, we tested whether ML-IAP could play a role in chaperone-mediated C-RAF degradation. We knocked down ML-IAP in Colo829 using siRNAs and checked for the kinetics of C-RAF degradation upon treatment with 17-AAG, which is a geldanamycin analog. Treatment with 17-AAG inhibits Hsp90 function in vivo and thereby triggers C-RAF degradation via proteasomes. Interestingly, degradation of C-RAF with 17-AAG was reduced at every time point when ML-IAP levels were depleted with siRNAs (Fig. 3A). These data demonstrated that ML-IAP functionally synergizes with the Hsp90 quality control system to regulate C-RAF protein stability in these cells. As the basal C-RAF levels are increased upon ML-IAP depletion, we tested whether ML-IAP regulated C-RAF polyubiquitination at endogenous levels. We transfected Colo829 cells with ML-IAP siRNAs, treated with MG132 for 6 h prior to lysis, and then immunoprecipitated C-RAF. Western blot analyses of the immunoprecipitated samples show a decrease in the polyubiquitinated smear of C-RAF in ML-IAP knockdown cells as compared with control cells (Fig. 3B). These experiments reveal that ML-IAP promotes C-RAF ubiquitination to regulate its protein homeostasis.
FIGURE 3.
ML-IAP is required for chaperone-mediated C-RAF ubiquitination. A, knockdown of ML-IAP was performed using siRNA in Colo829 cells and then treated with 17AAG for various time points. ML-IAP knockdown leads to an increase in C-RAF levels as compared with siControl cells for every time point that was monitored by Western blot analysis. B, ML-IAP was knocked down using siRNA in Colo829 cells. 48 h after transfection, cells were treated with MG132 for 6 h, and then endogenous C-RAF was immunoprecipitated (IP) as stated under “Experimental Procedures.” C-RAF ubiquitination was observed by Western blot analysis.
Loss of ML-IAP Activates the MAPK Cascade and Promotes Cell Migration in a MAPK-dependent Manner
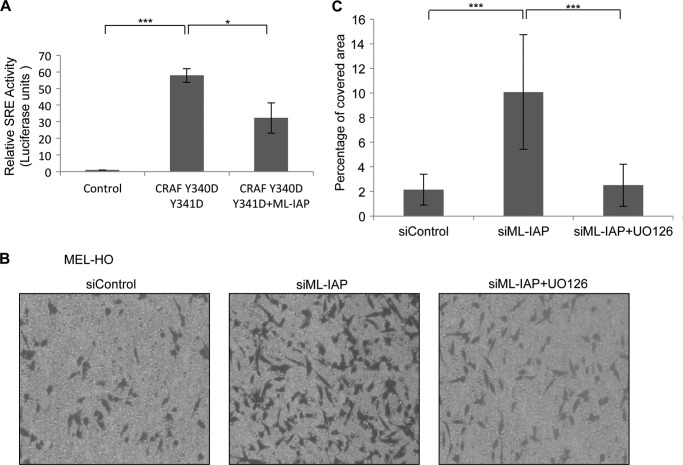
To test whether ML-IAP depletion-mediated C-RAF increase can lead to the activation of the classical MAPK cascade and increased ERK1/2 activity, we performed SRE (serum-responsive element) reporter assays. We used a 293T overexpression system where we transiently transfected the ELK1-GAL4 transactivator plasmid and reporter gene plasmid with a synthetic GAL4 promoter, GAL4-luciferase, with C-RAF DD with or without ML-IAP. The luciferase activity was monitored using the dual-luciferase reporter assay system. We observed a more than 50-fold increase in SRE activity upon expression of C-RAF DD, and this activity is significantly reduced upon coexpression of ML-IAP with C-RAF DD (Fig. 4A). Further, MEL-HO cells were stably transduced with lentiviruses carrying the SRE-responsive luciferase reporter constructs, and the cells were subsequently transfected with two different siRNAs targeted against ML-IAP. The cells were harvested in lysis buffer 48 h after transfection, and the activity of firefly luciferase was measured as a reporter for detecting the activation of the MAPK pathway. We detected a modest but significant increase in the SRE activity upon knockdown of ML-IAP in comparison to the controls (supplemental Fig. S4A). We used EGF stimulation as positive control and MEK inhibitor UO126 as a negative control (supplemental Fig. S4B).
FIGURE 4.
Loss of ML-IAP activates the MAPK cascade and promotes cell migration in MAPK-dependent manner. A, luciferase assay was performed using 293T overexpression system. The ELK1-GAL4 transactivator plasmid and reporter gene plasmid with a synthetic GAL4 promoter, GAL4-luciferase, were transiently transfected with C-RAF DD with or without ML-IAP, and the luciferase activity was monitored as mentioned under “Experimental Procedures.” B, a transwell migration assay was performed using MEL-HO cells with control as well as ML-IAP siRNAs 48 h after transfection and also with and without UO126 treatment overnight. C, quantification of transwell migration assay in B is graphically represented. Student's t test was performed to check for significance. *, p < 0.05; **, p < 0.01; ***, p < 0.005).
We wanted to test the functional significance of ERK1/2 activation upon ML-IAP depletion and checked for the migration ability of MEL-HO cells. We performed a transwell migration assay with cells transfected with control or ML-IAP siRNAs. ML-IAP-depleted MEL-HO cells exhibited an enhanced transwell migration (Fig. 4, B and C). To check whether the high migration index observed in ML-IAP depleted cells is dependent on MAPK activation, the cells were pretreated with MEK1/2 inhibitor UO126. As expected, the migration of ML-IAP-depleted MEL-HO cells were blocked by treatment with UO126 (Fig. 4, B and C). These data demonstrated that depletion of ML-IAP enhances cell migration in a MAPK-dependent manner.
IAPs Interact with Each Other to Form Heteromeric Complexes
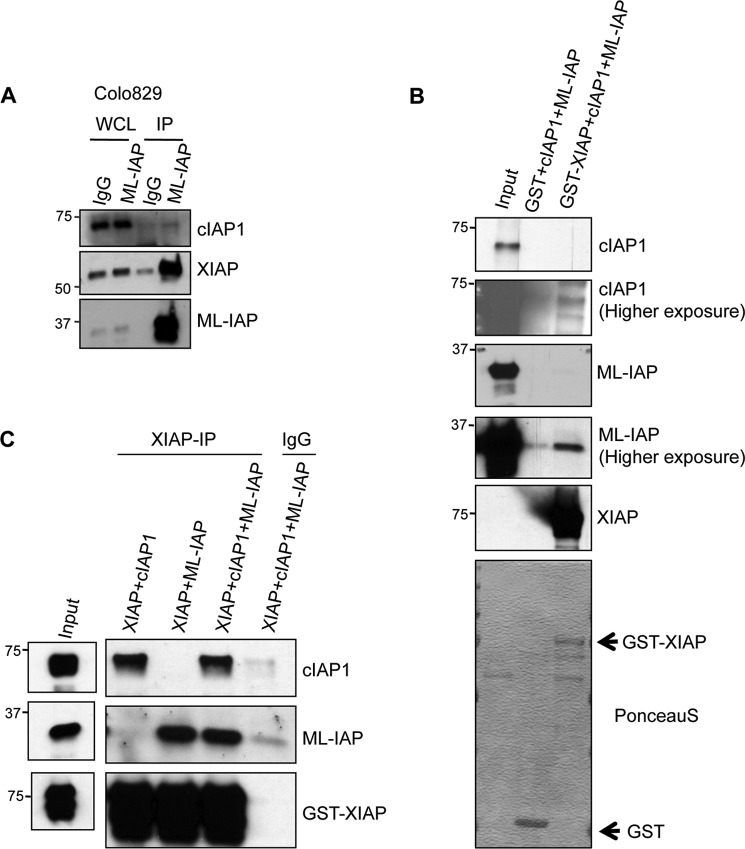
Knockdown of individual IAPs (XIAP, cIAP1, cIAP2, as well as ML-IAP) leads to a similar increase in the levels of C-RAF in various cell lines. These data suggested that these IAPs might form a functional heteromeric complex to regulate C-RAF stability. Further, previous studies have shown that IAPs exist in heteromeric protein complexes (IAPosomes) in the cytosol of various tumor cell lines (40). To directly test whether ML-IAP exist in complex with other IAPs at endogenous levels, we immunoprecipitated ML-IAP from Colo829 cells and checked for the presence of coprecipitating IAPs. As expected, we could detect significant amounts of XIAP and trace amounts of cIAP1 in ML-IAP immunoprecipitates, suggesting that ML-IAP exists in an IAP-IAP heteromeric complex in these cells (Fig. 5A). We then checked whether ML-IAP can directly interact with other IAPs by employing purified proteins. GST pull-down experiments confirmed that cIAP1 and ML-IAP could directly form a complex with GST-XIAP (Fig. 5B). As IAPs exhibit some background binding to GST beads, we repeated the experiments with an immunoprecipitation strategy. These experiments reaffirmed that cIAP1 and ML-IAP can directly interact with XIAP and that, when mixed together, they can be precipitated together, suggesting the formation of an IAP·IAP complex between these IAPs (Fig. 5C). These results confirmed that IAPs can directly interact with each other and that ML-IAP can directly interact with XIAP.
FIGURE 5.
IAPs interact with each other to form heteromeric complexes. A, ML-IAP was immunoprecipitated (IP) from Colo829 cells, and the coprecipitating cIAP1 and XIAP was analyzed by Western blot analyses. WCL, whole cell lysates. B, ML-IAP and His-cIAP1 interacts with GST-XIAP. The interaction between GST-XIAP and cIAP1 or ML-IAP or both was monitored with a GST pull-down assay. The proteins were detected by Western blot analyses. Arrow indicates GST tagged XIAP or GST proteins. C, purified GST-XIAP was immunoprecipitated from RIPA buffer containing His-cIAP1 or ML-IAP or both, and the coprecipitated proteins were monitored by Western blot analyses.
DISCUSSION
ML-IAP/Livin is a single BIR domain containing IAP and has been shown to resist apoptosis to regulate tumor cell survival (1). ML-IAP is overexpressed in various solid and liquid tumors with prognostic significance. Because ML-IAP expression has been significantly correlated with the tumor stage, it has turned out to be an attractive target for cancer therapy, and inhibition of ML-IAP by various strategies is pursued as an adjunct to chemotherapy.
This study identifies another role of ML-IAP in regulating MAPK signaling and cell migration. Our previous observations unveiled a role of cellular IAPs and XIAP in MAPK pathway regulation by affecting the stability of C-RAF kinase. We extend our observations to a single BIR domain containing IAP to observe if a single BIR domain IAP can bind to C-RAF and promote its degradation via the protein quality control machinery. Modeling studies revealed that the BIR domain of ML-IAP closely resembled the XIAP BIR2 domain, and our current data suggest that C-RAF preferentially binds to type II BIR domains and not to type I BIR domains (supplemental Fig. S1). Further, a combination of the BIR2 and BIR3 domains significantly enhanced the interaction between IAPs and C-RAF (supplemental Fig. S1). In these lines, it is interesting to point out that residues Asp-138 and Asp-120 of ML-IAP, which contribute to Smac binding, are also exploited for the interaction between ML-IAP and C-RAF, although the mutation of these residues in ML-IAP failed to completely abrogate interaction with C-RAF. These data are consistent with the observations that treatment of HeLa cells with cell-permeable Smac-N7-peptides led to stabilization of C-RAF protein in HeLa cells (34). The domain(s) of C-RAF responsible for driving IAP interaction is/are currently unclear, although kinase activity seems to enhance the interaction between XIAP and C-RAF in vitro (34). On the IAP front, we observed that the RING domain of ML-IAP seems to limit the interaction between C-RAF and ML-IAP (Fig. 1F). Loss of the RING domain facilitated stronger interaction between ML-IAP and C-RAF. Although BIR domains have been shown to regulate the activity of RING domains (41), it is interesting to detect a role for the RING domain in regulating the BIR binding activity. We have also observed that mutation of Zn2+-coordinating residues in ML-IAP leads to a strong destabilization of the ML-IAP protein, possibly by triggering the RING activity.3 Further, ML-IAP autoubiquitinates itself, and the RING domain might impair the binding of BIR domain to its putative partners by ubiquitinating the BIR domain. For instance, XIAP autoubiquitination sites are shown to be present in the BIR3 domain, contributing to its turnover (42).
IAPs directly bind to C-RAF, and it is tempting to propose that the interaction between IAPs and C-RAF is highly dynamic and dependent on various factors, including the activation status of C-RAF in vivo. Our experiments with RAF inhibitors revealed that C-RAF inactivation leads to destabilization of this kinase, and, in fact, recent studies from the laboratory of Cathrin Pritchard demonstrated that C-RAF autophosphorylates at Ser-621, which is crucial for 14-3-3 binding, failing which, the kinase is degraded via proteasomes (39). Consistently, the phosphorylation of Ser-621 is increased in IAP-depleted cells. PP5, on the other hand, has been shown to dephosphorylate C-RAF at Ser-338, leading to the direct inactivation of this kinase (43, 44). We propose that interaction with IAPs adds another layer of regulation for this kinase through ubiquitination and proteasomal degradation (Fig. 6). C-RAF, upon phosphorylation at Ser-259 and Ser-621, binds to 14-3-3 which, in turn, leads to the stabilization of the kinase in the cytosol. Indeed, phosphorylation of C-RAF at these residues is increased in IAP-depleted cells. These data suggest that there could be a potential competition between 14-3-3 and IAPs for binding to C-RAF in the cytosol, which could then decide the fate of C-RAF kinase in cells (Fig. 6). Clearly, the spatiotemporal dynamics of IAP-C-RAF interaction deserves further investigation. Although IAPs failed to regulate the protein levels of other RAF isoforms, A-RAF and B-RAF, they might still influence the activity of these kinases indirectly by influencing the dimer formation between C-RAF and other isoforms.
FIGURE 6.
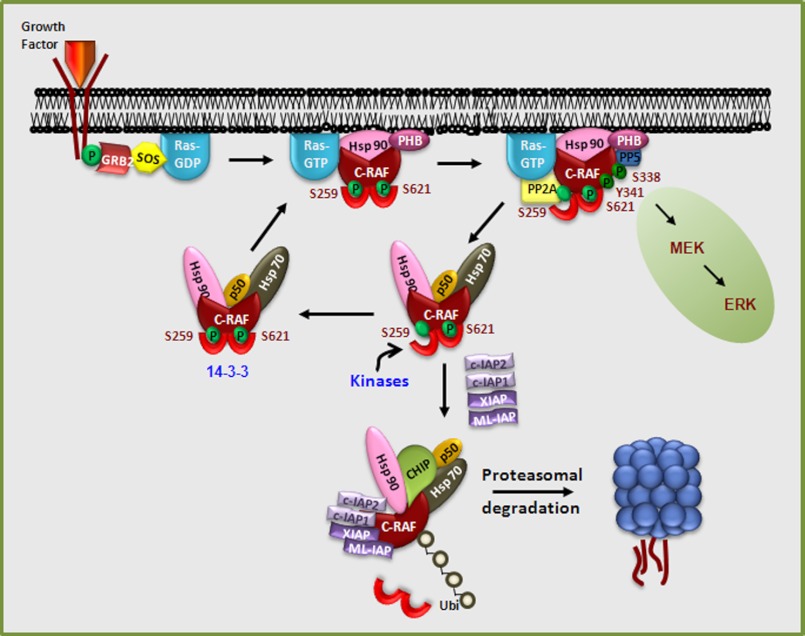
Spatiotemporal dynamics of IAP-C-RAF interaction. Growth factor binding to its receptor activates Ras, which, in turn, recruits C-RAF to the plasma membrane. Prohibitin assists in the displacement of 14-3-3 bound to C-RAF, leading to dephosphorylation of Ser-259 by PP2A, which facilitates stronger RAS binding and membrane localization, leading to activation. PP5 directly dephosphorylates Ser-338, leading to inactivation of C-RAF. Phosphorylation at Ser-259 and Ser-621 leads to 14-3-3 binding, which leads to stabilization and localization of C-RAF in the cytosol. Binding of IAP·IAP complexes could possibly displace 14-3-3 from C-RAF, leading to polyubiquitination and degradation of C-RAF. Polyubiquitination of C-RAF mediated by IAP binding adds another layer of regulation for this kinase. Inactivation of IAPs would stabilize C-RAF kinase, thus enhancing MAPK activation.
RNAi-mediated depletion of multiple IAPs led to an identical increase in C-RAF levels (supplemental Fig. 2). This could primarily be attributed to the formation of a functional IAP·IAP heteromeric complex (IAPosomes), which in turn regulate C-RAF stability. Previous studies have shown that IAPs exist in multimeric protein complexes and that they can heteromerize via their BIR domains and RING domains (5, 40). In these lines, we detected that cIAP1, XIAP, and ML-IAP coprecipitate at endogenous levels in Colo829 cells (Fig. 5). Further, we present evidence that ML-IAP can directly bind to XIAP and that XIAP-cIAP1 and ML-IAP can directly form an IAP·IAP complex in the absence of other proteins (Fig. 5). As several IAPs are required to regulate C-RAF homeostasis, we propose that IAP·IAP complexes rather than individual IAPs are functional in vivo. Currently, we are testing the possibility whether an IAP·IAP complex might function as a multisubunit E3 ubiquitin ligase. IAPs also cross-regulate each other, and loss of one IAP often leads to an increase in the protein levels of another IAP, which could also contribute to the lack of any major development phenotypes in single IAP-deficient mice (45). Further, the effects could also be attributed to species and cell type-specific differences. We have shown previously that the RING domain of XIAP is not required for promoting the polyubiquitination of C-RAF kinase in vivo and that XIAP recruits the cochaperone E3 ligase CHIP to the Hsp90·C-RAF complex (34). Our current data with ML-IAP also suggest that IAPs synergize with the chaperone machinery to regulate C-RAF levels. In fact, cIAP1 has been shown to be associated with Hsp90 during cellular differentiation (46). Further, silencing of CHIP expression has also led to an increase in C-RAF levels in these melanoma cell lines (data not shown).
Finally, we observed that silencing of ML-IAP promotes MAPK activation and cell migration, as revealed by the SRE reporter assays and transwell migration experiments. Although the activation of MAPK upon ML-IAP depletion was subtle, the effect was significant enough to drive the migration of these cells. These observations are consistent with previous reports from our group substantiating the fact that knockdown of IAPs using various strategies promotes cell migration in a MAPK/Rac1-dependent manner (35). ML-IAP has turned out to be a strong candidate for targeting cancer progression. As IAP antagonists are in clinical trials, these observations are important, as they shed further light into the biology of IAPs, which allow us to adroitly administer these promising therapeutic drugs. Finally, our results revealed that ML-IAP functions in concert with other IAPs, in heteromeric complexes, to regulate MAPK activation and tumor cell migration.
Supplementary Material
Acknowledgments
We thank Richard Marais for kindly providing various melanoma cell lines, Genentech for providing ML-IAP antibody, and Walter Kolch for the GAL-4 plasmids. We also thank Carrie Anderson and Conny Pyko for assistance with protein production, Masato Akutsu for advice on PyMOL software, and Taner Dogan for his advice on migration assays.
This work was supported by Emmy Noether Program Grant RA1739/1-1 from the Deutsche Forschungs Gemeinschaft (to K. R.).

This article contains supplemental Figs. S1–S4.
K. Rajalingam and T. K. Oberoi-Khanuja, unpublished observations.
- IAP
- inhibitor of apoptosis
- ML-IAP
- melanoma inhibitor of apoptosis
- BIR
- baculoviral inhibitor of apoptosis repeat
- cIAP
- cellular inhibitor of apoptosis
- XIAP
- X-linked inhibitor of apoptosis
- SRE
- serum-responsive element
- RAS
- Rat Sarcoma
- Raf
- rapidly growing fibrosarcoma.
REFERENCES
- 1. Vucic D., Stennicke H. R., Pisabarro M. T., Salvesen G. S., Dixit V. M. (2000) ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr. Biol. 10, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 2. Srinivasula S. M., Ashwell J. D. (2008) IAPs. What's in a name? Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaux D. L., Silke J. (2005) IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell Biol. 6, 287–297 [DOI] [PubMed] [Google Scholar]
- 4. Vaux D. L., Silke J. (2005) IAPs. The ubiquitin connection. Cell Death Differ. 12, 1205–1207 [DOI] [PubMed] [Google Scholar]
- 5. Silke J., Kratina T., Chu D., Ekert P. G., Day C. L., Pakusch M., Huang D. C., Vaux D. L. (2005) Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc. Natl. Acad. Sci. U.S.A. 102, 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gyrd-Hansen M., Darding M., Miasari M., Santoro M. M., Zender L., Xue W., Tenev T., da Fonseca P. C., Zvelebil M., Bujnicki J. M., Lowe S., Silke J., Meier P. (2008) IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-κB as well as cell survival and oncogenesis. Nat. Cell Biol. 10, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blankenship J. W., Varfolomeev E., Goncharov T., Fedorova A. V., Kirkpatrick D. S., Izrael-Tomasevic A., Phu L., Arnott D., Aghajan M., Zobel K., Bazan J. F., Fairbrother W. J., Deshayes K., Vucic D. (2008) Ubiquitin binding modulates IAP antagonist-stimulated proteasomal degradation of c-IAP1 and c-IAP2(1). Biochem. J. 417, 1–3 [DOI] [PubMed] [Google Scholar]
- 8. Augello C., Caruso L., Maggioni M., Donadon M., Montorsi M., Santambrogio R., Torzilli G., Vaira V., Pellegrini C., Roncalli M., Coggi G., Bosari S. (2009) Inhibitors of apoptosis proteins (IAPs) expression and their prognostic significance in hepatocellular carcinoma. BMC Cancer 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun J. G., Liao R. X., Zhang S. X., Duan Y. Z., Zhuo W. L., Wang X. X., Wang Z. X., Li D. Z., Chen Z. T. (2011) Role of inhibitor of apoptosis protein Livin in radiation resistance in nonsmall cell lung cancer. Cancer Biother. Radiopharm. 26, 585–592 [DOI] [PubMed] [Google Scholar]
- 10. Wu H. H., Wu J. Y., Cheng Y. W., Chen C. Y., Lee M. C., Goan Y. G., Lee H. (2010) cIAP2 up-regulated by E6 oncoprotein via epidermal growth factor receptor/phosphatidylinositol 3-kinase/AKT pathway confers resistance to cisplatin in human papillomavirus 16/18-infected lung cancer. Clin. Cancer Res. 16, 5200–5210 [DOI] [PubMed] [Google Scholar]
- 11. Gyrd-Hansen M., Meier P. (2010) IAPs. From caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat. Rev. Cancer 10, 561–574 [DOI] [PubMed] [Google Scholar]
- 12. Zender L., Spector M. S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S. T., Luk J. M., Wigler M., Hannon G. J., Mu D., Lucito R., Powers S., Lowe S. W. (2006) Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Xu J., Ju S., Ni H., Zhu J., Wang H. (2010) Livin gene plays a role in drug resistance of colon cancer cells. Clin. Biochem. 43, 655–660 [DOI] [PubMed] [Google Scholar]
- 14. Gazzaniga P., Gradilone A., Giuliani L., Gandini O., Silvestri I., Nofroni I., Saccani G., Frati L., Aglianò A. M. (2003) Expression and prognostic significance of LIVIN, SURVIVIN and other apoptosis-related genes in the progression of superficial bladder cancer. Ann. Oncol. 14, 85–90 [DOI] [PubMed] [Google Scholar]
- 15. Crnkovic-Mertens I., Wagener N., Semzow J., Grone E. F., Haferkamp A., Hohenfellner M., Butz K., Hoppe-Seyler F. (2007) Targeted inhibition of Livin resensitizes renal cancer cells towards apoptosis. Cell Mol. Life Sci. 64, 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kempkensteffen C., Hinz S., Christoph F., Krause H., Koellermann J., Magheli A., Schrader M., Schostak M., Miller K., Weikert S. (2007) Expression of the apoptosis inhibitor livin in renal cell carcinomas. Correlations with pathology and outcome. Tumour Biol. 28, 132–138 [DOI] [PubMed] [Google Scholar]
- 17. Tanabe H., Yagihashi A., Tsuji N., Shijubo Y., Abe S., Watanabe N. (2004) Expression of survivin mRNA and livin mRNA in non-small-cell lung cancer. Lung Cancer 46, 299–304 [DOI] [PubMed] [Google Scholar]
- 18. Qiuping Z., Jei X., Youxin J., Wei J., Chun L., Jin W., Qun W., Yan L., Chunsong H., Mingzhen Y., Qingping G., Kejian Z., Zhimin S., Qun L., Junyan L., Jinquan T. (2004) CC chemokine ligand 25 enhances resistance to apoptosis in CD4+ T cells from patients with T-cell lineage acute and chronic lymphocytic leukemia by means of livin activation. Cancer Res. 64, 7579–7587 [DOI] [PubMed] [Google Scholar]
- 19. Kim D. K., Alvarado C. S., Abramowsky C. R., Gu L., Zhou M., Soe M. M., Sullivan K., George B., Schemankewitz E., Findley H. W. (2005) Expression of inhibitor-of-apoptosis protein (IAP) livin by neuroblastoma cells. Correlation with prognostic factors and outcome. Pediatr Dev. Pathol. 8, 621–629 [DOI] [PubMed] [Google Scholar]
- 20. Yuan B., Ran B., Wang S., Liu Z., Zheng Z., Chen H. (2011) siRNA directed against Livin inhibits tumor growth and induces apoptosis in human glioma cells. J. Neurooncol. 107, 81–87 [DOI] [PubMed] [Google Scholar]
- 21. Ma L., Huang Y., Song Z., Feng S., Tian X., Du W., Qiu X., Heese K., Wu M. (2006) Livin promotes Smac/DIABLO degradation by ubiquitin-proteasome pathway. Cell Death Differ. 13, 2079–2088 [DOI] [PubMed] [Google Scholar]
- 22. Vucic D., Franklin M. C., Wallweber H. J., Das K., Eckelman B. P., Shin H., Elliott L. O., Kadkhodayan S., Deshayes K., Salvesen G. S., Fairbrother W. J. (2005) Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor. Implications for Smac-dependent anti-apoptotic activity of ML-IAP. Biochem. J. 385, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vucic D., Deshayes K., Ackerly H., Pisabarro M. T., Kadkhodayan S., Fairbrother W. J., Dixit V. M. (2002) SMAC negatively regulates the anti-apoptotic activity of melanoma inhibitor of apoptosis (ML-IAP). J. Biol. Chem. 277, 12275–12279 [DOI] [PubMed] [Google Scholar]
- 24. Dynek J. N., Chan S. M., Liu J., Zha J., Fairbrother W. J., Vucic D. (2008) Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 68, 3124–3132 [DOI] [PubMed] [Google Scholar]
- 25. Yuan D., Liu L., Gu D. (2007) Transcriptional regulation of livin by β-catenin/TCF signaling in human lung cancer cell lines. Mol. Cell Biochem. 306, 171–178 [DOI] [PubMed] [Google Scholar]
- 26. Dasgupta A., Peirce S. K., Findley H. W. (2009) MycN is a transcriptional regulator of livin in neuroblastoma. Oncol. Rep. 22, 831–835 [DOI] [PubMed] [Google Scholar]
- 27. Wang R., Lin F., Wang X., Gao P., Dong K., Zou A. M., Cheng S. Y., Wei S. H., Zhang H. Z. (2008) Silencing Livin gene expression to inhibit proliferation and enhance chemosensitivity in tumor cells. Cancer Gene. Ther. 15, 402–412 [DOI] [PubMed] [Google Scholar]
- 28. Wang H., Tan S. S., Wang X. Y., Liu D. H., Yu C. S., Bai Z. L., He D. L., Zhao J. (2007) Silencing livin gene by siRNA leads to apoptosis induction, cell cycle arrest, and proliferation inhibition in malignant melanoma LiBr cells. Acta Pharmacol. Sin. 28, 1968–1974 [DOI] [PubMed] [Google Scholar]
- 29. Kyriakis J. M. (2007) The integration of signaling by multiprotein complexes containing Raf kinases. Biochim. Biophys. Acta 1773, 1238–1247 [DOI] [PubMed] [Google Scholar]
- 30. Rajalingam K., Schreck R., Rapp U. R., Albert S. (2007) Ras oncogenes and their downstream targets. Biochim. Biophys. Acta 1773, 1177–1195 [DOI] [PubMed] [Google Scholar]
- 31. Kyriakis J. M., App H., Zhang X. F., Banerjee P., Brautigan D. L., Rapp U. R., Avruch J. (1992) Raf-1 activates MAP kinase-kinase. Nature 358, 417–421 [DOI] [PubMed] [Google Scholar]
- 32. Wellbrock C., Karasarides M., Marais R. (2004) The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 33. Downward J. (2003) Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 34. Dogan T., Harms G. S., Hekman M., Karreman C., Oberoi T. K., Alnemri E. S., Rapp U. R., Rajalingam K. (2008) X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat. Cell Biol. 10, 1447–1455 [DOI] [PubMed] [Google Scholar]
- 35. Oberoi T. K., Dogan T., Hocking J. C., Scholz R. P., Mooz J., Anderson C. L., Karreman C., Meyer zu Heringdorf D., Schmidt G., Ruonala M., Namikawa K., Harms G. S., Carpy A., Macek B., Köster R. W., Rajalingam K. (2012) IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 31, 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dumaz N., Hayward R., Martin J., Ogilvie L., Hedley D., Curtin J. A., Bastian B. C., Springer C., Marais R. (2006) In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 66, 9483–9491 [DOI] [PubMed] [Google Scholar]
- 37. Light Y., Paterson H., Marais R. (2002) 14–3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell Biol. 22, 4984–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dhillon A. S., Meikle S., Yazici Z., Eulitz M., Kolch W. (2002) Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noble C., Mercer K., Hussain J., Carragher L., Giblett S., Hayward R., Patterson C., Marais R., Pritchard C. A. (2008) CRAF autophosphorylation of serine 621 is required to prevent its proteasome-mediated degradation. Mol. Cell 31, 862–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajalingam K., Sharma M., Paland N., Hurwitz R., Thieck O., Oswald M., Machuy N., Rudel T. (2006) IAP-IAP complexes required for apoptosis resistance of C. trachomatis-infected cells. PLoS Pathog. 2, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dueber E. C., Schoeffler A. J., Lingel A., Elliott J. M., Fedorova A. V., Giannetti A. M., Zobel K., Maurer B., Varfolomeev E., Wu P., Wallweber H. J., Hymowitz S. G., Deshayes K., Vucic D., Fairbrother W. J. (2011) Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 334, 376–380 [DOI] [PubMed] [Google Scholar]
- 42. Shin H., Okada K., Wilkinson J. C., Solomon K. M., Duckett C. S., Reed J. C., Salvesen G. S. (2003) Identification of ubiquitination sites on the X-linked inhibitor of apoptosis protein. Biochem. J. 373, 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 44. von Kriegsheim A., Pitt A., Grindlay G. J., Kolch W., Dhillon A. S. (2006) Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat. Cell Biol. 8, 1011–1016 [DOI] [PubMed] [Google Scholar]
- 45. Conze D. B., Albert L., Ferrick D. A., Goeddel D. V., Yeh W. C., Mak T., Ashwell J. D. (2005) Posttranscriptional down-regulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell Biol. 25, 3348–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Didelot C., Lanneau D., Brunet M., Bouchot A., Cartier J., Jacquel A., Ducoroy P., Cathelin S., Decologne N., Chiosis G., Dubrez-Daloz L., Solary E., Garrido C. (2008) Interaction of heat-shock protein 90 beta isoform (HSP90 β) with cellular inhibitor of apoptosis 1 (c-IAP1) is required for cell differentiation. Cell Death Differ. 15, 859–866 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.