Background: TTF-1, an amplified lung oncogene, also suppresses lung cancer metastasis, whereas the occludin phenotype in lung carcinomas is poorly understood.
Results: TTF-1 transactivates occludin and occludin expression, reversing metastatic characteristics of lung cancer cells.
Conclusion: Transcriptional regulation of occludin by TTF-1 negates metastatic properties of lung carcinoma cells.
Significance: This study describes a novel extension of the multipronged anti-metastatic activity of TTF-1.
Keywords: Lung Cancer, Metastasis, Migration, Tight Junctions, Transcription Factors, NKX2–1, TTF-1, Claudin-1, Occludin
Abstract
The thyroid transcription factor 1 gene (TTF-1 or NKX2–1) is essential to lung development; however, it is also a critical factor in lung cancer. TTF-1 is amplified in lung cancers, suggesting that it is a gain-of-function lung oncogene. Conversely, TTF-1 counters epithelial to mesenchymal transition in cell-based studies and inhibits progression of primary lung adenocarcinomas to metastases in an animal model of lung adenocarcinomas. The unifying theory regarding TTF-1 is that it exhibits both pro-oncogenic and anti-metastatic function depending on the cellular context. Occludin is the first discovered constituent of the epithelial tight junction; in recent years, a functional role of occludin as a tumor suppressor has begun to emerge. Here, we demonstrate that TTF-1 transactivated the expression of the epithelial tight junction molecules occludin (OCLN) and claudin-1 (CLDN1). We show that transcriptional activation occurred through a direct interaction of TTF-1 with the OCLN and CLDN1 promoters. Furthermore, in cells that lack TTF-1, exogenous TTF-1 expression dampened the inhibitory effect of TGF-β on occludin and claudin-1 content. Using cells derived from a genetically engineered mouse model of lung adenocarcinomas, we observed that silenced TTF-1 expression down-regulated occludin, which we supported with additional siRNA experiments. Finally, TTF-1 knockdown conferred human lung cancer cells resistance to anoikis, and expression of occludin restored cellular sensitivity to anoikis. Overexpression of occludin impeded migration and induced anoikis in lung carcinoma cells. Collectively, these data suggest that TTF-1 transcriptionally regulates occludin, which represents another avenue of TTF-1-mediated metastasis suppression.
Introduction
The thyroid transcription factor 1 gene (TTF-1 or NKX2–1) is essential to fetal lung development (1); however, it is also a critical player in lung tumorigenesis. TTF-1 is a member of the homeodomain-containing transcription factor family. Homeodomains are protein motifs that bind to specific bases and are critical in development and differentiation (2). TTF-1 is located on human chromosome 14q13.3, which can undergo recurrent gene amplification in lung cancers (3–6). The presence of TTF-1 in a lung cancer-associated amplicon suggests an oncogenic function for TTF-1. Indeed, several lines of experimental evidence support this concept. Kendall et al. (3) detected a multigenic amplicon at 14q13.3 in which three genes (TTF-1, NKX2–8, and PAX9) were co-amplified. Enforced co-expression of any two of the three co-amplified transcription factor genes increased lung epithelial proliferation (3). More recently, analysis of mouse models of lung adenocarcinomas revealed an enrichment of a TTF-1-regulated protein set in the plasmas of these animals (7). Furthermore, ectopic expression of TTF-1 due to genetic rearrangement was detected in a subset of T cell acute lymphoblastic leukemia (8). Finally, TTF-1 activates the expression of the receptor tyrosine kinase-like orphan receptor 1 (ROR1), which is critical to balancing the actions of the PI3K-AKT pathway and p38 in lung adenocarcinomas (9). Taken together, the data reported in these studies are consistent with an oncogenic role of TTF-1.
Conversely, recent reports also suggest that TTF-1 suppresses lung adenocarcinoma progression and metastasis. Immunohistochemical analysis of human lung adenocarcinomas demonstrates that TTF-1 positivity is associated with a better clinical outcome compared with patients with TTF-1-negative tumors (10–13). Further, TTF-1 counters epithelial to mesenchymal transition (EMT)3 through activation of the E-cadherin (CDH1) promoter and a subsequent increase in E-cadherin protein content, thus impeding transforming growth factor-β (TGF-β)-induced EMT and associated EMT phenotypes (14). Moreover, in a lentiviral Cre-driven genetically engineered mouse model of lung adenocarcinomas, analysis of non-metastatic and metastatic tumors revealed that loss of Ttf-1 expression was required for primary lung tumor cells to metastasize (15). Finally, the chromosomal region at 14q13.3 containing the TTF-1 amplicon can also undergo allelic loss in lung cancers (16). Clearly, the function of TTF-1 in cancers is multifaceted and context-dependent.
The tight junction (TJ) complex is the most apical junction complex in the cell and contributes to the regulation of paracellular flux. TJs are composed of integral membrane proteins, such as occludin, the claudins, tricellulin, and junctional adhesion molecules along with the cytosolic, membrane-associated zonula occludens (ZO) family members, which link the membrane-bound proteins to the actin cytoskeleton (17). Although a great deal of attention has been given to TJ proteins for their role in regulating barrier function, there is a rekindled interest in studying the role of TJs in cell proliferation control and transformation. Genetic ablation of occludin in mice leads to mucus cell hyperplasia (18). Further, silencing occludin increases proliferation rates in ARPE-19 cells (19), and phosphorylation of occludin regulates the G2/M transition in Madin-Darby canine kidney cells (20). Occludin also contributes to directional migration in epithelial cells (21). Moreover, RAF-1-mediated transformation of rat salivary epithelial cells leads to the transcriptional silencing of occludin through Slug (22). However, introduction of human occludin sufficiently rescues the transformed phenotype (23). Finally, Osanai et al. (24) found that occludin expression retards migration in the B16F10 (murine melanoma) cell line and induces anoikis in HeLa (human cervical cancer), B16F10, AC2M2 (metastatic murine breast cancer), and MCF7 (human breast cancer) cell lines. Furthermore, stable expression of occludin in B6F10 and AC2M2 cells, followed by injection into the craniolateral thorax and mammary fat pad, respectively, reduces metastasis to the lung (24). These data clearly indicate that occludin is critical to metastasis suppression; however, the functional consequence of occludin expression on metastatic characteristics in lung carcinoma cells has not been explored.
The claudin family as a whole is emerging as important players in multiple cancer types (25, 26). Intuitively, it seems that the expression of claudins would decrease during cellular transformation and cancer progression. However, the current literature indicates that the expression alterations of claudins vary and are cancer-specific (25, 26). For example in colon cancer, claudin-1 expression increases with the progression of colon carcinomas and was found to regulate cellular transformation and metastatic behavior of primary cancer cells (27), but in lung cancer, claudin-1 appears to be down-regulated in lung adenocarcinomas (28) and to suppress invasion and metastasis (29). Analysis of CLDN1 mRNA content and immunohistochemical reactivity in two independent lung cancer patient populations demonstrates a correlation between low claudin-1 and shorter overall survival in patients with lung adenocarcinomas (29). Other claudin family members, such as claudin-3 and claudin-4, are frequently up-regulated in ovarian cancer and may represent novel biomarkers for this disease (30). Although lower expression of claudin-2 was seen in breast and prostatic carcinomas, expressions of claudin-1 and claudin-7 increase in the cervical neoplasia (31, 32). Via genome-wide RNA expression profiling, a new claudinlow subtype of breast cancer was recently identified (33). This group of claudinlow breast tumors shows low expression of many claudin genes and has features of stem cells and EMT (34). Clearly, the claudin family of 27 members harbors TJ proteins of diverse cancer phenotypes.
In this study, we set out to examine the connection between TTF-1 and TJ proteins. Here, we show that TTF-1 transactivates OCLN and CLDN1 through a direct interaction with respective promoters. Expression of TTF-1 in a premalignant human lung cell line also induced the expression of occludin and claudin-1. This in turn affects the net cellular responses to TGF-β. Intriguingly, only occludin expression paralleled that of TTF-1 in cell lines derived from a mouse model of primary lung adenocarcinomas, and RNA interference experiments supported these findings. Finally, functional studies in lung carcinoma cell lines demonstrated that occludin expression reversed cellular activities toward metastasis. Our data connect TTF-1 to epithelial TJ and support occludin as a mediator of the anti-metastatic activity of TTF-1.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmid Constructs
NCI-H441, A549, NCI-H1299, and Beas-2B cells were acquired from the ATCC. Mouse TnonMet cells were generated by Winslow et al. (15). NCI-H441, A549, and NCI-H1299 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and penicillin and streptomycin. Beas-2B cells were maintained in Keratinocyte serum-free medium supplemented with human recombinant EGF, bovine pituitary extract, and penicillin and streptomycin. Mouse TnonMet cells were maintained in DMEM supplemented with 10% fetal bovine serum and penicillin and streptomycin. All cells were cultured at 37 °C at 6% CO2. The OCLN promoter was cloned per Felinski et al. (35). The OCLN-GFP fusion cDNA was created in the laboratory of Dr. David Antonetti using the two restriction sites (XhoI/SalI) of the vector pMaxFP-Green-C. The CLDN1 and SpB (surfactant protein B) promoters were amplified from human genomic DNA using PCR and were cloned into pGL4.1 Basic vectors. TTF-1 was amplified from a human cDNA library and was cloned into the pcDNA3.1 vector. Deletion mutants of TTF-1 were created using a PCR strategy where primers directed at nucleotides flanking the region to be deleted were used for amplification. Subsequent amplicons were digested and religated to loop out the desired region. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene) according to the manufacturer's protocol. Primer sequences are listed in the supplemental Table S1.
RNA Interference and Chemicals
All small interfering RNAs (siRNAs) were purchased from Dharmacon: siTTF-1(oligo#1) (catalog no. D-019105–03) and siTTF-1(oligo#2) (catalog no. D-019105–04). TGF-β1 was obtained from R&D Systems. Doxycycline was purchased from MP Biomedicals.
Transfections
All transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Transient transfection efficiency was monitored by GFP expression and was estimated to be ≥90%. Stable pools (Beas-2B cells, TTF-1 doxycycline-inducible) were created by transfecting cells followed by G418 selection.
Luciferase Assays
Cells were seeded in 96-well plates and transfected as indicated the next day. The day following reporter transfections, luciferase assays were performed using the Dual-Glo luciferase assay (Promega). Luminescence values were quantified using a Glomax 96 microplate luminometer (Promega). Relative luciferase activities were normalized for transfection efficiency by dividing Firefly luciferase values by Renilla luciferase values.
Chromatin Immunoprecipitation (ChIP)
NCI-H441 cells were lysed in ChIP buffer (150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.5% Nonidet P-40, 50 mm Tris, pH 7.4). Chromatin shearing was performed using a Bioruptor (Diagenode). Whole cell lysates were immunoprecipitated using 5 μg of the indicated antibody. Real-time PCR was performed with SYBR Green detection using a Step One plus real-time PCR system (Applied Biosystems). The percent immunoprecipitated was calculated as follows: (immunoprecipitated signal/input signal) × 100. Primer sequences are listed in the supplemental Table S1.
mRNA Quantitation
Total RNA was harvested using the RNeasy total RNA kit (Qiagen). Approximately 300 ng of total RNA was reverse transcribed as described previously (36). Reactions were performed using TaqMan reagents and normalized to β-actin. Primer sequences are listed in the supplemental Table S1.
Protein Harvest and Immunoblotting
Cells were harvested using membrane-solubilizing buffer (100 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.2% SDS, 2 mm EDTA, 10 mm HEPES, pH 7.5). For Western blot analysis, 5–20 μg of whole cell lysate was fractionated using PAGE and transferred to a nitrocellulose membrane. Blocked membranes were incubated with antibodies (TTF-1 (1:1000 (Santa Cruz Biotechnology, Inc.) or 1:2000 (Dako)); Occludin (Invitrogen), 1:1000; Claudin-1 (Invitrogen), 1:1000; E-cadherin (Cell Signaling), 1:1000; β-tubulin (Cell Signaling), 1:1000; Hsp90 (BD Transduction Laboratories), 1:2000) overnight, and proteins were detected with appropriated HRP-conjugated secondary antibodies (1:3000; Thermo Scientific).
Wound Healing Assay
Transfected cells were grown to a monolayer and wounded with a pipette tip. Cells were imaged using a Nikon Eclipse TS 100 microscope (×4 objective) each day as indicated, and the width of the wound was measured using NIS Element D 3.0 software (Nikon). For each biological replicate, three distances were measured and averaged. The percentage of coverage was determined as follows: ((starting width − measured width)/starting width) × 100.
Anoikis Assay
Anoikis was assayed by plating cells, 24 h post-transfection, in wells coated with polyHEMA (20 mg/ml, resuspended in 95% ethanol). Coated wells were washed three times with 1× PBS to remove residual ethanol. Transfected cells were seeded, and cell death was monitored using trypan blue exclusion the following day. For each biological replicate, a minimum of 100 cells were scored.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism version 5 software by performing Student's t test (one control and one experimental group), revised one-way analysis of variance followed by Tukey's post-test (multiple comparisons) or two-way analysis of variance (time course). Data presented represent the mean ± S.D. of one representative experiment that was repeated at least twice. Data with p < 0.05 were considered to be statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
TTF-1 Transactivates the OCLN and CLDN1 Promoters through a TTF-1 Binding Element
A recent study demonstrated that in A549 cells, TTF-1 expression activates the E-cadherin gene (CDH1) promoter and subsequently increases E-cadherin intracellular staining (14); however, nothing was described with regard to TTF-1 and TJ protein expression. Therefore, we tested if TTF-1 could induce the protein expression of occludin and claudin-1. To accomplish this, we chose A549 cells because of its lack of endogenous TTF-1 expression and the fact that it is a cell host commonly used to characterize TTF-1 biology (14). Transient expression of TTF-1 in A549 cells increased occludin and claudin-1 protein level but did not affect ZO-1 protein content (Fig. 1A).
FIGURE 1.
TTF-1 transactivates the OCLN and CLDN1 promoters through a TTF-1 binding element. A, A549 cells were transfected with TTF-1 and harvested for protein 48 h later. Western blot analysis of ZO-1, occludin, claudin-1, TTF-1, and Hsp90 (loading control) are shown. B, schematic of the OCLN and CLDN1 promoters where the putative TBE is depicted in white (top). The putative TBE in OCLN and CLDN1 was compared with the consensus TBE (indicated with an asterisk) and mutated to a KpnI restriction site (underlined). C and D, luciferase assays of the OCLN and CLDN1 promoters using EV, wild-type TTF-1 (WT), and mutated TBE (TBEmut) constructs. ***, EV versus WT, p < 0.001; §, WT versus TBEmut and EV versus TBEmut, p < 0.001; @, WT versus TBEmut, p < 0.001, and EV versus TBEmut, not significant; n = 6. Error bars, S.D.
Considering that TTF-1 expression increased occludin and claudin-1 protein, we posited that this was likely through transactivation of their promoters. In order to identify potential TTF-1 binding sites, we performed an in silico analysis of the OCLN and CLDN1 promoters using PROMO, an algorithm used for identifying putative transcription factor binding sites (37, 38). Based on the PROMO prediction, we identified a potential TTF-1 binding element (TBE) in both the OCLN and the CLDN1 promoters. In order to strengthen the argument that these sites could function as TBEs, we compared them to known TBEs. Intriguingly, our potential TBEs had 64% identity (Fig. 1B) to the consensus TBE (GNNCACTCAAG, designated TBE*) described previously (39). Therefore, we mutated these sites within the TBE (Fig. 1B, underlined sequence). To test if the wild type promoters could be transactivated by TTF-1 and if mutation of the TBE (TBEmut) would alter the TTF-1 transactivation of the OCLN and CLDN1 promoters, we co-expressed control (empty pcDNA3.1 expression vector) or TTF-1 plasmid along with empty luciferase reporter vector (EV), OCLN/CLDN1 (WT) promoters, and OCLN/CLDN1 (TBEmut) promoters in A549 cells. TTF-1 transactivated the OCLN and CLDN1 WT promoters; however, transactivation of both OCLN and CLDN1 TBEmut by TTF-1 was reduced by ∼50% (Fig. 1, C and D). Taken together, we conclude that the PROMO-identified TBE does contribute to the TTF-1 transactivation of the OCLN and CLDN1 promoters.
To further investigate the TTF-1 regulation of occludin and claudin-1, we altered two amino acids simultaneously within the homeodomain of TTF-1 known to dictate DNA binding specificity, Q50K and Y54M (40–42), and performed luciferase assays using the OCLN and CLDN1 promoters (amino acid numbers describe the position relative to their location in the homeodomain). A known target of TTF-1, the surfactant protein B (SpB) promoter (43), was included as a reference. Protein expression of wild-type TTF-1 and TTF-1(Q50K/Y54M) was verified by Western blot (Fig. 2A). Previous mutagenesis work suggests that the Lys-50/Met-54 pair in a homeodomain would severely reduce its capacity of discriminating different DNA sequences (42). We thus speculated that the homeodomain double point mutant of TTF-1 might lose transactivation activities toward specific substrate DNAs. Indeed, WT TTF-1 activated the SpB promoter, whereas TTF-1(Q50K/Y54M) failed to do so (Fig. 2B). Surprisingly, TTF-1(Q50K/Y54M) retained wild-type activity toward the OCLN promoter reporter (Fig. 2C), whereas it exhibited a reduction in transactivating CLDN1 promoter reporter by ∼50% of wild-type TTF-1 (Fig. 2D). These observations show that the TTF-1-directed transcriptional regulations of pulmonary surfactant proteins and TJ molecules are mechanistically different and can be uncoupled through sequence manipulation of TTF-1.
FIGURE 2.

A TTF-1 DNA binding domain point mutant attenuates the transactivation of SpB yet differentially regulates CLDN1 and OCLN transactivation properties. A, Western blot analysis of the expression of TTF-1(WT) and TTF-1(Q50K/Y54M). A549 cells were transfected with the indicated TTF-1 mutants along with the SpB (B), OCLN (C), and CLDN1 (D) promoters, and luciferase assays were performed the following day (n = 12). Error bars, S.D.
Effects of TTF-1 Overexpression on Expression of Occludin and Claudin-1 following TGF-β Treatment
TGF-β promotes EMT and is known to disrupt the cellular junction by suppressing expression of junctional proteins, such as occludin and claudin-1 (44). In view of our finding that TTF-1 up-regulates TJ molecules, we surmised that TTF-1 may “dampen” the suppressive action of TGF-β on occludin and claudin-1. To test this hypothesis, we created a human lung cell system in which the expression of TTF-1 is subject to doxycycline induction. The system was based in a premalignant lung epithelial cell line, Beas-2B (45). Like A549 cells, Beas-2B cells do not express TTF-1 endogenously (46), thus maximizing the potential downstream signaling derived from the TTF-1 transgene. Furthermore, since the viral oncogene-immortalized Beas-2B cells are non-malignant, it provides a cellular background mimicking “normal” lung epithelia. Beas-2B cells were stably transfected with plasmids containing a doxycycline response element and vector alone (rtTA(EV)) or inducible TTF-1 (rtTA(TTF-1)) to perform our studies. Initially, we examined the TJ mRNA content as a consequence of TTF-1 expression. We found that turning on the expression of TTF-1 without TGF-β treatment increased OCLN and CLDN1 mRNA by ∼3- and 5.5-fold, respectively (Fig. 3, A and B). Treatment with TGF-β alone in the absence of TTF-1 induction (rtTA(EV)) resulted in a reduction of OCLN and CLDN1 mRNA (Fig. 3, A and B), which is consistent with the literature (44). However, turning on TTF-1 expression in the presence of TGF-β restored OCLN mRNA to the control level and increased CLDN1 mRNA to 4-fold over the control (Fig. 3, A and B). Further, Western blot analysis of occludin and claudin-1 revealed that changes at the protein level paralleled the observations with their mRNAs (Fig. 3C). In summary, these data corroborate our findings in A549 cells and demonstrate that TTF-1 alleviates the suppressive action conferred by TGF-β on the OCLN and CLDN1 expression.
FIGURE 3.
Effects of TTF-1 overexpression on expression of occludin and claudin-1 following TGF-β treatment of Beas-2B cells. Beas-2B cells were stably transfected with plasmids containing the doxycycline response element and vector alone (rtTA(EV)) or inducible TTF-1 (rtTA(TTF-1)). Cells were treated with TGF-β for 48 h and then doxycycline was added in the presence or absence of TGF-β for an additional 24 h. A and B, total RNA was isolated and analyzed by reverse transcription real-time PCR using TaqMan probes specific to OCLN, CLDN1, and GAPDH. OCLN and CLDN1 values were normalized to GAPDH. C, cells were harvested for Western blot analysis of occludin, claudin-1, TTF-1, and β-tubulin (loading control) (n = 3). Error bars, S.D.
As an independent means to confirm our findings, we took on a loss-of-function approach by silencing TTF-1 expression using pooled siRNAs for 3 days in a human lung cancer cell line expressing a high level of endogenous TTF-1 (NCI-H441). Three days after siRNA transfection, we co-expressed either the OCLN or CLDN1 promoter reporters and added TGF-β, and we performed luciferase assays the following day. Contrary to its suppressive action on OCLN and CLDN1, TGF-β in this cell system resulted in higher activities of OCLN and CLDN1 promoter reporters (supplemental Fig. S1). Nevertheless, TTF-1 knockdown compromised the TGF-β stimulation of TJ promoter activities (supplemental Fig. S1), demonstrating the requirement of TTF-1 for this phenotype. In conclusion, both gain- and loss-of-function studies support the notion that TTF-1 drives transcription of occludin and claudin-1 in the absence or presence of TGF-β.
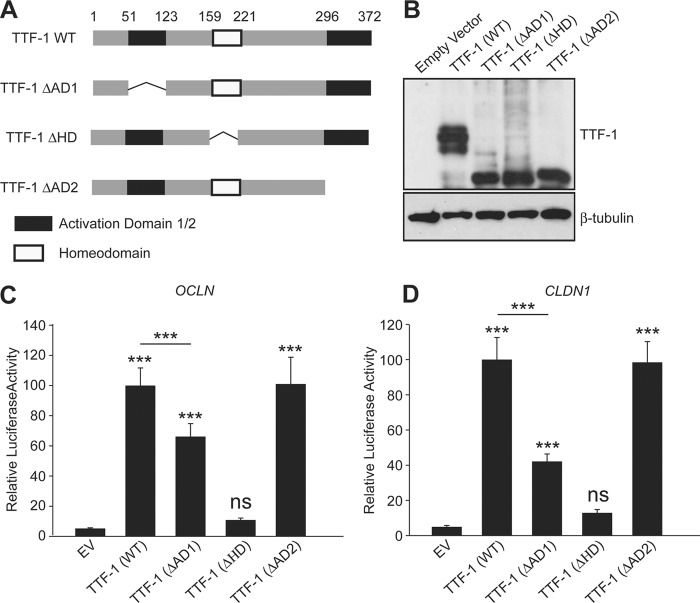
The Activation Domain-1 and Homeobox Domain of TTF-1 Are Required for TTF-1 Activation of OCLN and CLDN1
An earlier study identified two separate domains of TTF-1 involved in transcriptional activation (47); therefore, we wished to determine if the two activation domains were differentially utilized for TTF-1-mediated OCLN and CLDN1 transactivation. To this end, we created plasmids encoding TTF-1 mutants lacking the activation domain 1 (ΔAD1), homeodomain (ΔHD), or activation domain 2 (ΔAD2) as illustrated in Fig. 4A. Western blot analysis revealed that truncating domains did not affect the protein expression of TTF-1 (Fig. 4B). Via the luciferase assays using the TTF-1 domain deletions, we found that, as expected, the TTF-1 homeodomain was absolutely required for the transactivation of the OCLN, CLDN1, and SpB promoters (Figs. 4, C and D, and supplemental Fig. S2). Interestingly, although TTF-1(ΔAD1) exhibited a weakened ability to transactivate the OCLN and CLDN1 promoters, TTF-1(ΔAD2) did not (Fig. 4, C and D). This differential transactivation requirement of AD1 and AD2 of TTF-1 was not detected with the control SpB promoter reporter because both TTF-1 ΔAD1 and ΔAD2 increased SpB promoter activation (supplemental Fig. S2). These data substantiate promoter transactivation of OCLN and CLDN1 by TTF-1 and indicate that TTF-1 regulates OCLN and CLDN1 in a manner distinct from its control of SpB.
FIGURE 4.
The homeodomain and activation domain 1 are required for complete activation of the OCLN and CLDN1 promoters. A, schematic representation of TTF-1 indicating the activation domain 1 (black), homeodomain (white), and activation domain 2 (black). B, Western blot analysis of A549 cells transiently transfected with the domain deletion mutants. Western blots of TTF-1 and β-tubulin (loading control) are shown. C and D, luciferase assays using the OCLN and CLDN1 promoters in combination with the domain deletion mutants as indicated (ns, data not significant; n = 12). Error bars, S.D.
TTF-1 Directly Binds to the OCLN and CLDN1 Promoters
To determine whether TTF-1 can directly bind to the OCLN and CLDN1 promoters, we performed ChIP using lysates from NCI-H441 cells. The positions of the primer pairs used for PCR amplification are described in Fig. 5A. A polyclonal antibody directed against TTF-1 was used to pull down DNA, and the PCR analyses of ChIP-derived material for the presence of OCLN and CLDN1 promoters are shown in Fig. 5B. Importantly, very little DNA was precipitated using a normal rabbit IgG control (comparison of TTF-1- versus IgG-precipitated DNA revealed an ∼80- and 245-fold enrichment for the OCLN and CLDN1 promoters, respectively). To confirm the reliability of the ChIP experiments, we also included a negative control (GAPDH exon 1; Fig. 5B) and a positive control that detected the SpB promoter (supplemental Fig. S3). Comparison of anti-TTF-1 versus rabbit IgG precipitated DNA found less than 1-fold (0.67-fold) chromatin enrichment for GAPDH and ∼160-fold for SpB. These data suggest that TTF-1 directly binds to the OCLN and CLDN1 promoters.
FIGURE 5.

TTF-1 directly interacts with the OCLN and CLDN1 promoters. A, positions of the primers used in the real-time PCRs. B, real-time PCR analysis of DNA precipitated by anti-TTF-1 (catalog no. H190, Santa Cruz Biotechnology, Inc.) or IgG. PCRs were performed using primers directed against the OCLN and CLDN1 promoters or GAPDH exon 1 (n = 3). Error bars, S.D.
Occludin, but Not Claudin-1, Is Down-regulated in TTF-1-silenced Cells
Next, we took on a loss-of-function approach to determine if TTF-1 silencing affects occludin and claudin-1 protein levels. We addressed this issue using two independent strategies. Initially, we analyzed tumor cell lines derived from a mouse model of lung adenocarcinomas that are non-metastatic in nature (TnonMet) (15). It is known that stable suppression of Ttf-1 via a shRNA (pLKO-shTtf-1) in TnonMet cells would increase the cellular metastatic potential in animal-based assays (15). We thus examined the occludin and claudin-1 protein content in the TnonMet cells stably transfected with either pLKO-shTtf-1 or pLKO-shLuc (a control shRNA directed against luciferase) by immunoblot analysis and found that the occludin level decreased in the Ttf-1 knockdown cells (pLKO-shTtf-1; Fig. 6A) in comparison with the control (pLKO-shLuc) cells. Interestingly, claudin-1 expression was constant under both conditions (Fig. 6A).
FIGURE 6.
Occludin, but not claudin-1, is down-regulated in TTF-1-silenced cells. A, non-metastatic (TnonMet) cells transfected with pLKO-shLuc (394T4bc37) or pLKO-shTtf-1 (394T4E1) were analyzed by Western blot for ZO-1, occludin, claudin-1, TTF-1, and Hsp90 (loading control). B, NCI-H441 cells were transfected with scrambled control siRNA (siScr) or siRNA directed against TTF-1 (siTTF-1) and harvested for protein 72 h later. Western blot analysis of ZO-1, occludin, claudin-1, TTF-1, and Hsp90 (loading control) are shown. C, luciferase assay of the OCLN promoter reporters using siScr and siTTF-1 in NCI-H441 cells (n = 12). Error bars, S.D.
We were perplexed by the finding that Ttf-1 loss down-regulated occludin but not claudin-1 in the murine lung cancer cells; therefore, we silenced TTF-1 using two siRNA oligonucleotides in the TTF-1-positive human NCI-H441 cells. We show that treatment with two individual siRNAs against TTF-1 resulted in a loss of occludin protein (Fig. 6B), without altering ZO-1 expression. However, only one of the two siTTF-1s (i.e. oligo#2) elicited a loss of claudin-1 expression (Fig. 6B). We attributed the claudin-1 decrease associated with siTTF-1(oligo#2) as a nonspecific/off-target effect to siTTF-1(oligo#2) because the observation was not shared by both siRNAs against TTF-1. Furthermore, in the luciferase reporter assay, TTF-1 knockdown yielded an ∼50% reduction of OCLN promoter activity (Fig. 6C) while having a less pronounced effect on the CLDN1 promoter (∼35% decrease by siTTF-1(oligo#1) and ∼25% decrease by siTTF-1(oligo#2); Supplemental Fig. S4). Collectively, the preponderance of the evidence implies that OCLN, but not CLDN1, is down-regulated following TTF-1 knockdown.
Functional Impact of Occludin on Migration and Anoikis of Lung Carcinoma Cells
TTF-1 is now recognized as an anti-metastasis protein in lung adenocarcinomas (15, 48), and the notion that occludin is able to counter metastatic phenotypes has been documented in other cell types (24). Therefore, we set out to determine if occludin is able to impede migration of human lung cancer cells. To accomplish this, GFP-expressing vector alone (Control) or an occludin-GFP fusion-expressing plasmid (Occludin) was transiently transfected into A549 cells (protein expression of occludin and tagged occludin is shown in Supplemental Fig. S5A). The following day, confluent monolayers were wounded. Importantly, cells were maintained in 1% serum to minimize potential effects of differential proliferation rates. Each day, wounds of control and occludin-expressing cells were captured (Fig. 7A). Over a 3-day time course, control cells migrated to near full wound closure; however, occludin expression significantly impeded the migration rate (Fig. 7B).
FIGURE 7.
Occludin expression reduces migration and induces anoikis in lung carcinoma cells. Confluent monolayers of A549 cells transiently transfected with a control plasmid expressing GFP only or a plasmid expressing occludin-GFP fusion protein were wounded. Bright field microscopy (A) and quantification (B) were performed each day over a 3-day time course. Scale bar, 200 μm. C, control (GFP vector) or occludin-GFP-transfected A549 and NCI-H1299 cells were seeded on polyHEMA-coated wells, and cell death was quantified 24 h later using a trypan blue exclusion assay (migration assay, n = 9; anoikis assay, n = 4). Error bars, S.D.
Anoikis is a detachment-induced cell death process that cancer cells, including A549 cells, are able to circumvent (49). We hypothesized that occludin may induce anoikis in lung carcinoma cells. To extend our investigation beyond A549 cells, we included a second human lung cancer cell line (i.e. NCI-H1299) in our studies. As in A549 cells, the NCI-H1299 cell line does not express endogenous TTF-1 and exhibits resistance to anoikis (50), hence affording us a window to detect the hypothesized pro-anoikis activity of occludin. We seeded cells transfected with GFP vector (Control) or occludin-GFP (Occludin) in wells coated with polyHEMA to prevent attachment (protein expression of occludin and tagged occludin in NCI-H1299 cells is shown in supplemental Fig. S5B). The following day, cells were collected and analyzed for cell death. We found that occludin expression increased cell death in NCI-H1299 and A549 cells by nearly 2- and 3-fold, respectively, compared with control cells (Fig. 7C). Our control cell death percentage values were nearly identical to those reported previously (24). Importantly, under the normal conditions where cells were cultured in wells with attachable surface, occludin expression in either NCI-H1299 or A549 cells did not enhance cell death significantly (supplemental Fig. S6).
The metastasis-suppressive activity of TTF-1 suggests that knocking down TTF-1 expression in lung cancer cells would endow cells with resistance to anoikis. To this end, we monitored the anoikis sensitivities of NCI-H441 cells transfected with the two individual siTTF-1s. The validity of the siTTF-1s in repressing TTF-1 was confirmed by the experiments shown in Fig. 6B. In line with the notion that TTF-1 has anti-metastatic activity, both TTF-1 knockdown cells were desensitized to anoikis (Fig. 8A). Importantly, expression of occludin (as occludin-GFP fusion) in the TTF-1 knockdown cells restored cellular sensitivity to anoikis, with the siTTF-1(oligo#1) knockdown cells showing a more robust reversal by occludin than that observed in the siTTF-1(oligo#2) knockdown cells. Collectively, these data suggest that occludin sensitizes cells to anoikis and is capable of reversing the anti-anoikis behavior of TTF-1low human lung cancer cells.
FIGURE 8.

Occludin reverses TTF-1 knockdown-induced resistance to anoikis; diagram summarizing known mediators of the anti-metastatic activity of TTF-1. A, occludin (occludin-GFP)- or GFP (control)-transfected NCI-H441 cells were seeded on polyHEMA-coated wells, and cell death was quantified 24 h later using a trypan blue exclusion assay (n = 3). B, I, Winslow et al. (15) discovered that TTF-1 suppresses lung cancer metastasis in a mouse model of lung adenocarcinomas, and HMGA2 mediates this activity. II, the findings of Saito et al. (14) identified E-cadherin as a target of TTF-1 and linked this to suppression of EMT. Hosono et al. (48) implicated MYBPH as a mediator of the metastasis-suppressive function of TTF-1. The data in this report show that occludin is a direct target of TTF-1. TTF-1 is needed for maintaining occludin expression. Finally, occludin expression is sufficient to reverse metastatic phenotypes of lung carcinoma cells. Error bars, S.D.
DISCUSSION
Previously, it was shown that TTF-1 activates the CDH1 promoter (14); however, that study did not address the possibility that TTF-1 also activates TJ gene expression. Here, we provide the first evidence that TTF-1 regulates the expression of TJ proteins (i.e. occludin and claudin-1). Moreover, we demonstrated that TTF-1 alleviates the known suppression of OCLN and CLDN1 by TGF-β. Using a model system in which TTF-1 inhibits the progression of primary lung cancers to metastases, we detected a clear association of the relationship between TTF-1 and occludin in the conversion of a non-metastatic lung cancer cell to a metastatic one. Finally, we provide cellular phenotypes that address the importance of occludin expression in relation to metastatic characteristics.
Traditionally, TJ proteins have been thought of as static proteins whose purpose is to regulate barrier function. However, recent studies show that these proteins can also contribute to cell cycle control, transformation, migration, invasion, and metastasis (20, 21, 23, 29). Specifically, claudin-1 has a demonstrated role in regulating invasion, migration, and metastasis (29). In terms of occludin, several reports describe it as a regulator of the cell cycle as well as reversing transformation, contributing to epithelial cell migration (51, 52), and combating metastatic properties in melanoma, breast, and cervical cancer cell lines (24), yet little is known about occludin in regard to lung cancer progression. Our observations suggest that alterations in occludin content may accompany the process of switching from a non-metastatic to metastatic phenotype in lung cancers. Specifically, we show that occludin expression is able to suppress the migratory potential of a lung carcinoma cell. Cell migration is well documented in the metastatic process. Of equal importance, we found that expression of occludin induced anoikis. Anoikis resistance is a hallmark of metastatic malignancies. As the cell leaves the primary tumor environment, it must avoid the detachment-induced cell death. Indeed, in this study, TTF-1 knockdown conferred to human lung cancer cells resistance to anoikis. This observation is consistent with the reported anti-metastatic function of TTF-1 (15). Because transfection of an occludin-expressing plasmid reversed the anoikis resistance caused by TTF-1 knockdown, we suggest that the TTF-1/occludin connection is functionally important to how lung cancer cells respond to the stress induced by loss of attachment substrate. Clearly, our data demonstrate that expression of occludin imparts anti-metastatic characteristics and provide evidence that occludin loss is not merely a casualty in the metastatic process. Although in this study, we did not observe a change in the steady-state expression level of ZO-1, this observation does not rule out the possibility that ZO-1 may undergo redistribution in localization. Considering the established activities of ZO-1 in cell cycle control (53, 54) and nuclear translocation (55), we are interested in investigating the role of ZO-1 as a mediator downstream to the TTF-1/occludin signaling axis in the future. Although our data indicate that loss of TTF-1 does not appear to concurrently cause claudin-1 loss in lung cancer cells, this does not mean that claudin-1 is functionally insignificant in lung cancer progression. Additional work will be needed to untangle this important issue.
To explore the mechanism of OCLN and CLDN1 transactivation by TTF-1, we altered TTF-1 in two different manners: (i) mutating two amino acids (Q50K/Y54M) within the homeodomain and (ii) removing functional domains individually (ΔAD1, ΔHD, or ΔAD2). Based on an earlier study (42), we predicted that TTF-1(Q50K/Y54M) would suffer from a pronounced loss in DNA binding specificity. Although, we did not conduct a comprehensive analysis of the DNA binding spectrum of TTF-1(Q50K/Y54M), the promoter reporter assays of this study implicate that TTF-1(Q50K/Y54M) did exhibit altered DNA binding preferences (OCLN > CLDN1 > SpB). This is intriguing in that the two mutated homeodomain residues are known to be involved in discriminating nucleotides at the 3′ end of substrate DNAs (56, 57). Thus, a mutant TTF-1 protein harboring altered residues at these two positions may differ from WT TTF-1 in favoring DNA sequences with certain 3′ ends. The last two nucleotides at the 3′ termini of the presumed TBE sequences that we identified in the promoters of OCLN and CLDN1 are CC (OCLN) and CG (CLDN1), respectively (Fig. 1B). We speculate that TTF-1(Q50K/Y54M) favors a TBE ending in CC based on the higher OCLN promoter reporter activity transactivated by the mutant TTF-1. The second strategy of domain deletion identified that AD2 was dispensable for promoter transactivation of both OCLN and CLDN1. This is perhaps not unexpected because most proteins known to interact with TTF-1 do so via the AD1 of TTF-1 (1). Additional studies are required to understand how TTF-1 differentially regulates TJ molecules versus other known target genes.
To study the interplay between TTF-1 and TGF-β, we took on two different systems to characterize transactivation of TJ genes by TTF-1 in the presence of TGF-β. The first system was based in Beas-2B cells with inducible TTF-1 expression (Fig. 3). In this system, the readout was the mRNA and protein level of endogenous OCLN and CLDN1. The observations were that TGF-β decreased the expression of both TJ genes as expected and that the degree of the TGF-β-induced suppression of TJ genes was tapered by TTF-1. Subsequently, we moved into a different cell system (i.e. the NCI-H441 cells with high endogenous TTF-1 expression). The readout in this system was the activities of OCLN and CLDN1 promoter reporters, and we detected that TGF-β induced higher promoter activities of both TJ genes (supplemental Fig. S1). This was somewhat unexpected in view of our reported finding that in NCI-H441 cells, TGF-β represses TTF-1 expression by up-regulating miR-365, which targets TTF-1 (36). There are several putative explanations to reconcile the opposite observations of TGF-β effects on TJ gene transcription between the two cell systems (e.g. the sub-full-length promoters used in the reporter assay may act differently from the native loci). Nevertheless, knocking down endogenous TTF-1 expression in NCI-H441 inhibited the TGF-β-dependent promoter responses of OCLN and CLDN1 (supplemental Fig. S1). Although different readouts were monitored in the two cell systems, the data collectively point to the requirement of TTF-1 in transactivating TJ genes.
The fact that TTF-1 is now recognized as having both pro-oncogenic and anti-metastatic roles is not without precedent. The frequently amplified human MYC oncogene was recently shown to possess anti-metastasis activity by repressing integrin subunits (58). To address the question of how our data relate to metastasis, we analyzed tumor cell lines derived from a mouse model of lung adenocarcinomas (15). We chose this system because Ttf-1 suppresses the progression of primary lung cancers to metastases in this model system. After intratracheal administration of lentiviral vectors expressing the Cre-recombinase, KrasG12D/+; p53flos/flox mice developed multifocal lung adenocarcinoma. Some of the primary lung tumors eventually led to macroscopic metastases to the draining lymph nodes, pleura, kidneys, heart, adrenal glands, and liver. Because lentiviruses integrate stably into the genome, the integration site was a unique molecular identifier, allowing primary tumors to be unambiguously linked to their related metastases. Gene expression profiling of two types of primary lung tumors (non-metastatic (TnonMet) and metastatic (TMet)) indicated that Ttf-1 was consistently and significantly depressed in the primary tumors (TMet) with clonally related metastases. Further, re-expression of Ttf-1 in TMet cells led to fewer tumors and a reduction in the ability of the cells to grow in an anchorage-independent assay, whereas silencing of Ttf-1 in TnonMet cells resulted in an increased ability of cells to grow in anchorage-independent assays and a higher number of liver nodules in an intrasplenic implantation assay (15). In this study, the non-metastatic Ttf-1-positive TnonMet cells, upon stable knockdown of Ttf-1 (which enhances metastasis (15)), exhibited reduced occludin expression but not that of claudin-1. Similarly, in a TTF-1-positive human lung cancer cell line (NCI-H441), TTF-1 knockdown only down-regulated occludin expression, with less pronounced effects on claudin-1 expression. The TTF-1/CLDN1 relationship that we observed is reminiscent of the reported action of TTF-1 on E-cadherin in that TTF-1 up-regulates E-cadherin expression, but TTF-1 knockdown does not reduce E-cadherin level (14). In parallel, we examined the expression of occludin and claudin-1 in the TMet cells in which stable expression of Ttf-1 has been reintroduced (15). Intriguingly, occludin and claudin-1 were expressed in neither the control (pMSCV-Puro) nor the Ttf-1-re-expressing TMet cells (pMSCV-Ttf-1),4 suggesting that re-expression of Ttf-1 in the TMet cells is not sufficient to reactivate the TJ protein expression. Perhaps the TMet cells may have constitutively active Snail, which is known to repress junctional molecules (59). Unbiased gene expression analysis and functional studies by Winslow et al. (15) identified HMGA2 as a downstream mediator of the anti-metastasis activity of Ttf-1. In view of our data and the recent discovery that a downstream transcriptional target of TTF-1, MYBPH, was found to inhibit cell motility and metastasis (48), we propose that the metastasis-suppressive activity of TTF-1 is multipronged with contributions from a number of factors (i.e. the TJ proteins, HMGA2, and MYBPH).
In summary, our results identify a novel regulatory mechanism of OCLN and CLDN1 gene expression mediated by TTF-1, thus connecting a pivotal lung developmental and cancer factor (TTF-1) to the tight junction proteins. In Fig. 8B, we present schematics to summarize the observations of this and other studies. Given the critical role of TTF-1 in orchestrating fetal lung development, we suggest that the cooperation between TTF-1 and TJs may be essential to pulmonary development. In terms of lung cancer, this study sharpens our understanding of how mechanistically occludin comes to be lost during lung cancer progression, a process by which the pulmonary cells must lose their contacts in order to leave the primary tumor site. We believe that the data reported herein may set a stage for future work searching for a means to intervene in the metastatic process.
Supplementary Material
Acknowledgment
We thank Dr. Sinisa Dovat for access to the Bioruptor.
This work was supported, in whole or in part, by National Institutes of Health Grant CA127547 (to D. M.).

This article contains supplemental Table S1 and Figs. S1–S6.
E. A. Runkle and D. Mu, unpublished results.
- EMT
- epithelial to mesenchymal transition
- TJ
- tight junction
- TBE
- TTF-1 binding element
- polyHEMA
- poly(2-hydroxyethyl methacrylate).
REFERENCES
- 1. Maeda Y., Davé V., Whitsett J. A. (2007) Transcriptional control of lung morphogenesis. Physiol. Rev. 87, 219–244 [DOI] [PubMed] [Google Scholar]
- 2. Briscoe J., Pierani A., Jessell T. M., Ericson J. (2000) A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445 [DOI] [PubMed] [Google Scholar]
- 3. Kendall J., Liu Q., Bakleh A., Krasnitz A., Nguyen K. C., Lakshmi B., Gerald W. L., Powers S., Mu D. (2007) Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 16663–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwei K. A., Kim Y. H., Girard L., Kao J., Pacyna-Gengelbach M., Salari K., Lee J., Choi Y. L., Sato M., Wang P., Hernandez-Boussard T., Gazdar A. F., Petersen I., Minna J. D., Pollack J. R. (2008) Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 27, 3635–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka H., Yanagisawa K., Shinjo K., Taguchi A., Maeno K., Tomida S., Shimada Y., Osada H., Kosaka T., Matsubara H., Mitsudomi T., Sekido Y., Tanimoto M., Yatabe Y., Takahashi T. (2007) Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 67, 6007–6011 [DOI] [PubMed] [Google Scholar]
- 6. Weir B. A., Woo M. S., Getz G., Perner S., Ding L., Beroukhim R., Lin W. M., Province M. A., Kraja A., Johnson L. A., Shah K., Sato M., Thomas R. K., Barletta J. A., Borecki I. B., Broderick S., Chang A. C., Chiang D. Y., Chirieac L. R., Cho J., Fujii Y., Gazdar A. F., Giordano T., Greulich H., Hanna M., Johnson B. E., Kris M. G., Lash A., Lin L., Lindeman N., Mardis E. R., McPherson J. D., Minna J. D., Morgan M. B., Nadel M., Orringer M. B., Osborne J. R., Ozenberger B., Ramos A. H., Robinson J., Roth J. A., Rusch V., Sasaki H., Shepherd F., Sougnez C., Spitz M. R., Tsao M. S., Twomey D., Verhaak R. G., Weinstock G. M., Wheeler D. A., Winckler W., Yoshizawa A., Yu S., Zakowski M. F., Zhang Q., Beer D. G., Wistuba II, Watson M. A., Garraway L. A., Ladanyi M., Travis W. D., Pao W., Rubin M. A., Gabriel S. B., Gibbs R. A., Varmus H. E., Wilson R. K., Lander E. S., Meyerson M. (2007) Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taguchi A., Politi K., Pitteri S. J., Lockwood W. W., Faça V. M., Kelly-Spratt K., Wong C. H., Zhang Q., Chin A., Park K. S., Goodman G., Gazdar A. F., Sage J., Dinulescu D. M., Kucherlapati R., Depinho R. A., Kemp C. J., Varmus H. E., Hanash S. M. (2011) Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer cell 20, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Homminga I., Pieters R., Langerak A. W., de Rooi J. J., Stubbs A., Verstegen M., Vuerhard M., Buijs-Gladdines J., Kooi C., Klous P., van Vlierberghe P., Ferrando A. A., Cayuela J. M., Verhaaf B., Beverloo H. B., Horstmann M., de Haas V., Wiekmeijer A. S., Pike-Overzet K., Staal F. J., de Laat W., Soulier J., Sigaux F., Meijerink J. P. (2011) Integrated transcript and genome analyses reveal NKX2–1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer cell 19, 484–497 [DOI] [PubMed] [Google Scholar]
- 9. Yamaguchi T., Yanagisawa K., Sugiyama R., Hosono Y., Shimada Y., Arima C., Kato S., Tomida S., Suzuki M., Osada H., Takahashi T. (2012) NKX2–1/TITF1/TTF-1-induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 21, 348–361 [DOI] [PubMed] [Google Scholar]
- 10. Anagnostou V. K., Syrigos K. N., Bepler G., Homer R. J., Rimm D. L. (2009) Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J. Clin. Oncol. 27, 271–278 [DOI] [PubMed] [Google Scholar]
- 11. Barletta J. A., Perner S., Iafrate A. J., Yeap B. Y., Weir B. A., Johnson L. A., Johnson B. E., Meyerson M., Rubin M. A., Travis W. D., Loda M., Chirieac L. R. (2009) Clinical significance of TTF-1 protein expression and TTF-1 gene amplification in lung adenocarcinoma. J. Cell. Mol. Med. 13, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berghmans T., Paesmans M., Mascaux C., Martin B., Meert A. P., Haller A., Lafitte J. J., Sculier J. P. (2006) Thyroid transcription factor 1. A new prognostic factor in lung cancer. A meta-analysis. Ann. Oncol. 17, 1673–1676 [DOI] [PubMed] [Google Scholar]
- 13. Tang X., Kadara H., Behrens C., Liu D. D., Xiao Y., Rice D., Gazdar A. F., Fujimoto J., Moran C., Varella-Garcia M., Lee J. J., Hong W. K., Wistuba I. I. (2011) Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC. Implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 17, 2434–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saito R. A., Watabe T., Horiguchi K., Kohyama T., Saitoh M., Nagase T., Miyazono K. (2009) Thyroid transcription factor-1 inhibits transforming growth factor-β-mediated epithelial-to-mesenchymal transition in lung adenocarcinoma cells. Cancer Res. 69, 2783–2791 [DOI] [PubMed] [Google Scholar]
- 15. Winslow M. M., Dayton T. L., Verhaak R. G., Kim-Kiselak C., Snyder E. L., Feldser D. M., Hubbard D. D., DuPage M. J., Whittaker C. A., Hoersch S., Yoon S., Crowley D., Bronson R. T., Chiang D. Y., Meyerson M., Jacks T. (2011) Suppression of lung adenocarcinoma progression by Nkx2–1. Nature 473, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harris T., Pan Q., Sironi J., Lutz D., Tian J., Sapkar J., Perez-Soler R., Keller S., Locker J. (2011) Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin. Cancer Res. 17, 690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsukita S., Yamazaki Y., Katsuno T., Tamura A. (2008) Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 27, 6930–6938 [DOI] [PubMed] [Google Scholar]
- 18. Schulzke J. D., Gitter A. H., Mankertz J., Spiegel S., Seidler U., Amasheh S., Saitou M., Tsukita S., Fromm M. (2005) Epithelial transport and barrier function in occludin-deficient mice. Biochim. Biophys. Acta 1669, 34–42 [DOI] [PubMed] [Google Scholar]
- 19. Phillips B. E., Cancel L., Tarbell J. M., Antonetti D. A. (2008) Occludin independently regulates permeability under hydrostatic pressure and cell division in retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 49, 2568–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Runkle E. A., Sundstrom J. M., Runkle K. B., Liu X., Antonetti D. A. (2011) Occludin localizes to centrosomes and modifies mitotic entry. J. Biol. Chem. 286, 30847–30858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du D., Xu F., Yu L., Zhang C., Lu X., Yuan H., Huang Q., Zhang F., Bao H., Jia L., Wu X., Zhu X., Zhang X., Zhang Z., Chen Z. (2010) The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev. Cell 18, 52–63 [DOI] [PubMed] [Google Scholar]
- 22. Wang Z., Wade P., Mandell K. J., Akyildiz A., Parkos C. A., Mrsny R. J., Nusrat A. (2007) Raf 1 represses expression of the tight junction protein occludin via activation of the zinc finger transcription factor slug. Oncogene 26, 1222–1230 [DOI] [PubMed] [Google Scholar]
- 23. Li D., Mrsny R. J. (2000) Oncogenic Raf-1 disrupts epithelial tight junctions via down-regulation of occludin. J. Cell Biol. 148, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osanai M., Murata M., Nishikiori N., Chiba H., Kojima T., Sawada N. (2006) Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res. 66, 9125–9133 [DOI] [PubMed] [Google Scholar]
- 25. Morin P. J. (2005) Claudin proteins in human cancer. Promising new targets for diagnosis and therapy. Cancer Res. 65, 9603–9606 [DOI] [PubMed] [Google Scholar]
- 26. Singh A. B., Sharma A., Dhawan P. (2010) Claudin family of proteins and cancer. An overview. J. Oncol. 2010, 541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dhawan P., Singh A. B., Deane N. G., No Y., Shiou S. R., Schmidt C., Neff J., Washington M. K., Beauchamp R. D. (2005) Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Invest. 115, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paschoud S., Bongiovanni M., Pache J. C., Citi S. (2007) Claudin-1 and claudin-5 expression patterns differentiate lung squamous cell carcinomas from adenocarcinomas. Mod. Pathol. 20, 947–954 [DOI] [PubMed] [Google Scholar]
- 29. Chao Y. C., Pan S. H., Yang S. C., Yu S. L., Che T. F., Lin C. W., Tsai M. S., Chang G. C., Wu C. H., Wu Y. Y., Lee Y. C., Hong T. M., Yang P. C. (2009) Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 179, 123–133 [DOI] [PubMed] [Google Scholar]
- 30. Rangel L. B., Agarwal R., D'Souza T., Pizer E. S., Alò P. L., Lancaster W. D., Gregoire L., Schwartz D. R., Cho K. R., Morin P. J. (2003) Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res. 9, 2567–2575 [PubMed] [Google Scholar]
- 31. Lee J. W., Lee S. J., Seo J., Song S. Y., Ahn G., Park C. S., Lee J. H., Kim B. G., Bae D. S. (2005) Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol. Oncol. 97, 53–59 [DOI] [PubMed] [Google Scholar]
- 32. Soini Y. (2005) Expression of claudins 1, 2, 3, 4, 5, and 7 in various types of tumors. Histopathology 46, 551–560 [DOI] [PubMed] [Google Scholar]
- 33. Hennessy B. T., Gonzalez-Angulo A. M., Stemke-Hale K., Gilcrease M. Z., Krishnamurthy S., Lee J. S., Fridlyand J., Sahin A., Agarwal R., Joy C., Liu W., Stivers D., Baggerly K., Carey M., Lluch A., Monteagudo C., He X., Weigman V., Fan C., Palazzo J., Hortobagyi G. N., Nolden L. K., Wang N. J., Valero V., Gray J. W., Perou C. M., Mills G. B. (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 69, 4116–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perou C. M. (2011) Molecular stratification of triple-negative breast cancers. Oncologist 16, 61–70 [DOI] [PubMed] [Google Scholar]
- 35. Felinski E. A., Cox A. E., Phillips B. E., Antonetti D. A. (2008) Glucocorticoids induce transactivation of tight junction genes occludin and claudin-5 in retinal endothelial cells via a novel cis-element. Exp. Eye Res. 86, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qi J., Rice S. J., Salzberg A. C., Runkle E. A., Liao J., Zander D. S., Mu D. (2012) MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle 11, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farré D., Roset R., Huerta M., Adsuara J. E., Roselló L., Albà M. M., Messeguer X. (2003) Identification of patterns in biological sequences at the ALGGEN server. PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M. M. (2002) PROMO. Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 39. Guazzi S., Price M., De Felice M., Damante G., Mattei M. G., Di Lauro R. (1990) Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 9, 3631–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Damante G., Fabbro D., Pellizzari L., Civitareale D., Guazzi S., Polycarpou-Schwartz M., Cauci S., Quadrifoglio F., Formisano S., Di Lauro R. (1994) Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 22, 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fabbro D., Tell G., Leonardi A., Pellizzari L., Pucillo C., Lonigro R., Formisano S., Damante G. (1996) In the TTF-1 homeodomain the contribution of several amino acids to DNA recognition depends on the bound sequence. Nucleic Acids Res. 24, 3283–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pellizzari L., Tell G., Fabbro D., Pucillo C., Damante G. (1997) Functional interference between contacting amino acids of homeodomains. FEBS Lett. 407, 320–324 [DOI] [PubMed] [Google Scholar]
- 43. Bohinski R. J., Di Lauro R., Whitsett J. A. (1994) The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Cell. Biol. 14, 5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Medici D., Hay E. D., Goodenough D. A. (2006) Cooperation between snail and LEF-1 transcription factors is essential for TGF-β1-induced epithelial-mesenchymal transition. Mol. Biol. Cell 17, 1871–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reddel R. R., Ke Y., Gerwin B. I., McMenamin M. G., Lechner J. F., Su R. T., Brash D. E., Park J. B., Rhim J. S., Harris C. C. (1988) Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 48, 1904–1909 [PubMed] [Google Scholar]
- 46. Hsu D. S., Acharya C. R., Balakumaran B. S., Riedel R. F., Kim M. K., Stevenson M., Tuchman S., Mukherjee S., Barry W., Dressman H. K., Nevins J. R., Powers S., Mu D., Potti A. (2009) Characterizing the developmental pathways TTF-1, NKX2–8, and PAX9 in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 106, 5312–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Felice M., Damante G., Zannini M., Francis-Lang H., Di Lauro R. (1995) Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J. Biol. Chem. 270, 26649–26656 [DOI] [PubMed] [Google Scholar]
- 48. Hosono Y., Yamaguchi T., Mizutani E., Yanagisawa K., Arima C., Tomida S., Shimada Y., Hiraoka M., Kato S., Yokoi K., Suzuki M., Takahashi T. (2012) MYBPH, a transcriptional target of TTF-1, inhibits ROCK1 and reduces cell motility and metastasis. EMBO J. 31, 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei L., Yang Y., Yu Q. (2001) Tyrosine kinase-dependent, phosphatidylinositol 3′-kinase, and mitogen-activated protein kinase-independent signaling pathways prevent lung adenocarcinoma cells from anoikis. Cancer Res. 61, 2439–2444 [PubMed] [Google Scholar]
- 50. Kumar S., Park S. H., Cieply B., Schupp J., Killiam E., Zhang F., Rimm D. L., Frisch S. M. (2011) A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol. Cell. Biol. 31, 4036–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Capaldo C. T., Nusrat A. (2009) Cytokine regulation of tight junctions. Biochim. Biophys. Acta 1788, 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin T. A., Jiang W. G. (2009) Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 1788, 872–891 [DOI] [PubMed] [Google Scholar]
- 53. Balda M. S., Garrett M. D., Matter K. (2003) The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sourisseau T., Georgiadis A., Tsapara A., Ali R. R., Pestell R., Matter K., Balda M. S. (2006) Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell. Biol. 26, 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gottardi C. J., Arpin M., Fanning A. S., Louvard D. (1996) The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc. Natl. Acad. Sci. U.S.A. 93, 10779–10784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Damante G., Pellizzari L., Esposito G., Fogolari F., Viglino P., Fabbro D., Tell G., Formisano S., Di Lauro R. (1996) A molecular code dictates sequence-specific DNA recognition by homeodomains. EMBO J. 15, 4992–5000 [PMC free article] [PubMed] [Google Scholar]
- 57. Hanes S. D., Brent R. (1991) A genetic model for interaction of the homeodomain recognition helix with DNA. Science 251, 426–430 [DOI] [PubMed] [Google Scholar]
- 58. Liu H., Radisky D. C., Yang D., Xu R., Radisky E. S., Bissell M. J., Bishop J. M. (2012) MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat. Cell Biol. 14, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vincent T., Neve E. P., Johnson J. R., Kukalev A., Rojo F., Albanell J., Pietras K., Virtanen I., Philipson L., Leopold P. L., Crystal R. G., de Herreros A. G., Moustakas A., Pettersson R. F., Fuxe J. (2009) A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-β-mediated epithelial-mesenchymal transition. Nat. Cell Biol. 11, 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.