Background: O-GlcNAcylation regulates cellular processes such as transcription, signal transduction, metabolism, and cell cycle.
Results: O-GlcNAcylation is elevated in CRC metastatic clone. OGA silencing resulted in the acquisition of a fibroblast-like morphology, growth retardation, and alteration of gene expression.
Conclusion: O-GlcNAcylation affects gene transcription, metabolism, and proliferation.
Significance: This research supports O-GlcNAcylation involvement in various aspects of tumor cell physiology.
Keywords: Cancer, Cell Metabolism, Gene Expression, O-GlcNAc, O-GlcNAcylation, O-GlcNAcase
Abstract
O-Linked β-N-acetylglucosamine (O-GlcNAc) glycosylation is a regulatory post-translational modification occurring on the serine or threonine residues of nucleocytoplasmic proteins. O-GlcNAcylation is dynamically regulated by O-GlcNAc transferase and O-GlcNAcase (OGA), which are responsible for O-GlcNAc addition and removal, respectively. Although O-GlcNAcylation was found to play a significant role in several pathologies such as type II diabetes and neurodegenerative diseases, the role of O-GlcNAcylation in the etiology and progression of cancer remains vague. Here, we followed O-GlcNAcylation and its catalytic machinery in metastatic clones of human colorectal cancer and the effect of OGA knockdown on cellular phenotype and on the transcriptome. The colorectal cancer SW620 metastatic clone exhibited increased O-GlcNAcylation and decreased OGA expression compared with its primary clone, SW480. O-GlcNAcylation elevation in SW620 cells, through RNA interference of OGA, resulted in phenotypic alterations that included acquisition of a fibroblast-like morphology, which coincides with epithelial metastatic progression and growth retardation. Microarray analysis revealed that OGA silencing altered the expression of about 1300 genes, mostly involved in cell movement and growth, and specifically affected metabolic pathways of lipids and carbohydrates. These findings support the involvement of O-GlcNAcylation in various aspects of tumor cell physiology and suggest that this modification may serve as a link between metabolic changes and cancer.
Introduction
O-Linked β-N-acetylglucosaminylation (O-GlcNAcylation),2 i.e. the attachment of N-acetylglucosamine (GlcNAc) to serine and threonine residues via a β-O-linkage, is a novel post-translational modification of nuclear and cytoplasmic proteins that is analogous to phosphorylation. Dynamic and reversible, O-GlcNAcylation occurs in response to alterations in cellular metabolic conditions and to internal and environmental triggers (e.g. hormones, growth factors, stresses, and nutrients). Thus far, only two enzymes have been found to regulate O-GlcNAcylation, O-GlcNAc transferase (OGT) and N-acetyl d-glucosaminidase (OGA), which catalyze O-GlcNAc addition and removal, respectively. O-GlcNAcylation may regulate protein activity, including protein-protein interactions, localization, transcription, and degradation (1, 2). Interestingly, the majority of O-GlcNAcylated proteins are also known to undergo phosphorylation, and in many cases, the two modifications were found to be reciprocal (3, 4).
A myriad of O-GlcNAcylated proteins have been identified, including metabolic enzymes, translation and transcription factors, cytoskeletal proteins, signaling components, oncogenic products, and tumor suppressor proteins (5). Moreover, O-GlcNAcylation functions as a nutrient sensor because its precursor, UDP-GlcNAc, is positioned at the nexus of the metabolic pathways of glucose, fatty acids, nucleic acids, and nitrogen (6). Thus, proper O-GlcNAcylation seems to be crucial for cellular processes affecting cell function, growth, and survival. Indeed, aberrant patterns of O-GlcNAcylation on key proteins have been documented in various pathologies such as type II diabetes and cardiovascular and neurodegenerative diseases (22), and alterations in O-GlcNAcylation were found to contribute to tumorigenesis (7). In human colon cancer cell lines, increased UDP-GlcNAc levels led to disruptions of cell growth and differentiation (8). Overexpression of both OGT and OGA disrupted mitosis (possibly by disrupting cyclin expression), and in the case of OGT, overexpression also increased polyploidy, a characteristic of cancer cells (9). Increased O-GlcNAcylation and OGT levels were reported in various types of cancer, including colon, lung, and breast cancers (7, 10, 11), and were found to correlate with the progression of tumorigenesis (10–12). O-GlcNAcylation was also found to directly affect key proteins involved in tumorigenesis. The tumor suppressor p53 is mutated in numerous cancers (13), including human colorectal adenocarcinoma (14). O-GlcNAcylation of the p53 at Ser-149, a mutational hot spot in various cancers (such as intestinal adenocarcinoma), was found to regulate its activity and stability (15). Similarly, O-GlcNAc also modifies the transactivation domain of the c-Myc transcription factor, a product of the c-Myc proto-oncogene, at a known phosphorylation site (Thr-58), which is the main mutational hot spot in lymphomas (16, 17).
Colorectal cancer (CRC) is one of the most widespread cancers in the developed world, and its leading cause of death is metastasis (18). Although it is well established that the phosphorylation of signaling proteins is altered during cancer development and progression, the data concerning O-GlcNAcylation and cancer are rather poor, and only in recent years has this issue been addressed. In a few cancer types (i.e. colon, breast, lung, and liver), O-GlcNAcylation was found to be enhanced, especially in advanced tumors, and in some cases it was linked to cellular features relevant to metastasis, e.g. invasion and migration (7, 10, 11, 19).
In this study, we seek to shed light on the role of O-GlcNAcylation in cancer cell physiology and progression using a model of primary colorectal adenocarcinoma and lymph node metastatic clones (SW480 and SW620, respectively) that originated from the same patient. As such, we have characterized O-GlcNAcylation and its catalytic machinery in the two cell lines and investigated the effect of OGA silencing on the SW620 cell phenotype. In addition, for the first time a gene expression signature was generated, using cDNA microarray analysis, for OGA silencing.
EXPERIMENTAL PROCEDURES
Reagents
The human colon adenocarcinoma cell lines, SW480 and SW620, were purchased from the American Type Culture Collection (ATCC). DMEM and fetal bovine serum were obtained from Invitrogen. Penicillin/streptomycin, trypsin-EDTA, and an ECL kit were purchased from Biological Industries (Beit-Haemek, Israel); VersoTM cDNA kit was from Thermo Fisher Scientific Inc., and TransIT-LT1 transfection kit was from Mirus. Anti-O-GlcNAc-specific antibodies (CTD110.6 and RL2, mouse monoclonal) were purchased from Abcam, and anti-OGA and OGT antibodies were generously provided by Prof. Gerald W. Hart, The Johns Hopkins University. All other antibodies were purchased from Santa Cruz Biotechnology. Protease inhibitor mixture was purchased from Calbiochem. Protein assay reagent was obtained from Bio-Rad. Thiamet-G (TMG) was kindly donated by Prof. Gerald W. Hart.
Cell Culture
The human colon adenocarcinoma primary and metastatic cell lines, SW480 and SW620, respectively, were derived from the colon and lymph node of the same patient (1 year later). Cells were grown at 37 °C under a humidified atmosphere with 5% CO2 in complete DMEM (supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) and were used for up to 35 passages in vitro. Cells transfected with pSUPERretro plasmid were grown with the addition of 0.4 μg/ml puromycin.
Cytogenetic Analysis
The karyotype was prepared according to Mandhal (20). Briefly, cells in the log phase of growth were arrested at metaphase by colcemid, subjected to hypotonic treatment, fixed, mounted onto slides, and stained by Giemsa for G-bands. From each culture, 15 metaphase spreads were analyzed. G-banded karyotypes were analyzed according to the International System for Human Cytogenetic Nomenclature (21).
Soft Agar Colony Forming Assay
To monitor anchorage-independent growth, 104 cells (validated for viability by trypan blue staining and counted using a hemocytometer) were suspended in 1.5 ml of DMEM containing 0.5% low melting point agarose and overlaid on 2 ml of 0.6% agarose in DMEM coating 35-mm dishes. Each cell line was seeded in triplicate and assayed six different times. Following agarose solidification, the dishes were incubated at 37 °C. After 14 days, colonies were stained with 0.05% crystal violet, counted, and photographed. For each dish, ≥3 (1 cm2) fields were counted. Statistical analysis was performed using a paired Student's t test.
Protein Extraction and Subcellular Fractionation
Cells at 80% confluency were washed twice with ice-cold PBS, harvested by scraping into chilled PBS containing 1 mm PMSF, and centrifuged at 1000 × g for 5 min. For whole-cell lysate, cells were suspended with RIPA buffer (10 mm Tris-HCl, pH 8.0, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, and protease inhibitor mixture), homogenized by passing the solution through a 21-gauge needle, and kept on ice for 1 h. After centrifugation at 15,000 × g for 15 min at 4 °C, the supernatant was stored at −70 °C. For subcellular fractionation, cells (20 × 106) were washed twice with ice-cold PBS, pelleted, and resuspended in ice-cold hypotonic buffer containing 10 mm Hepes-KOH, pH 7.5, 20 mm KCl, 1.5 mm MgCl2, 0.5 mm DTT, and protease inhibitor mixture. The lysate was homogenized as described above, incubated on ice for 30 min, and subsequently centrifuged at 4 °C for 15 min at 16,000 × g. The supernatant was saved as the cytosolic fraction. The pellet was then resuspended in hypertonic buffer (20 mm Hepes-KOH, pH 7.5, 1.5 mm MgCl2, 25% glycerol, 420 mm NaCl, 0.2 mm EDTA, 1 mm DTT and protease inhibitor mixture), homogenized, and incubated on ice for 30 min. The sample was then centrifuged (15 min, 16,000 × g, 4 °C), and the supernatant was saved as the nuclear fraction. Protein concentration was determined by Bradford assay (Bio-Rad). Western blot analysis with anti-HSP90 and anti-poly(ADP-ribose) polymerase (PARP)-specific antibodies was used to confirm the purity of the cytosolic and nuclear fractions, respectively.
Isolation of O-GlcNAcylated Proteins
Equal amounts of whole-cell lysates (300 μg of protein/sample) were incubated overnight at 4 °C, with rotation, with 300 μl of succinylated wheat germ agglutinin in a solution containing 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 2 mm MnCl2, 1% Triton X-100, 50 mm Tris, pH 5.8. Unbound proteins were then removed by three washes with Tris-buffered saline (150 mm NaCl, 10 mm Tris, pH 7.5) (TBS), and bound O-GlcNAcylated proteins were eluted with TBS containing 0.3 m GlcNAc.
Western Blotting and Densitometric Analysis
Equal amounts of whole-cell lysates and cytosolic and nuclear extracts (25–30 μg of protein/lane) were separated on 10% SDS-PAGE and electroblotted onto nitrocellulose membranes. The membranes were blocked for 1 h with TBS containing 0.1% (v/v) Tween 20 and supplemented with 4% bovine serum albumin (BSA) or with 5% nonfat dry milk. The membranes were then probed overnight at 4 °C with the anti-O-GlcNAc primary antibodies CTD110.6 and RL2 (1:1000) and anti-OGA (1:1000), OGT (1:5000), HSP90 (1:500), PARP (1:1500), IκB-β (1:800), caveolin-1 (1:1200), β-catenin (1:1000), and E-cadherin (1:800). Next, the membranes were incubated for 1 h with the appropriate HRP-conjugated secondary antibody, and protein cross-reactive bands were detected using an ECL kit. Each immunoblot was repeated at least three times (using different protein extracts), and relative protein levels and immunoreactive intensities (normalized according to the actin level) were quantified using Quantity One (Bio-Rad) software and presented as the mean ± S.E. of adjusted volume intensity × mm2. Statistical analysis was performed using the nonparametric Kruskal-Wallis test (p ≤ 0.05) with the SPSS software.
OGA Silencing by Short Interfering RNA
RNA interference using the short hairpin RNA (shRNA)-expressing vector pSUPERretro (Oligoengine, Seattle, WA) was performed to generate a stable SW620 cell line specifically suppressed for the expression of OGA. The shRNA-encoding sequence was created by two complementary oligonucleotides, each containing the 19–22 nucleotides that target the OGA sequence, followed by a short spacer and an antisense sequence of the target. The following sequence was eventually used for the gene silencing in the cells (after verification of the silencing efficiency using real time PCR, as described below): GATCCCCAGAATATGAGATAGAGTTCATTTCAAGAGAATGAACTCTATCTCATATTCTTTTTTA. The OGA-shRNA-encoding sequence was cloned into the BglII and HindIII sites of the pSUPERretro plasmid that contains a puromycin resistance gene. The plasmid constructs were transformed into DH5α Escherichia coli, and selected positive clones were verified by DNA sequencing. A pSUPERretro plasmid without a silencing sequence served as a control for the OGA-silencing plasmid. In addition, SW620 cells carrying a pSUPERretro plasmid containing an L1CAM-silencing sequence (22), kindly donated by Prof. Avri Ben-Ze'ev from the Weizmann Institute of Science, were used as a reference in some experiments.
Cell Transfection
The pSUPER plasmids, with or without the silencing inserts (the latter serving as control), were transfected into SW620 cells grown in 6-cm diameter plates (Greiner Bio-One, Austria) at a density of 1 × 105 cells/plate. Transfection was performed in a medium containing 10% PBS and 2.5 μg of DNA using the TransIT-LT1 transfection kit according to the manufacturer's protocol. Forty eight hours after transfection, the medium was replaced by a selection medium containing 1.2 μg/ml puromycin (Invivogen). The medium was refreshed every other day until resistant colonies became visible. All individual colonies from the same transfection were pooled and re-plated in 24-well plates to yield an oligoclonal puromycin-resistant cell population. OGA silencing was verified by quantitative real time reverse-transcription PCR (qRT-PCR).
RNA Extraction and Quantitative Real Time RT-PCR
Total RNA was extracted using EZ-RNA total RNA isolation kit according to the manufacturer's instructions. The VersoTM cDNA kit was used to obtain cDNA by reverse transcribing 1 μg of total RNA. Relative quantification of OGA and OGT transcript levels was obtained by real time RT-PCR using the universal ProbeLibrary assay (Roche Applied Science). Gene-specific intron-spanning primers and their matching probes were designed using the ProbeFinder Software (Roche Applied Science). Relative quantification of OGA and OGT transcription levels was obtained using the following primers: OGA_F, 5′-TGTGGTGGAAGGATTTTATGG-3′, and OGA_R, 5′-TCATCTTTTGGGGCATACAAG-3′; OGT_F, 5′-TTAGAGTTCCCAGGGTTTGAAG-3′, and OGT_R, 5′-ATGCTCTGGAGGGCTTGAG-3′. The OGA and OGT primers were added to the FastStart Universal Probe Master (Rox) (Roche Diagnostics) and the universal ProbeLibrary Probes 50 or 74, respectively (Roche Diagnostics). The 18 S gene was used as a normalizing gene using the following primers: 18 S_F, 5′-GGAGAGGGAGCCTGAGAAAC-3′, and 18S_R, 5′-TCGGGAGTGGGTAATTTGC-3, and the 74 probe (Roche Diagnostics). Reactions were performed using the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Statistical analysis was done using the nonparametric Kruskal-Wallis test (p ≤ 0.05) with the SPSS software.
cDNA Microarray and Data Analysis
RNA was extracted from the siOGA-SW620 (silenced cells) and the pS-SW620 control cells (as described above) and treated with the TURBO DNA-freeTM kit (Applied Biosystems, Foster City, CA). For each cell line, three different samples were prepared, with each sample used for a single chip (i.e. each of the two cell lines had three chips). From each sample, 300 ng of total RNA was amplified and hybridized to GeneChip® Human Gene 1.0 ST arrays (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer's instructions. These arrays assay for the expression of about 28,869 well annotated genes with 764,885 distinct probes.
Data analysis was performed using the Partek® Genomics SuiteTM at the Bioinformatics Core Facility, National Institute of Biotechnology in the Negev, Ben-Gurion University of the Negev. Microarray CEL files were read into Partek and preprocessed by Robust Multiarray Averaging (23) using default parameters, followed by quantile normalization and probe summarization using median polish. Expression signals were generated and displayed on a log 2 scale. Signal distribution plots and principal component analysis of the normalized samples were examined to assess the quality of the microarrays. Statistical hypothesis testing to identify differentially expressed genes between siOGA-SW620 and pS-SW620 (control) was performed using Student's t test. The resulting p values per gene were further adjusted for multiple testing by step-up false discovery rate according to Benjamini and Hochberg (24). Fold change values are represented on a linear scale, where positive and negative values indicate up- and down-regulated expression, respectively. Differentially expressed genes were defined as those having absolute expression signals >5 in at least one of the arrays, false discovery rate adjusted p values <0.05 and fold change values either below or above 1.3. The 1302 genes passing the cutoff criteria were assessed for functional annotations and subjected to over-representation analysis to identify biologically annotated canonical pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database by Ingenuity Pathway Analysis (IPA).
RESULTS
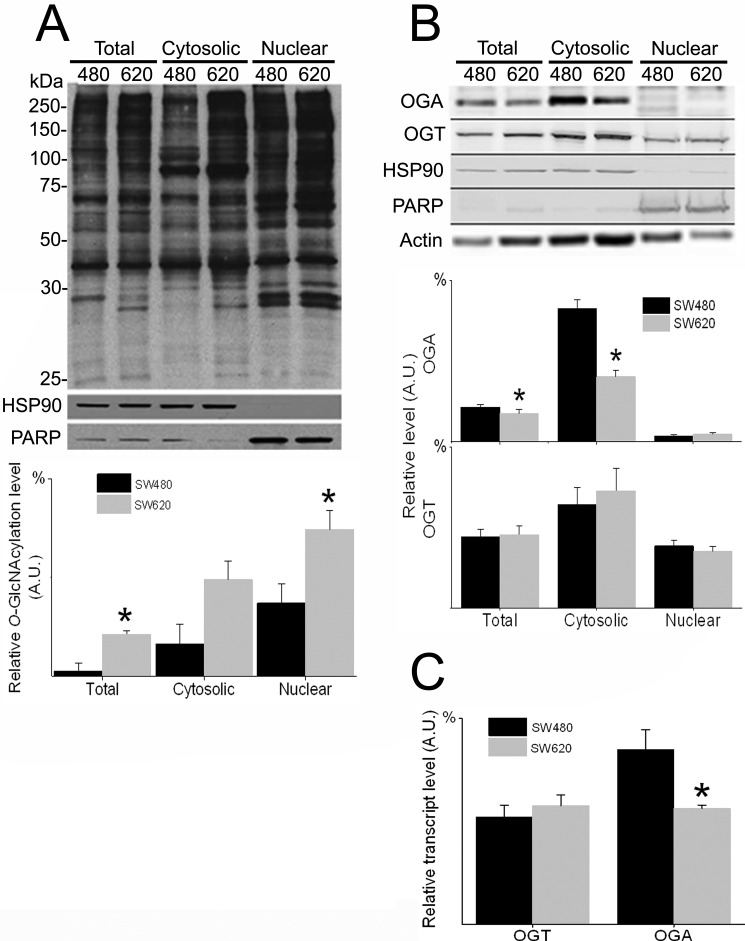
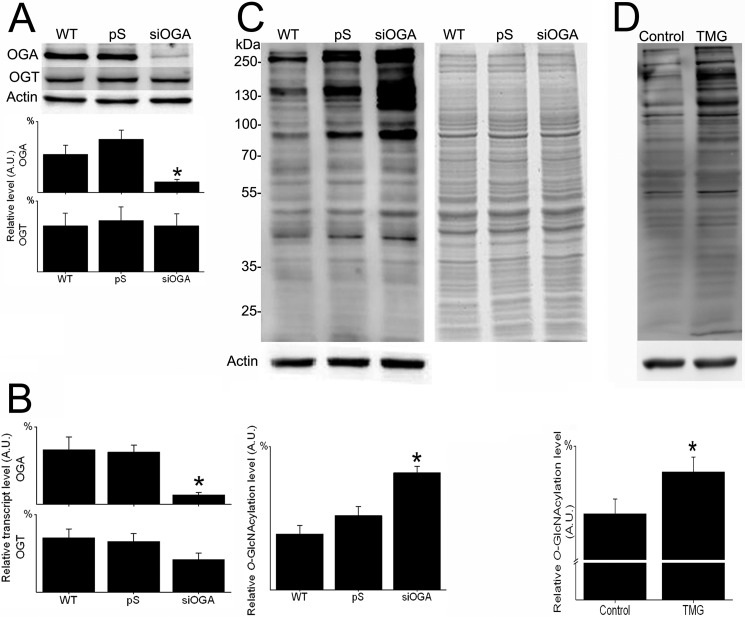
To elucidate the significance of O-GlcNAcylation in cancer cell physiology, we studied the primary SW480 CRC clone, its lymph node metastatic clone SW620, and an OGA-silenced subclone. O-GlcNAcylation and its enzymatic machinery were evaluated in the two WT CRC clones by immunoblotting and by qRT-PCR (Fig. 1). Compared with the SW480 primary clone, the SW620 lymph node metastatic clone consistently displayed a higher, statistically significant (p <0.05) O-GlcNAcylation level for both total protein extract and nuclear protein fraction (Fig. 1A). In an alternative approach, O-GlcNAcylated proteins were isolated from the two cell lines by using a specific O-GlcNAc-binding lectin, succinylated wheat germ agglutinin, and resolved on SDS-PAGE. As shown in supplemental Fig. 1, both Coomassie Blue staining and immunoblotting using the anti-O-GlcNAc antibody RL2 (other than the anti-O-GlcNAc antibody CTD110.6 used in Fig. 1A) confirmed the significantly higher (p <0.05) content of O-GlcNAcylated proteins in SW620 cells than in SW480 cells. In agreement with these findings, OGA was found to be significantly reduced (p <0.05) at both transcription and protein levels in SW620 cells compared with SW480 cells (Fig. 1, B and C). However, no significant difference between the clones was found for OGT. OGA-silenced SW620 cells (siOGA) displayed a reduction of >70% in enzyme expression in both transcription and translation levels (as shown by immunoblotting and qRT-PCR) compared with the control cell line pS (Fig. 2, A and B, respectively). No corresponding alteration in OGT expression was observed. As expected, OGA silencing was accompanied by a significant elevation of >50% (p <0.05) in the cellular O-GlcNAcylation level (Fig. 2C). To confirm that manipulation of OGA directly affects the cellular O-GlcNAcylation level, the selective potent inhibitor of OGA, TMG, was applied. TMG-treated SW620 cells showed a significantly higher level (p <0.05) of protein O-GlcNAcylation compared with nontreated control SW620 cells (Fig. 2D).
FIGURE 1.
O-GlcNAcylation is elevated and OGA is down-regulated in metastatic CRC clone compared with primary clone. Total, cytosolic, and nuclear proteins were purified from primary SW480 and metastatic SW620 cells and analyzed for levels of O-GlcNAcylation, OGA, and OGT. A, upper panel, representative Western blots using anti-O-GlcNAc (CTD110.6), HSP90, and PARP antibodies. HSP90 and PARP were used as cytosolic and nuclear fraction markers, respectively. Lower panel, quantitative analysis of the relative O-GlcNAcylation levels in the various fractions of the two CRC clones. B, upper panel, representative Western blots using anti-OGA, OGT, HSP90, PARP, and actin antibodies. Actin was used to evaluate protein quantity. Lower panel, quantitative analysis of relative OGA and OGT levels. C, relative OGA and OGT transcript levels in the CRC clones as quantified by real time RT-PCR. All quantitative results (A.U., arbitrary units) are expressed as the mean ± S.E. of at least three repeats. Asterisks indicate significant difference between clones (p ≤ 0.05).
FIGURE 2.
OGA silencing and selective pharmacological inhibition elevate O-GlcNAcylation in CRC cells. Total proteins were extracted from SW620 cells (WT) and from SW620 carrying the pSUPER plasmid, with or without an OGA-silencing sequence (pS and siOGA, respectively), and analyzed for levels of OGA, OGT, and O-GlcNAcylation. A, upper panel, representative Western blots using anti-OGA, OGT, and actin antibodies. Lower panel, quantitative analysis of relative OGA and OGT levels in the various cell lines. B, relative OGA and OGT transcript levels in the various cell lines, as quantified by real time RT-PCR. C, upper panel, representative Western blots using anti-O-GlcNAc and anti-actin antibodies (left panel) and the corresponding Coomassie Blue-stained gel (right panel). Lower panel, quantitative analysis of the relative O-GlcNAcylation levels in the various cell lines. D, total proteins were extracted from SW620 cells (control) and from SW620 cells treated with 25 μm TMG for 24 h (TMG) and analyzed for the level of O-GlcNAcylation. Upper panel, representative Western blots using anti-O-GlcNAc (RL2) and anti-actin antibodies. Lower panel, quantitative analysis of the relative O-GlcNAcylation level in the two samples. All quantitative results (A.U., arbitrary units) are expressed as the mean ± S.E. of at least three repeats. Asterisks indicate significant difference between clones (p ≤ 0.05).
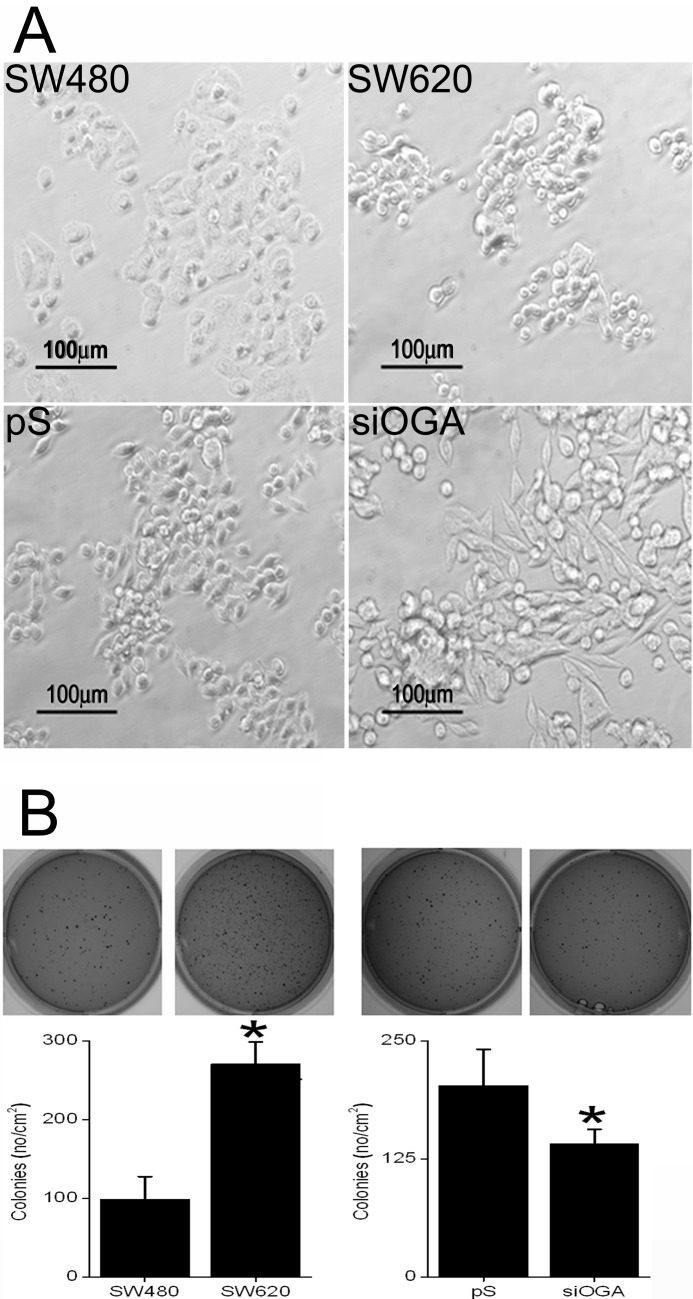
The effect of O-GlcNAcylation on CRC cell phenotype was also examined by following the morphology and growth of the various clones. As shown in Fig. 3A, the SW480 cells showed a typical epithelial type morphology characterized by a tendency to form cohesive groups, whereas SW620 cells were spherical or spindle-shaped (fibroblast-like) and less cohesive (25). The latter mixed population of cells was also found in the SW620 subclones, pS and siOGA. Yet although most of the WT SW620 cells were small and spherical with diameters of ∼10–20 μm and only some were elongated or spindle-shaped (17–20 μm long), the pS spindle-shaped cells, at >30 μm in length, were slightly larger. Moreover, in the siOGA clone, cells were predominantly spindle-shaped and significantly more elongated, with many measuring 40–70 μm in length (Fig. 3A). When grown in soft agar, the SW620 clone exhibited a significantly higher tendency, by ∼3-fold (p ≤ 0.01), for autonomous colony formation than the SW480 clone (Fig. 3B, left panel). Conversely, the OGA-silenced cells displayed growth retardation, manifested by the formation of >30% (p ≤ 0.01) fewer colonies and by their smaller sizes compared with control cells (Fig. 3B, right panel).
FIGURE 3.
OGA silencing alters the morphology and reduces the anchorage-independent growth of CRC cells. A, representative images of the CRC clones morphology. Note spindle-shaped cells in siOGA. B, colony formation in soft agar of the CRC clones. For each cell line, triplicates of 104 cells were seeded, and 14 days later they were stained with crystal violet, counted, and photographed. Upper panel, representative images of colony-bearing agar dishes. Lower panel, quantification of colony formation. SW480, primary CRC clone; SW620, metastatic CRC clone; pS, SW620 subclone carrying an empty pSUPER plasmid; siOGA, SW620 subclone carrying an OGA silencing sequence. All quantitative results are expressed as the mean ± S.E. of at least six different repeats. Asterisks indicate significant difference between clones (p ≤ 0.01).
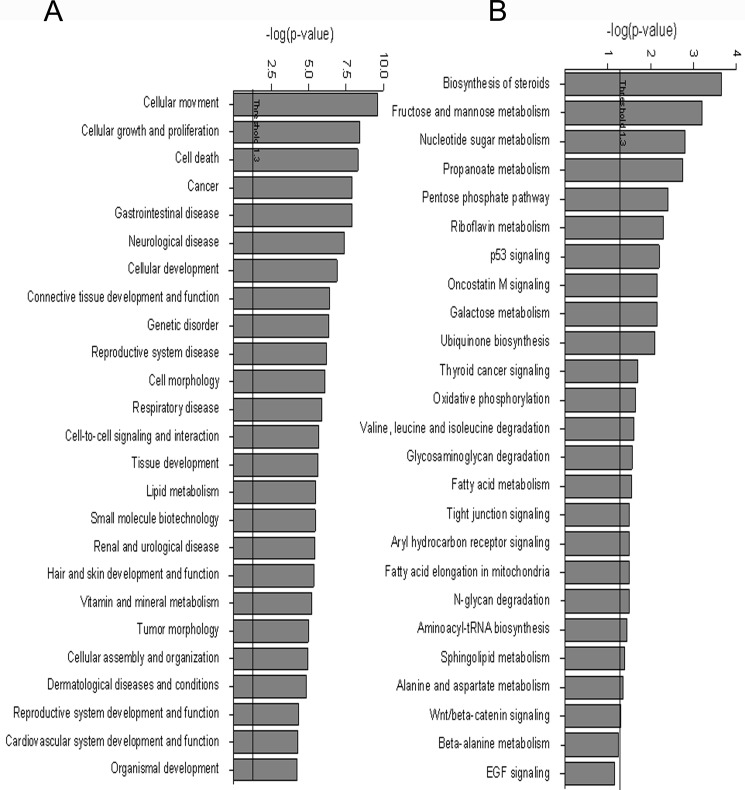
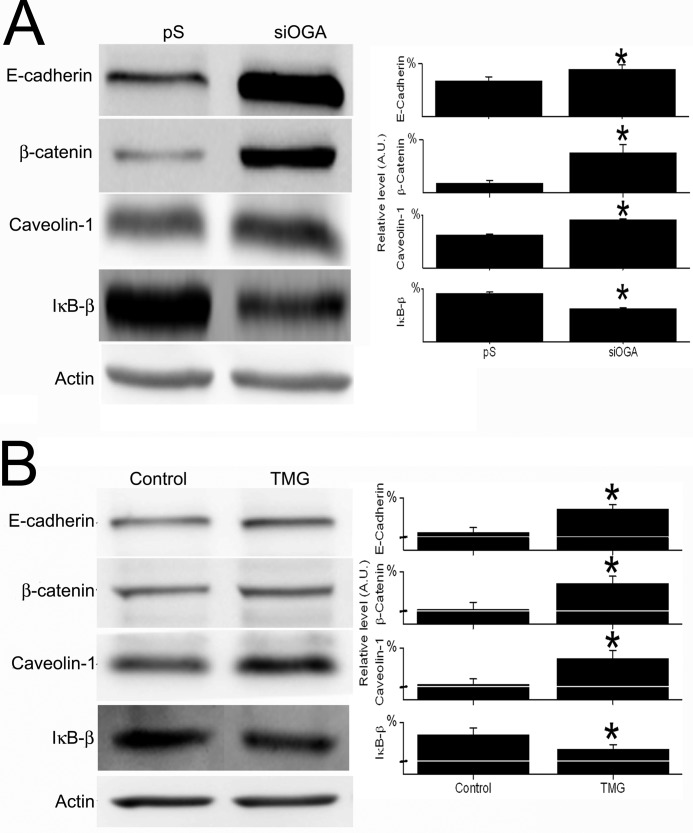
Genome-wide expression profiling was used to generate a signature of OGA silencing and to monitor functional alterations due to silencing. Initial analysis of cDNA microarray data revealed 1302 genes that were differently regulated (by ≥1.3-fold, p <0.05) in siOGA-SW620 cells versus in their control pS-SW620 cells, with 651 up-regulated and 651 down-regulated genes (supplemental data). Functional annotation and enrichment of canonical pathways were explored using the IPA software. Of the 1302 molecules examined, 1229 were mapped (73 remained unmapped); of these 1067 were network eligible. The IPA summary is shown in Table 1 and Fig. 4, and selected up- and down-regulated genes of special interest (see below) are detailed in Table 2. Selected networks, including a network centered around the OGA gene (MGEA5), are shown in Fig. 6. Immunoblot analysis validated the microarray data for four representative proteins (Table 2 and Fig. 5) as follows: E-cadherin, caveolin-1, β-catenin, and IκB-β encoded by the CDH1, CAV1, CTNNB1, and NFKBIB genes, respectively. Microarray analysis indicated up-regulation of the first three genes and down-regulation of the fourth (by ∼2.55-, ∼1.5-, ∼1.9-, and ∼1.42-fold, respectively) in siOGA cells compared with pS cells (supplemental data) and immunoblot results were consistent (Fig. 5). This distinct protein expression pattern of the siOGA cells was further validated by a comparison with SW620 cells carrying a pSUPER plasmid containing a L1CAM-silencing sequence (siL1-SW620) (see “Experimental Procedures”). Compared with siL1-SW620 cells, siOGA cells showed a significantly (p <0.05) higher level of O-GlcNAcylation, as well as higher levels of E-cadherin, caveolin-1, and β-catenin, and a lower level of IκB-β (supplemental Fig. 2). In addition, to confirm that the manipulation of OGA did indeed affect protein expression, the level of these four proteins was examined in SW620 cells following treatment with the OGA-specific inhibitor, TMG. As shown in Fig. 5B, the pharmacological inhibition of OGA showed the same, statistically significant (p <0.05), effect on protein expression as OGA silencing.
TABLE 1.
Summary of IPA performed for cDNA microarray analysis of the siOGA-SW620 and pS-SW620 clones
FIGURE 4.
OGA silencing affects the transcriptome of SW620 metastatic CRC clone. Control pS-SW620 cells and OGA-silenced SW620 cells were subjected to cDNA microarray analysis. Gene Ontology-based over-representation for the microarray dataset was performed with the IPA software. Over-represented genes were categorized according to their functions (A), and canonical pathways were annotated using the KEGG database (B).
TABLE 2.
Selected up- and down-regulated genes in the siOGA clone categorized according to cellular function
Genes names in boldface indicate that the protein is modified by O-GlcNAc (based on previous data).
| Function | Up-regulated genes | Down-regulated genes |
|---|---|---|
| Growth factors and insulin signaling | IGF2, IGFBP5, IGFBP4, INS-IGF2, IGFBPL1, FGFR1, FGFR4, FGF13, CPE, TGFBR2, TGFBR3 | IGF2BP3, FGFR1OP, FGFBP1, FGF9, TGFBI, TGFA |
| Transcription | JUN, PRDX4, PIAS3, UXT, SREBF2, MED14, BCOR, TFDP1, TCF19, BACH1, CREB3L2 | T, TP53, TP53BP1, WWTR1, NFKBIB, TBP, GTF2F2, TCF25, USF2, CNOT3, CNOT1, HSF1, GTF3A, ELF1, HSF2, JDP2, PIAS1, TAF1, BRMS1, TAF5L, GTF2IRD1 |
| Cell growth, differentiation, and proliferation | CCNG2, CDK2, CDK5, CDKAL1, CDCA4, CDCA7, GDF11, PDCD6IP, PLCG1, TMSL2, PROM1, RPS6KA3, S100A2, S100A3, S100A16, GADD45A | CDK10, CCND1, GAS6, AKT1S1, ITPKB, CDCA7L, TPD52L1, RB1 |
| Cell-cell interaction and motility | GABRA2, TNC, LPXN, PCDHB5, FERMT2, CD24, SERPINE2, MCAM, ACTG1, DST, MYO10, KRT81, CDH1, CD99L2, ITGB7, ITGA2, CLDN1, CLDN12, CD302, FBLN1, LIMS3, ABLIM1, ABLIM3, FLNA, FLNB, TSPAN4, TSPAN12, ITGB5, EPHA4, EPHB6, EPHB3, THBS1, TUBA1A, TUBA4A, VSIG1, RAP2A, S100A6, FN1, LAMB1, L1CAM, TIMP1, VIM, TIMP2, CYFIP2, CEACAM1, EFNB2, AHNAK2, FSCN1 | CLDN7, MMP14, PLAU, ADAMTS17, ADAM19, ADRM1, CELSR1, ST14, EFNB1, TIMP3, THSD1, ARPC4, ACTN4, KRT6C |
| Cell signaling | TSPAN5, ERBB3, ARAF, BRAF, ELK1, GEFT, MAP3K3, RASA3, FZD10, ICK, HIPK2, JAK1, CTNNB1, CTNNBIP1, KREMEN1, MAMLD1, CAV1, PAK1, RHOB, LRP1, LRP4, PLCG1, DAB2, LRIG1 | FZD6, WNT11, SFRP5, CDCA7L, CAPN1, RAB27A |
| Cell death | CYFIP2, CASP2, CMTM8, PDCD6IP, SERPINB9 | AIFM1, AIFM2, CIAPIN1, BCL2L13, BCL2L1, BNIP2, PDCD2, AVEN, TRADD, NOTCH2 |
| Metabolism and biosynthesis | PDK1, G6PD, PHKA2, PDHA1, GK, TGDS, LIPC, STARD4, PLA2G16, SC4MOL, AZGP1, GLUL, ALDOC, FBP1, NSDHL, ANGPTL2, DHCR7, LSS, FDPS, FDFT1, PECR, ACSS2, PPAP2B, HMGCR, HMGCS1 | GNE, GPT2, PFKM, PRKAR1B, PHKB, GALK2 |
| Immune response | IL6R, IL8, IL17RD, IL23A, CXCL6, SLAMF9, IFNGR2 | SIGIRR, IL20RA, CXCR3, IGBP1, HIVEP2 |
FIGURE 6.
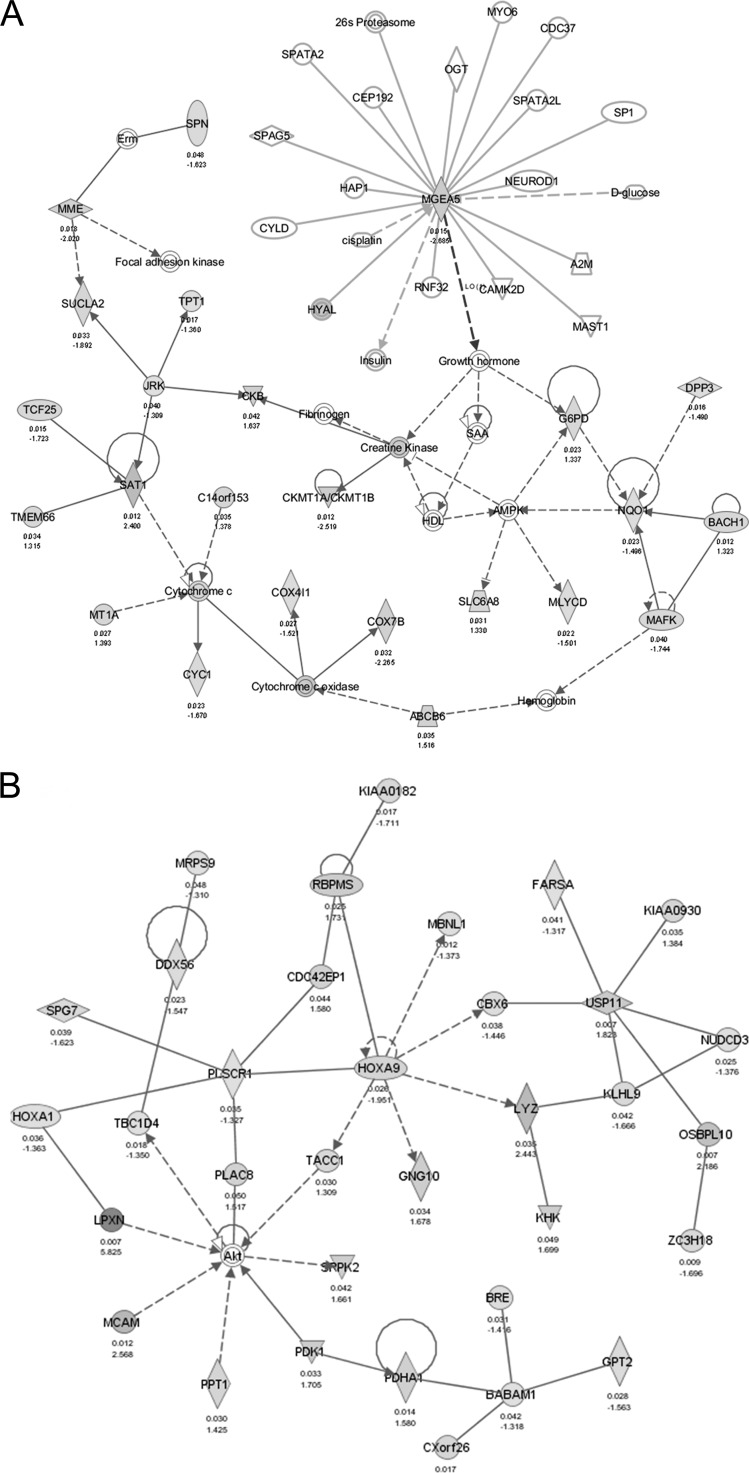
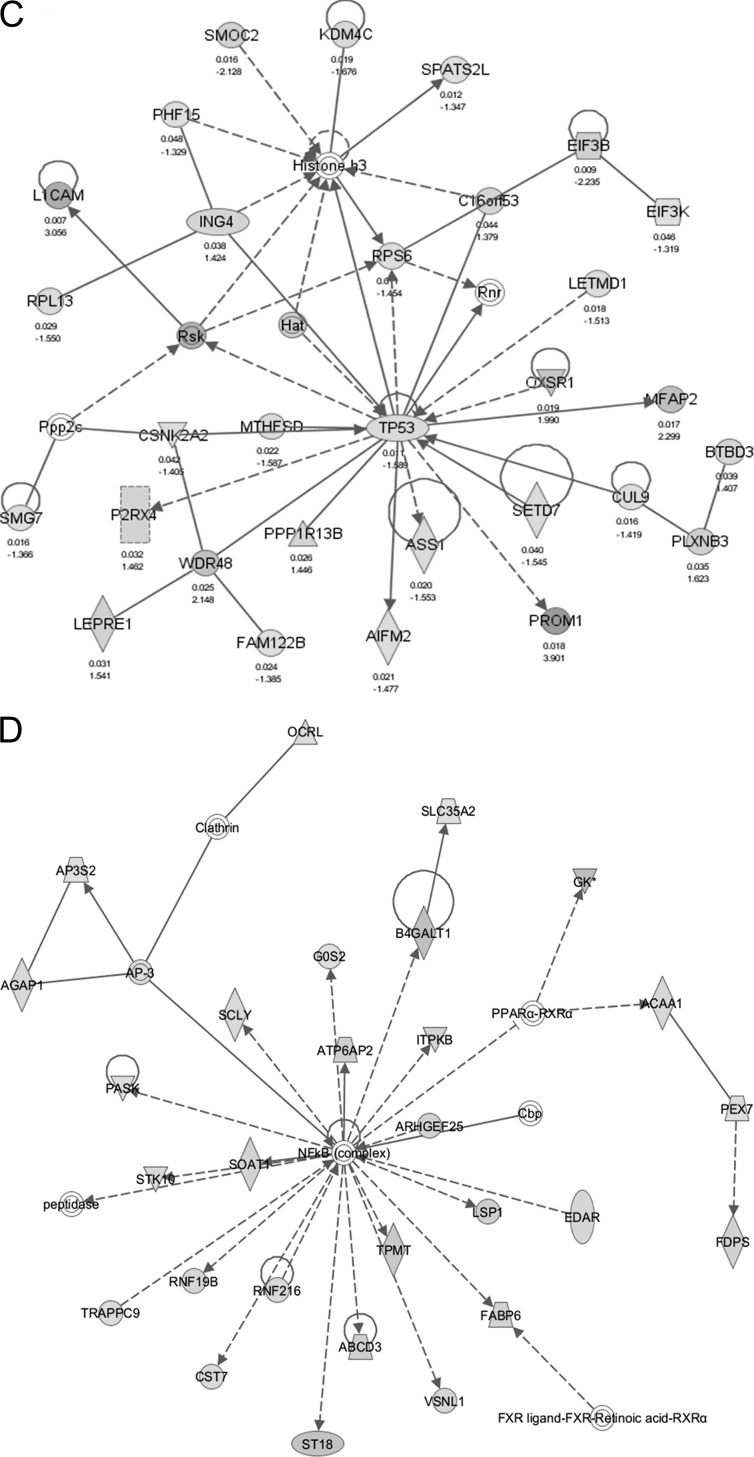
IPA of genes correlated with OGA silencing. Red and green (online) indicate up- and down-regulated genes, respectively, following OGA silencing. Solid and broken lines indicate direct and indirect interactions, respectively, between molecules. A, network centered around MGEA5, the OGA gene, indicating its up- and downstream interactions. B, top network number 1 (see Table 1), focusing on lipid metabolism, post-translation modifications, and small molecule biochemistry. C, top network number 5, focusing on cancer, protein synthesis, cellular function, and maintenance. D, top network number 7, focusing on carbohydrate metabolism, molecular transport, and nucleic acid metabolism.
FIGURE 5.
Experimental validation of microarray data for four selected proteins. Total proteins were extracted from pS-SW620 and siOGA-SW620 cells and analyzed for the levels of four proteins that showed altered regulation by cDNA microarray assay. A, left panel, representative Western blots using anti-E-cadherin, β-catenin, caveolin-1, IκB-β, and actin antibodies. Right panel, quantitative analysis of the protein levels in the two cell lines. B, total proteins were extracted from SW620 cells (control) and from SW620 cells treated with 25 μm TMG for 24 h (TMG) and analyzed for the level of the above-mentioned proteins. Left panel, representative Western blots. Right panel, quantitative analysis of the relative protein levels in the two samples. All quantitative results (A.U., arbitrary units) are expressed as the mean ± S.E. of at least three repeats. Asterisks indicate significant difference between clones (p ≤ 0.05).
The top five biological functions altered by OGA silencing were cellular movement, cellular growth and proliferation, cell death, cancer, and gastrointestinal diseases (Fig. 4A and Table 1). Cellular development and morphology and cell-to-cell signaling and interaction were also altered but to lesser extents. Indeed, the morphological changes observed in the siOGA cells (Fig. 3) coincide with these results. Among the altered genes, some were related to actin reorganization and to cell migration and invasion, e.g. those encoding ARPC4, β-actinin, fascin, filamin, LIM proteins, AHNAK2, eIFs, S100 proteins, and claudins (Table 2). Other altered genes encode proteins, including those for the cytoskeleton (actin γ1, tubulin, and dystonin), motors (myosin X), structure (keratin 6C), ECM (tenascin C, fermitin family homolog 2, serpin peptidase inhibitor clade E, and tissue inhibitor of metalloproteinases), membrane and scaffolding (E-cadherin, tetraspanins, caveolin-1, and claudins), and the oncostatin M signaling pathway involved in ECM remodeling, growth, and differentiation (JAK, SHC, and ELK1), to name but a few.
In addition, many of the altered genes in the OGA-silenced clone encode proteins that are involved in tissue development and cell-cell interactions (see Table 2). These included cell adhesion molecules such as MCAM, integrins β7, β5, and α2, and signaling proteins downstream, FAK, RHOA, Cdc42 effector proteins 1 and 4 that are involved in cytoskeletal organization and cell migration, and Eph receptors A4, B3, and B6, and their ligands, ephrin-B1 and -2, that mediate cell-cell communication and regulate cell shape and mobility. Interestingly, although the ECM-remodeling matrix metalloproteinases were not altered (except MMP14), their regulators TIMP1, -2, and -3 were altered. Other proteinases included the urokinase plasminogen activator, ADAM metallopeptidase (ADAMTS17 and ADAM19), thrombospondin 1 (THBS1 and THSD1), and cathepsin D. Motility-related proteins, including ECM components (fibronectin and laminin), and cytokines and their associated proteins such as TGF-β1, PDGFA, FGF, and IFNγR2 were also modified. As shown in Table 2, a few growth factors and some of their receptors and related proteins were also altered, as was IGF2 (and related proteins), which may also be involved in cell motility. Other genes found to be altered and potentially related to metastasis in CRC models (26) are those encoding MAGED2, peroxiredoxin 4, nucleolin, cyclin D1, caspase 2, IL8, S100A6, L1CAM, and annexin A5 (Table 2 and supplemental data).
The cDNA microarray data set was also analyzed, using the IPA software, for pathway over-representation to identify biologically annotated pathways (using the KEGG data base). The emerging top canonical pathways were lipid and carbohydrate pathways and included the biosynthesis of steroids and the metabolic pathways of fructose and mannose, nucleotide sugars, and propanoate and pentose phosphate (the latter pathway referred to as PPP) (Fig. 4B and Table 1). The metabolic pathways are further discussed, and selected genes involved in metabolism are shown in Table 2. In addition, the expression of several mitochondrial genes was altered as follows: 14 genes were related to mitochondrial malfunction (p = 0.0728) and 15 genes were connected with oxidative phosphorylation (complexes I, III, IV, and V) (p = 0.0223) (see Fig. 4B and supplemental data).
Cytogenetic analysis was performed to examine whether chromosomal changes might have occurred due to transfection in pS and siOGA cells. G-banding karyotype analysis of the WT SW620 clone and its subclones pS and siOGA revealed that the three cell lines have the same clone karyotype. However, although WT and pS cells shared the same karyotype, only the siOGA-SW620 cells showed a polyploid karyotype derived from the karyotype of the two other clones. The karyotype of the SW620 and pS-SW620 cells was as follows: 47∼50,X,−Y,−2,−3,−4,−5,−8,−8,−9,−10,−13,−16,−18,+X,+m1,+m2,+m3,+m4,+m5,+m6,+m7,+m8,+m9,+m10,+m11,+m12,+m13,+m14[cp15], whereas the karyotype of the siOGA-SW620 cells was as follows: 47∼50,X,−Y,−2,−3,−4,−5,−8,−8,−9,−10,−13,−16,−18,+X,+m1,+m2,+m3,+m4,+m5,+m6,+m7,+m8,+m9,+m10,+m11,+m12,+m13,+m14[cp3]/87∼97 idemX2[cp12].
DISCUSSION
O-GlcNAcylation plays a crucial role in cellular physiology (22) and is considered to be a sensor of the cellular state (23). Thus, its involvement in cancer-relevant processes (e.g. transcription, signal transduction, metabolism, and cell cycle) is garnering increasing attention. In this study, we monitored O-GlcNAcylation in metastatic CRC clones derived from the same patient and investigated its effect on cell physiology and genome-wide expression profile using a newly established subclone stably silenced for OGA expression.
The SW620 metastatic CRC clone exhibited higher levels of protein O-GlcNAcylation compared with the primary SW480 clone coinciding with the lower expression of OGA in the previous clone (Fig. 1). These findings are in accordance with previous reports describing enhanced O-GlcNAcylation in breast cancer metastatic lymph nodes compared with their primary tumor tissues and increased levels of O-GlcNAcylation and OGT in lung and colon tumor tissues compared with adjacent tissues (10, 11). Increased O-GlcNAcylation and elevated OGT expression were also found in several breast cancer cell lines (7). Moreover, lymph node metastases of ductal breast carcinomas were associated with a decreased OGA transcript level, and grade II and III tumors showed higher OGT expression and lower OGA expression compared with grade I tumors (12). Phenotypically, the two WT metastatic clones showed distinct cell morphologies and growth (Fig. 3). The SW480 CRC clone was characterized by a spreading epithelial-type morphology, although the SW620 clone displayed ovoid to elongated fibroblast-like morphology, indicating the cells maintained their original phenotypes and consistent with previous data (25, 27, 28). When grown in soft agar, the SW620 clone displayed enhanced growth manifested by higher colony formation ability than the SW480 clone (Fig. 3). Indeed, enhanced growth of the SW620 clone compared with the SW480 clone in both culture and agar was previously shown (29, 30). These data suggest that O-GlcNAcylation enhances anchorage-independent growth, an assumption supported by a previous report showing that colony formation by lung and colon cancer cells was reduced by OGT silencing and increased by OGA inhibition (10). Similarly, OGT silencing in breast cancer cells inhibited tumor growth in vitro and in vivo and decreased cell cycle progression (7). In several colorectal cancer cell lines, in which O-GlcNAcylation was elevated compared with normal cells, OGA inhibition accelerated cell proliferation (31).
Taken together, the above findings may indicate the involvement of O-GlcNAcylation in tumor progression and possibly metastasis. This issue was further investigated using the siOGA subclone, in which OGA expression was knocked down by >70% and the O-GlcNAcylation level was increased by >50% (Fig. 2). OGA-silenced cells displayed an altered phenotype, as manifested in their morphology and growth, and they exhibited an extensively altered gene expression profile. Although siOGA cells showed polyploidy, chromosomal analysis verified that they basically have the same clone karyotype as their control pS cells and the WT SW620 cells, with common gains and losses. Our findings are in keeping with previous reports that overexpression of OGT perturbs cytokinesis and promotes polyploidy (9, 32). Taken together, these findings further support the notion that the observed alterations in siOGA cells, specifically in their gene expression pattern, were indeed the result of OGA silencing. The phenomenon of polyploidy in siOGA cells will be addressed in a future study.
Compared with SW620 and pS cells, most of the siOGA cells acquired a distinct spindle-shaped, fibroblast-like morphology (Fig. 3) that strikingly coincides with the EMT. In a key cellular process in epithelial metastatic progression, in which cells acquire fibroblastic morphology and are characterized by increased migratory capacity, invasiveness, and production of ECM components (33), the EMT is enabled by the alteration of genes involved in actin cytoskeleton remodeling, cell polarity, and cell-cell interaction. The Wnt/β-catenin canonical pathway, specifically associated with CRC (34), and its major effectors β-catenin and E-cadherin, are considered integral components of the EMT. Membranous E-cadherin·β-catenin complexes are critical for the stability of epithelial cell-cell adhesions, although nuclear β-catenin can function as an oncogene. Integrin-mediated pathways, such as TGF-β signaling, are also involved in EMT (33, 35). Indeed, the current cDNA microarray and previous data support the notion of EMT in the siOGA cells. In these cells, the Wnt/β-catenin pathway was significantly enriched (18 molecules, p = 0.0388) (Fig. 4, Table 2, and supplemental data), and genes encoding key proteins of this and other signaling pathways (e.g. of growth factors) that control EMT were altered. These include the genes of TGFβ receptors 2 and 3, FGF receptors 1 and 4, β-catenin 1, and its targets the proto-oncogene cyclin D1 and L1CAM. The β-catenin gene, CTNNB1, was up-regulated by >1.9-fold (supplemental data). In accordance, it was recently shown that mice treated with glucose or glucosamine displayed increased O-GlcNAcylation and β-catenin levels in the colon (31). Other EMT-related proteins whose genes were altered are the Ras family GTPases RHOA and RHOB, phospholipase Cγ, IL8, Jun, and RPL13 (Table 2 and supplemental data). Interestingly, many proteins involved in cell morphology and motility are modified by O-GlcNAc (Table 2) (4, 36).
EMT is also characterized by the loss of epithelial markers, e.g. E-cadherin and claudin 1, and the gain of mesenchymal markers, e.g. vimentin and fibronectin (33, 35, 37). It was previously found that SW480 cells express E-cadherin and β-catenin but not vimentin, whereas the converse is true for SW620 cells. Yet claudin 1 is highly expressed in SW620 cells (33, 35, 37), demonstrating that the expected alteration of genes does not always occur. Here the OGA-silenced clone indeed displayed up-regulation of the vimentin and fibronectin genes and down-regulation of the claudin 7 gene, but the E-cadherin and claudin 1 and 12 genes were also found to be up-regulated (Table 2, Fig. 5, and supplemental data). Yet it is noteworthy that E-cadherin membrane localization is uncertain; it was previously reported that E-cadherin O-GlcNAcylation blocks its cell surface transport and its binding to p120 catenin, resulting in reduced intercellular adhesion (38). Considering OGA silencing, it is plausible that E-cadherin is O-GlcNAcylated, and as such, it could not stabilize cell-cell adhesions.
Metastases, detected in ∼33% of CRC patients, are responsible for the high CRC mortality rates. A vast number of proteins are required to enable metastasis (34). Here, we combined previous data with our microarray analysis and indicated the alteration of various metastasis-related genes, some specifically associated with CRC (Table 2) (26), in siOGA cells. These genes, detailed under “Results,” encode proteins involved in adhesion (e.g. integrins, E-cadherin), intracellular interaction (e.g. ephrins and Eph receptors), ECM (e.g. proteinases and tissue inhibitor of metalloproteinases), motility (e.g. fibronectin and laminin), and cell shape (e.g. actin, tubulin, and tetraspanins), as well as Gfs (e.g. TGFA and IGF2), and signaling proteins of relevant pathways (e.g. oncostatin M and Wnt/β-catenin). As mentioned above, a fibroblast-like appearance of cells may indicate metastatic progression. The SW620 cells, which showed such a morphology, displayed greater metastatic ability than SW480 cells (26–28). Thus, the metastatic ability of the OGA-silenced cells will be investigated in vivo in the future.
Although in the WT CRC clones O-GlcNAcylation seemed well correlated with cell growth, the OGA-silenced cells displayed growth retardation (compared with control cells) (Fig. 3). This growth inhibition is consistent with microarray results and with previous reports. Two main functions significantly altered in the siOGA cells were cellular growth, proliferation, and cell death (Fig. 4 and Table 1). Each function was associated with over 300 genes, among which were several key cell cycle genes, e.g. CDK2, CDK10, CDK5, CDCA4, CDCA7L, RASA3, and those of cyclins G2 and D1 (Table 2). As denoted above, cyclin D1 is targeted by β-catenin, whose gene transcription and signaling pathway were also altered (Table 2, Figs. 4B and 5, and supplemental data). Moreover, our current findings are consistent with a previous report showing that β-catenin O-GlcNAcylation reduced cyclin D1 expression (39). Thus, it seems that constant, extensive deregulation of this post-translational modification could affect the cell cycle. Indeed, such a tight relation was indicated by previous studies; O-GlcNAcylation was regulated in a cell cycle-dependent manner and O-GlcNAc elevation altered cyclin expression (9). In several cancer cell lines, increased O-GlcNAc resulted in growth defects linked to delayed G2/M progression, altered mitotic phosphorylation, and cyclin expression. OGA overexpression induced mitotic exit phenotype accompanied by delayed mitotic phosphorylation, altered cyclin expression, and disrupted nuclear organization. Moreover, increased UDP-GlcNAc slowed CRC cell growth and differentiation, O-GlcNAc reduction (by lowering UDP-GlcNAc) caused growth defects in fibroblasts, and galactosyltransferase injection or OGA inhibition resulted in damaged oocyte maturation (9). However, OGT overexpression in HeLa cells resulted in a polyploid phenotype with faulty cytokinesis, although its deletion was associated with delayed growth and increased death (9, 32, 40). In breast cancer cells, OGT silencing inhibited cell growth and decreased cell cycle progression (7). Together with our findings of a polyploid karyotype in siOGA cells, these data indicate that dynamic O-GlcNAc processing is a pivotal component of cell cycle regulation.
In addition, the growth inhibition of siOGA cells could be explained in light of the wide effect of OGA knockdown on cellular metabolism and function. Indeed, very high levels of O-GlcNAcylation in leukemia cells blocked signaling pathways that are key to rapid proliferation (37). The growth retardation of siOGA cells may be the result of their high susceptibility to nutritional stress and possibly enhanced glucose exhaustion; as mentioned above, despite the high O-GlcNAc level indicating an abundance of glucose, glycolysis was unchanged, although the PPP was altered. In fact, various glucose-related canonical pathways were altered (see Fig. 4 and supplemental data), suggesting the metabolic balance of the cells was impaired. For example, the galactose and fructose and mannose metabolism pathways, connected with glycolysis and gluconeogenesis, were significantly altered (p = 0.00652 and p = 0.000593, respectively), with most genes up-regulated (supplemental data). It is also possible that alterations of O-glycan biosynthesis genes and of glycosaminoglycan and N-glycan degradation pathways (p <0.03, see supplemental data) resulted in the accumulation of galactosamine, reported as hepatotoxin (38). In addition, the galactosyltransferase gene (B4GALT1), which encodes for the enzyme that caps O-glycosylation with a galactose residue, a process found to result in cell cycle arrest and apoptosis (35), was up-regulated in siOGA cells (by >2-fold). Alternatively, because mitochondrial failure may inhibit growth, as denoted under “Results,” the altered expression in the OGA-silenced cells of 29 genes related to mitochondrial malfunction and oxidative phosphorylation may explain the cell growth inhibition (supplemental data). Finally, in a finding (data not shown) that may indicate altered metabolism, it should also be noted that siOGA cell cultures displayed enhanced medium acidification even at low cell confluency.
The altered metabolism typically exhibited by cancer cells includes enhanced glycolysis, regardless of oxygen availability (i.e. “the Warburg effect”), and de novo fatty acid (FA) synthesis. Increased aerobic glycolysis facilitates the heightened use of available glucose and the diversion of glycolytic intermediates for the biosynthesis of macromolecules, i.e. lipids, proteins, and nucleic acids, to support increased proliferation. FAs are mainly used for membrane synthesis, and the regulation of oncoprotein transcription, transport, secretion, and activity (through signaling or as post-translational modifications) (41). In this study, microarray analysis revealed extensive metabolic changes in the OGA-silenced clone (Figs. 4 and 6 and Tables 1 and 2). The most prominent metabolic changes were elevated lipid pathway activity and altered carbohydrate pathways, specifically the PPP. Plausibly, the high cellular O-GlcNAc level indicates a high energetic level and the presence of hexosamine biosynthetic pathway intermediate products, thus activating the alternative pathway to glycolysis, the PPP. As a result, NADPH production for FA synthesis, ribose 5-phosphate production for nucleotide synthesis, and the formation of aromatic amino acids would be elevated. Indeed, the expression of Glucose-6-phosphate dehyrogenase, the first PPP enzyme, is elevated in OGA-silenced cells (Table 2). Additionally, the fructose metabolic pathway, in which most genes were up-regulated (Fig. 4B and Tables 1 and 2), produces trioses used for glycogen and FA synthesis or for glycolysis. The up-regulation of the gene of aldolase, which catalyzes glyceraldehyde-3-phosphate production, suggests that the previous pathway is used to “re-fuel” glycolysis. It should be noted that because O-GlcNAcylation is closely associated with cellular metabolism (6), the change of some metabolic genes in siOGA cells might have been the result of a compensatory effect. Nonetheless, other changes in siOGA cells (e.g. in their morphology and polyploidy) are less likely to have been caused by such an effect alone.
The wide ranging effects of OGA silencing on gene expression reflect its global effect on cellular proteins, specifically transcription factors (2), but it may also be related to the association of OGA with promoters and with microRNA that regulates various cellular processes (42), including EMT (43). In Caenorhabditis elegans, knock-outs of oga and ogt altered the life span and the expression of genes involved in aging, stress response, innate immunity, carbohydrate/lipid metabolism, amino acid metabolism, mitochondrial activity, and transcriptional regulation, among others (42).
Accumulating data suggest that metabolism and proliferation share regulatory pathways in cancer cells through various transcription and apoptotic factors, signaling molecules, oncogenes, and tumor suppressors (reviewed in Ref. 41). Two key pathways are those of p53 (controlling cell cycle and apoptosis and implicated in glycolytic and oxidative metabolism) and Akt (functioning in glucose, protein, and lipid metabolism and in cell proliferation and survival). In this study, genes encoding proteins of versatile pathways, e.g. PPARγ, HMG-CoA synthase, and reductase, were up-regulated in OGA-silenced cells, while genes of the Akt1 substrate 1, CPT1A, AIF1, AIF2, and p53 were down-regulated. In fact, the p53 signaling canonical pathway and various p53-associated proteins were significantly altered following OGA silencing (see Figs. 4 and 6 and supplemental data). Moreover, E2F1, the PPARγ coactivators 1α, Sp1, Sp3, c-Myc, AIF1, Akt, and p53 are all modified by O-GlcNAc (4, 36). Thus, O-GlcNAcylation seems to serve as a link that coordinates the metabolic response and cancer cell proliferation.
This is the first time the effect of OGA silencing on wide ranging gene expression in cancer cells has been shown. The dramatic change observed, of more than 1300 genes, indicates the central role of O-GlcNAcylation in the mediation of cellular processes, specifically metabolism and tumorigenic functions. The current findings indicate a link between O-GlcNAcylation and EMT in CRC clones. The increased O-GlcNAcylation, characteristic of cancer cells, induces the insulin resistance associated with type II diabetes (44). Thus, O-GlcNAcylation was previously proposed as a metabolic link between diabetes and cancer (6). In addition to supporting this notion, our findings of extensive carbohydrate and lipid metabolic changes following OGA silencing provide a possible link between obesity and cancer. Although a body of clinical evidence links lipid biosynthesis, obesity, and cancer development, the molecular mechanism underlying this linkage is unknown. The effect of OGA silencing on steroidogenesis and adipogenesis, concomitantly with cancer-related functions, provides a clue for a possible mechanism. The metastatic potential of the OGA-silenced clone will be further investigated by an in vivo xenograft assay.
Supplementary Material
Acknowledgments
We gratefully thank Prof. Allan Witztum and Inez Mureinik for styling the manuscript. We thank Dr. Vered Caspi and team (Bioinformatics Core Facility, National Institute of Biotechnology in the Negev, Ben-Gurion University) for analyzing the microarray data.
This work was supported by grants from the Israel Cancer Association and Israel Science Foundation Grant 1287/08.

This article contains supplemental Figs. S1–S13.
- O-GlcNAcylation
- O-linked β-N-acetylglucosaminylation
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- CRC
- colorectal cancer
- ECM
- extracellular matrix
- IPA
- ingenuity pathway analysis
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- OGA
- N-acetyl-β-d-glucosaminidase
- OGT
- O-GlcNAc transferase
- PARP
- anti-poly(ADP-ribose) polymerase
- PPP
- pentose phosphate pathway
- qRT-PCR
- quantitative real time reverse transcription-PCR
- TMG
- thiamet-G
- FA
- fatty acid.
REFERENCES
- 1. Zeidan Q., Hart G. W. (2010) The intersections between O-GlcNAcylation and phosphorylation. Implications for multiple signaling pathways. J. Cell Sci. 123, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zachara N. E., Hart G. W. (2002) The emerging significance of O-GlcNAc in cellular regulation. Chem. Rev. 102, 431–438 [DOI] [PubMed] [Google Scholar]
- 3. Butkinaree C., Park K., Hart G. W. (2010) O-Linked β-N-acetylglucosamine (O-GlcNAc). Extensive cross-talk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 [DOI] [PubMed] [Google Scholar]
- 5. Hart G. W. (1997) Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 66, 315–335 [DOI] [PubMed] [Google Scholar]
- 6. Slawson C., Copeland R. J., Hart G. W. (2010) O-GlcNAc signaling. A metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., Reginato M. J. (2010) Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29, 2831–2842 [DOI] [PubMed] [Google Scholar]
- 8. Wice B. M., Trugnan G., Pinto M., Rousset M., Chevalier G., Dussaulx E., Lacroix B., Zweibaum A. (1985) The intracellular accumulation of UDP-N-acetylhexosamines is concomitant with the inability of human colon cancer cells to differentiate. J. Biol. Chem. 260, 139–146 [PubMed] [Google Scholar]
- 9. Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. (2005) Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 10. Mi W., Gu Y., Han C., Liu H., Fan Q., Zhang X., Cong Q., Yu W. (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 1812, 514–519 [DOI] [PubMed] [Google Scholar]
- 11. Gu Y., Ande S. R., Mishra S. (2011) Altered O-GlcNAc modification and phosphorylation of mitochondrial proteins in myoblast cells exposed to high glucose. Arch. Biochem. Biophys. 505, 98–104 [DOI] [PubMed] [Google Scholar]
- 12. Krzeslak A., Forma E., Bernaciak M., Romanowicz H., Brys M. (2012) Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin. Exp. Med. 12, 61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hollstein M., Rice K., Greenblatt M. S., Soussi T., Fuchs R., Sørlie T., Hovig E., Smith-Sørensen B., Montesano R., Harris C. C. (1994) Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22, 3551–3555 [PMC free article] [PubMed] [Google Scholar]
- 14. Munro A. J., Lain S., Lane D. P. (2005) p53 abnormalities and outcomes in colorectal cancer. A systematic review. Br. J. Cancer 92, 434–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 16. Chou T. Y., Dang C. V., Hart G. W. (1995) Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 92, 4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou T. Y., Hart G. W., Dang C. V. (1995) c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 270, 18961–18965 [DOI] [PubMed] [Google Scholar]
- 18. Potter J. D. (1999) Colorectal cancer. Molecules and populations. J. Natl. Cancer Inst. 91, 916–932 [DOI] [PubMed] [Google Scholar]
- 19. Zhu Q., Zhou L., Yang Z., Lai M., Xie H., Wu L., Xing C., Zhang F., Zheng S. (2012) O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med. Oncol. 29, 985–993 [DOI] [PubMed] [Google Scholar]
- 20. Mandhal N. (1992) in Human Cytogenetics (Rooney D. E., Czepulkowsky B. H., eds) 2nd Ed., pp. 155–188, Oxford University Press, New York [Google Scholar]
- 21. Shaffer L. G., International Standing Committee on Human Cytogenetic, N., Slovak M. L., Campbell L. J. (2009) ISCN 2009: An International System for Human Cytogenetic Nomenclature, Karger, Basel, Switzerland [Google Scholar]
- 22. Gavert N., Ben-Shmuel A., Lemmon V., Brabletz T., Ben-Ze'ev A. (2010) Nuclear factor-κB signaling and ezrin are essential for L1-mediated metastasis of colon cancer cells. J. Cell Sci. 123, 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 24. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 25. Hewitt R. E., McMarlin A., Kleiner D., Wersto R., Martin P., Tsokos M., Stamp G. W., Stetler-Stevenson W. G. (2000) Validation of a model of colon cancer progression. J. Pathol. 192, 446–454 [DOI] [PubMed] [Google Scholar]
- 26. Wai P., Reddy S., Kuo P. (2008) Functional analysis of tumor metastasis: modeling colon cancer. Oncol. Rev. 2, 9–20 [Google Scholar]
- 27. Zirvi K. A., Keogh J. P., Slomiany A., Slomiany B. L. (1991) Transglutaminase activity in human colorectal carcinomas of differing metastatic potential. Cancer Lett. 60, 85–92 [DOI] [PubMed] [Google Scholar]
- 28. Katayama M., Nakano H., Ishiuchi A., Wu W., Oshima R., Sakurai J., Nishikawa H., Yamaguchi S., Otsubo T. (2006) Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg. Today 36, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 29. Duranton B., Holl V., Schneider Y., Carnesecchi S., Gossé F., Raul F., Seiler N. (2003) Polyamine metabolism in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). Amino Acids 24, 63–72 [DOI] [PubMed] [Google Scholar]
- 30. Provenzani A., Fronza R., Loreni F., Pascale A., Amadio M., Quattrone A. (2006) Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis 27, 1323–1333 [DOI] [PubMed] [Google Scholar]
- 31. Olivier S., Guinez C., Mir A. M., Perez-Cervera Y., Liu C., Michalski J. C., Lefebvre T. (2012) The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 302, E417–E424 [DOI] [PubMed] [Google Scholar]
- 32. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Extensive cross-talk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 34. Bogenrieder T., Herlyn M. (2003) Axis of evil. Molecular mechanisms of cancer metastasis. Oncogene 22, 6524–6536 [DOI] [PubMed] [Google Scholar]
- 35. Shankar J., Messenberg A., Chan J., Underhill T. M., Foster L. J., Nabi I. R. (2010) Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 70, 3780–3790 [DOI] [PubMed] [Google Scholar]
- 36. Zachara N. E., Hart G. W. (2006) Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761, 599–617 [DOI] [PubMed] [Google Scholar]
- 37. Buck E., Eyzaguirre A., Barr S., Thompson S., Sennello R., Young D., Iwata K. K., Gibson N. W., Cagnoni P., Haley J. D. (2007) Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol. Cancer Ther. 6, 532–541 [DOI] [PubMed] [Google Scholar]
- 38. Zhu W., Leber B., Andrews D. W. (2001) Cytoplasmic O-glycosylation prevents cell surface transport of E-cadherin during apoptosis. EMBO J. 20, 5999–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sayat R., Leber B., Grubac V., Wiltshire L., Persad S. (2008) O-GlcNAc-glycosylation of β-catenin regulates its nuclear localization and transcriptional activity. Exp. Cell Res. 314, 2774–2787 [DOI] [PubMed] [Google Scholar]
- 40. Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., Kim K. T., Ryu S. H., Suh P. G. (2012) O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 [DOI] [PubMed] [Google Scholar]
- 41. Fritz V., Fajas L. (2010) Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene 29, 4369–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Love D. C., Ghosh S., Mondoux M. A., Fukushige T., Wang P., Wilson M. A., Iser W. B., Wolkow C. A., Krause M. W., Hanover J. A. (2010) Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vosseller K., Wells L., Lane M. D., Hart G. W. (2002) Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.