Background: The CFTR chloride channel undergoes conformational changes during its gating cycle.
Results: H620Q mutation associated with increased channel Po, and the corrector/potentiator CFFT-001 both lead to similar conformational shifts in NBD1.
Conclusion: There is an intrinsic conformational equilibrium within NBD1 that is correlated with channel activity.
Significance: Conformational fluctuations within NBD1 are fundamental to CFTR regulation.
Keywords: ABC Transporter, CFTR, Drug Action, Nuclear Magnetic Resonance, Structural Biology, F508del, NBD1, Conformational Changes, Cystic Fibrosis, Folding Energetics
Abstract
Deletion of Phe-508 (F508del) in the first nucleotide binding domain (NBD1) of the cystic fibrosis transmembrane conductance regulator (CFTR) leads to defects in folding and channel gating. NMR data on human F508del NBD1 indicate that an H620Q mutant, shown to increase channel open probability, and the dual corrector/potentiator CFFT-001 similarly disrupt interactions between β-strands S3, S9, and S10 and the C-terminal helices H8 and H9, shifting a preexisting conformational equilibrium from helix to coil. CFFT-001 appears to interact with β-strands S3/S9/S10, consistent with docking simulations. Decreases in Tm from differential scanning calorimetry with H620Q or CFFT-001 suggest direct compound binding to a less thermostable state of NBD1. We hypothesize that, in full-length CFTR, shifting the conformational equilibrium to reduce H8/H9 interactions with the uniquely conserved strands S9/S10 facilitates release of the regulatory region from the NBD dimerization interface to promote dimerization and thereby increase channel open probability. These studies enabled by our NMR assignments for F508del NBD1 provide a window into the conformational fluctuations within CFTR that may regulate function and contribute to folding energetics.
Introduction
Cystic fibrosis (CF)4 is a genetic disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (1–3). CFTR encodes a 1480-residue integral membrane protein that functions as a chloride channel. As a member of the ABC transporter superfamily of proteins, CFTR is organized into two repeated units, each comprised of a membrane-spanning domain (MSD1 and 2) and a cytosolic nucleotide binding domain (NBD1 and -2) (supplemental Fig. S1). A large intrinsically disordered regulatory (R) region, unique to CFTR, connects NBD1 to MSD2. The first ∼30 residues of the R region found immediately C-terminal to NBD1 is called the regulatory extension (RE). Intracellular domains are formed by cytosolic extensions of the transmembrane helices and bridge the MSDs and the NBDs. Crystal structures of NBD1 (4), including the F508del variant, show a well conserved conformation of NBD1 that consists of three subdomains: the α-subdomain (the site of Phe-508), the β-subdomain (containing three anti-parallel β-strands), and a central ATP-binding core (Fig. 1B). A 34-residue disordered segment, called the regulatory insertion (RI), also unique to CFTR, is found between the first two β-strands of NBD1 (supplemental Fig. S1). The ATP-binding core is dominated by a central β-sheet (comprising strands S6, S7, S8, S3, S9, and S10). Two short helices, H8 and H9, at the C terminus of this NBD1 construct are arguably not part of the core NBD1, but can fold onto strands S3, S9, and S10 of this sheet (see Fig. 1B). Opening and closing of the CFTR channel involves phosphorylation of the R region and the RI by protein kinase A (5) (and potentially protein kinase C (6) and AMP kinase (7)), in combination with ATP binding and hydrolysis at the interface between NBD1 and NBD2. During the gating cycle, ATP molecules bind at the interface of NBD1 and NBD2, as shown in bacterial NBD homodimers (8). Conformational changes in the NBD1/NBD2 heterodimer are thought to be influenced by phosphorylation of the R region and RI (9, 10). It has been proposed that these ATP- and dimerization-induced conformational changes are relayed to the MSDs via the intracellular domains leading to pore opening and closing (10–12).
FIGURE 1.
Assignment of F508del NBD1 ΔRIΔRE. A, overlay of WT and F508del NBD1 15N-1H correlation spectra at 500 MHz. B, ribbon diagram of WT NBD1 ΔRIΔRE (PDB code 2PZE). The α- and β-subdomains and the ATP binding core are shown in blue, green, and gray, respectively. The C-terminal helices H8 and H9 are labeled. Red spheres represent the N atoms from residues not assigned in either WT or F508del (three red spheres representing residues Glu-391, Thr-390, and Pro-638 are obstructed from view). Yellow spheres are N atoms of residues not assigned in F508del. The orange sphere is the N atom of Phe-508. ATP is shown in cyan. The deletion site for the RI is indicated. C, using the same color coding as in B, these residues are shown in the amino acid sequence. In total, 82% of F508del NBD1 ΔRIΔRE has been assigned.
The most common and severe CF-causing mutation in the CFTR gene is deletion of Phe-508 (F508del) in NBD1 (13). The F508del mutation further exacerbates the normally inefficient folding and processing of CFTR, thereby targeting it to the endoplasmic reticulum-associated protein degradation pathway (14). Low temperature (15), glycerol addition (16), introduction of revertant (17) and solubilizing (18) mutations, and removal of the RI (19) can reduce this misprocessing and degradation, such that a small population of F508del CFTR molecules can escape the quality control mechanisms of the cell and traffic to the plasma membrane. However, once at the plasma membrane, these F508del CFTR molecules show a decreased open probability (20) and instability (21), eventually leading to endocytosis and degradation.
As potential therapies for CF, small-molecule compounds have been sought to address the underlying defects of F508del and G551D, a mutation that impairs gating. Pharmacological agents called “corrector” compounds aim to overcome processing/folding defects, allowing mutated CFTR to escape cellular degradation and increasing the number of CFTR molecules at the plasma membrane. Compounds may achieve this by enhancing folding, increasing stability, mimicking or affecting the levels of a molecular chaperone, or interrupting an interaction with the degradation machinery of the cell. A corrector compound, VX-809, has been shown to restore F508del CFTR processing in primary human bronchial epithelial cells isolated from patients homozygous for F508del (22, 23). “Potentiator” compounds increase the open probability of CFTR channels already transported to the plasma membrane. The compound VX-770 is a potentiator of G551D (24) and F508del-CFTR (25), as well as several other gating mutations (26, 27).
There has been success in the identification and discovery of correctors, potentiators, and dual corrector/potentiators (28), but their mechanisms of action on a molecular level including potential direct interactions with CFTR have not been characterized. Even in the cases of compounds for which physiological and biochemical data suggest a direct interaction with CFTR, the domain of the CFTR molecule to which particular compounds bind is not known. Such structural level data would not only give information about how a compound functions but also facilitate the design of improved therapeutic compounds. Details on the conformational effects of binding of compounds can also provide insights into the dynamic properties and the energetic landscape of the CFTR protein, known to be critical for channel function and folding (29, 30) A limitation, however, for these types of studies lies in the relatively poor solubility of most known CFTR modulator compounds in aqueous solution.
To probe the conformational effects of mutations and binding of small molecule modulators, we have applied a range of biophysical approaches to study H620Q and helix H9 variants of NBD1 and the interaction of the most soluble compound of a series of dual corrector/potentiator compounds provided by the Cystic Fibrosis Foundation, N-cyclohexyl-4-(6-methyl-3-pyridinyl) pyrimidine-2-amine, referred to here as CFFT-001. NMR backbone resonance assignments (82%) carried out on a human F508del NBD1 lacking the RI and ending at residue 646 (F508del NBD1 ΔRIΔRE) enable us to define conformational changes due to the H620Q mutation. We also present electrophysiology data demonstrating the dual corrector/potentiator activity of CFFT-001 for F508del CFTR, consistent with it interacting directly with CFTR, possibly at NBD1, the domain harboring the F508del mutation. Addition of CFFT-001 to F508del NBD1 ΔRIΔRE results in NBD1 conformational changes overlapping with those observed for the H620Q mutant protein that has higher open channel probability. Differential scanning calorimetry (DSC) data show a reduction in NBD1 thermal melting temperature for H620Q or in the presence of CFFT-001, demonstrating direct binding of the compound to a less thermostable conformation. Docking of the compound onto higher energy conformations of NBD1 accessed in replica exchange simulations describes possible compound binding modes consistent with the NMR and DSC data. Together, these orthogonal methods provide evidence for mutation-induced or compound-induced conformational changes within NBD1 and generate insights into the motional and energetic properties of CFTR relevant for channel activity and folding.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification (NMR and DSC Studies)
Human NBD1 (387–646, Δ405–436) constructs with or without Phe-508 and containing the H620Q mutation or deletion of helix H9(636–646) were expressed as His6-SUMO fusions at 16 °C in BL21(DE3) Codon Plus cells grown in minimal media with [15N]NH4Cl and/or [13C]glucose, for NMR studies, or LB for DSC studies, and purified as described previously (31, 32). Purified proteins were stored at 4 °C in buffer containing 12.5% glycerol before being exchanged into 50 mm sodium phosphate, pH 7.5, 150 mm NaCl, 5 mm ATP/MgCl2, 2% (v/v) glycerol, 2 mm DTT, 10% D2O with 0.05% sodium azide and 0.5% benzamidine for NMR or into 150 mm NaCl, 20 mm HEPES, pH 7.5, 10% glycerol, 10% ethylene glycol, 1 mm Tris(2-carboxymethyl)phosphine, 2 mm ATP, and 3 mm MgCl2 for DSC. The NMR buffer has been optimized for both stability and spectral line width, precluding the use of higher concentrations of glycerol and ethylene glycol used in the DSC buffer, which has been optimized for stability alone.
NMR Spectroscopy
All NMR experiments were carried out at 20 °C on a Varian 500 or 600 MHz spectrometer equipped with pulsed field gradients and either a triple resonance room temperature or cryo-probe, respectively. Data were processed and analyzed using NMRDraw (33), NMRView (34), and Sparky (35). Assignments were obtained using data from HNCO, HNCA, and HN(CO)CA experiments (36–38) as well as WT assignments.5 Combined N and H chemical shift changes in Hz were calculated using Equation 1.
 |
Origin of CFFT-001
The compound N-cyclohexyl-4-(6-methyl-3-pyridinyl) pyrimidine-2-amine (CFFT-001, supplemental Fig. S2) was synthesized originally in the CFTR modulator drug discovery program of EPIX Pharmaceuticals, Inc. A pharmacophore model was generated based on the previously identified corrector classes 1, 2, 3, and 5 (represented by corr-1c, corr-2b, corr-3c, and corr-5c) (39). The pharmacophore model generated with Catalyst software (Accelrys, San Diego, CA) also included a shape constraint to improve the spatial overlap between screened compounds and reference correctors. An EPIX in-house data base of ∼4 million unique commercially available compounds was filtered using a set of one-dimensional and two-dimensional chemical property descriptors derived from the selected compound classes, resulting in a focused library of ∼740,000 compounds. Screened compounds matching the pharmacophore model were subsequently filtered by the Catalyst fitness score. The score cutoff was selected by enrichment analysis and cutoff application reduced the library size to 2300 compounds. The remaining compounds were subsequently clustered by molecular similarity, and cluster representatives were selected based on the Catalyst fitness score. The structure activity relationship trends, which may be concluded from the reference compound data but could not be incorporated into the pharmacophore hypothesis for technical reasons, were used to prioritize compounds, as well as chemical novelty. 85 compounds were selected, purchased, and sent for biological testing, one of which was found to be weakly active in an FRT (Fischer rat thyroid) cell Ussing chamber assay. The synthesis of non-commercial analogs led to the discovery of CFFT-001 (68), an early member of the EPIX chemical series of dual-acting corrector/potentiator compounds.
Ussing Chamber Analysis
Base line-corrected short circuit current traces were analyzed from Ussing chamber recordings of transepithelial currents across monolayers of FRT cells that stably express F508del. Although all compound incubation was performed at 37 °C, the recordings were carried out at 27 °C, as described previously (40). This temperature was chosen to decrease the run-down of corrector effect by reducing internalization of CFTR. Cells were grown on Snapwell filters (Corning catalog no. 3801, Corning, NY), incubated for 24 h with negative control (0.3% DMSO), 10 or 30 μm CFFT-001, or 10 μm positive control C18 (69) (supplemental Fig. S2), which is distributed by the CF Foundation. Snapwell filters were then inserted into Physiologic Instruments Ussing chambers (Physiologic Instruments, Inc., San Diego, CA) for at least 20 min in 10 ml of buffer solution before CFTR was activated with forskolin.
Simulations and Docking
The crystal structure of F508del NBD1ΔRIΔRE (Protein Data Bank code 2PZF) (40), comprising residues 387–646 (Δ405–436), was prepared with Discovery Studio (41). Replica Exchange Molecular Dynamics (REMD) simulations were done using the Gromacs Molecular Dynamics package (42, 43) with the AMBER99SB-ILDN force field (44). The protein plus its bound ATP was submerged in TIP4P water in a rhombic dodecahedral box with an extra extension along each axis of the protein of 10 Å. Ions were added to the solution to make the system electrically neutral. The structure was minimized and equilibrated, first under NVT conditions for 100 ps and then under NPT conditions for an additional 100 ps. The REMD production phase was carried out under NPT conditions with a time step of 2 fs. REMD simulations consisted of 10 replicas covering a temperature range of 300 K to 313.6 K. The simulation of each replica was run for 2 ns. The temperature range was selected to give the same acceptance probability between all adjacent pairs over the entire temperature range based on known energy distributions for solvated proteins (45). The simulation was run with the leap-frog algorithm (46). Long range electrostatic interactions were computed using Particle Mesh Ewald electrostatics (47, 48). The cut-off for van der Waals and Coulomb interactions was set to 10 Å. Periodic boundary conditions were applied. The LINCS algorithm (49) was used to constrain bond lengths. The above procedure was repeated three times starting from different random seeds.
Selected conformations from the simulation, particularly those having frayed H8 and H9 helices and long distances between His-620 and Phe-640, were searched for potential binding sites based on the receptor cavities approach within Discovery Studio (41). Docking simulations of CFFT-001 to the potential binding site between S3/S9/S10 and H8/H9 in the different NBD1 conformations from the REMD were conducted using the flexible docking procedure (50) of Discovery Studio or using the induced fit protocol of Schrodinger (51–53). Flexible residues were selected to be those facing the binding site.
Differential Scanning Calorimetry
Protein samples of 0.5 ml were diluted to 15 ml in DSC buffer and concentrated to 0.5 ml at 4 °C using an Amicon Ultra-15 (10K) centrifugal filter. The procedure was repeated three times. CFFT-001 in 100% DMSO was added to the protein sample to create a 5% DMSO solution at the appropriate compound concentration. DSC was conducted with a VPCapillary DSC System (MicroCal Inc., GE Healthcare) using a scan rate of 2 K/min. A buffer-only heat capacity curve was subtracted from the protein curve, and data were analyzed with the instrument provided software. Repeated measurements for the H620Q F508del sample gave identical Tm values to the first decimal place; the general reproducibility of NBD1 Tm values is within ±0.3 °C.
Sequence Alignment
ABC family C sequences were collected with BLAST (54, 55) using one representative sequence for each of the 12 human ABC family C members (UniProt IDs P33527, Q92887, O15438, O15439, O15440, O95255, Q09428, O60706, Q5T3U5, Q96J66, and Q96J65), comparing them against the default non-redundant database (Aug. 18, 2011) using an E threshold of 1e− 200. The top 200 hits were retrieved, and for the non-CFTR targets, a match to the desired member was ensured by restricting to FASTA files containing any of the following keywords: “member N,” “protein N,” “mrp-N,” “MRP-N,” “mrpN,” or “MRPN,” where N is the member number of the target. All sequences were then filtered by manually removing duplicate sequences at 100% identity, truncated isoforms, sequences containing unsequenced residues (X), and sequences with less than 1400 residues. A total of 294 sequences were obtained, with 80 from CFTR. NBD1 sequences were isolated by aligning the full-length sequences in MUSCLE and extracting the alignment spanning human CFTR residues 388 to 680. These sequences were then realigned using MUSCLE. Sequence profiles were calculated for each of the 12 members by taking amino acid and gap frequencies for each position in the alignment.
Highly conserved residues in CFTR were defined by a 95% sequence identity threshold, and their specificity to CFTR was measured by measuring the frequency of the CFTR conserved residue in each of the other 11 family C members. A member was considered a potential match when it has 10% or greater identity to CFTR at that position.
RESULTS
Assignment of F508del Human NBD1 ΔRIΔRE
NMR studies were performed on F508del NBD1 lacking the RI and ending at residue 646 (NBD1 ΔRIΔRE: 387–646, Δ405–436) because of its enhanced solubility and stability (40). NBD1 ΔRIΔRE that is otherwise wild-type (referred to as WT) or additionally contains F508del can be concentrated to ∼1 mm and remains stable at 4 °C for several weeks. HSQC spectra of WT and F508del NBD1 ΔRIΔRE are overlaid in Fig. 1A. In agreement with crystallographic results (40), the similarity of the two spectra indicates that both share the same overall structure. However, differences in peak intensity within the individual spectra reflect significant conformational heterogeneity in both the WT and the F508del NBD1 ΔRIΔRE domains. WT assignments5 helped facilitate 82% assignment of backbone resonances for F508del NBD1 ΔRIΔRE. Unassigned residues were mapped onto the NBD1 structure and sequence (Fig. 1, B and C). Many of the unassigned residues are located in loops connecting helical segments. The inability to assign them is likely due to the dynamic nature of these loops leading to broadening of resonances.
H620Q Variant Reduces Helicity of H8 and H9 at the C Terminus of F508del NBD1 ΔRIΔRE
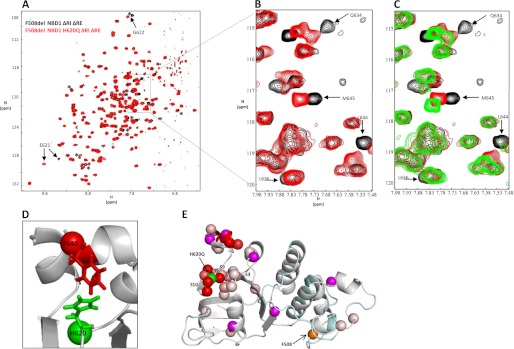
To address possible conformational changes within NBD1 that may be relevant to channel activity and/or misfolding, we focused on the C-terminal β-strands (S9 and S10) which have been shown to affect processing, activity and pharmacology of CFTR (56). In particular, an H620Q variant (in S9) originally identified in CF patients has an increased Po in single channels (56–58). Fig. 2, A and B, presents an overlay of NMR spectra for F508del NBD1 ΔRIΔRE and its H620Q variant (F508del H620Q NBD1 ΔRIΔRE). Arrows indicate a subset of peaks that shift upon H620Q mutation, including Glu-621, Gly-622, Gln-634, Leu-636, Ser-641, Leu-644, and Met-645. Mapping of the chemical shift changes (supplemental Fig. S3A) onto a ribbon diagram of the structure of WT NBD1 ΔRIΔRE (Fig. 2E) shows that in addition to effects adjacent to His-620 (Glu-621, Gly-622) there are changes in strands S3, S9, and S10. There are also significant peak shifts mapping to residues on the two short C-terminal helices (H8 and H9) that are immediately N-terminal to residues of the RE/R region in full-length CFTR. Strikingly, residues on both surfaces of these helices are affected with five of six of the H9 resonances perturbed in the variant. The general direction of the chemical shift changes (toward the center of the amide proton chemical shift range at ∼8.0 ppm) in H8 and H9 is consistent with a conformational equilibrium shift toward more disorder (supplemental Fig. S4A). Chemical shifts in the absence of the mutation demonstrate that these helices are in equilibrium with a coil conformation (supplemental Fig. S4C) (59) and the chemical shift perturbations for the residues of the H8/H9 helices upon replacing His-620 with a Gln reflect a change in the equilibrium further toward the coil conformation. The chemical shift perturbations expected for a full helix-to-coil transition are much larger than the observed shifts, indicating a small shift in the equilibrium. An examination of the structure reveals that the His-620 side chain in S9 interacts with the Phe-640 side chain in H9 (Fig. 2D). We propose that the helices are stabilized by binding to residues on strands S3, S9, and S10 and that mutation of His-620 disrupts the interaction between the surface of the β-strands and H8/H9, reducing the helical structure.
FIGURE 2.
H620Q variant reduces helicity of H8 and H9 at the C terminus of F508del NBD1 ΔRIΔRE. A, overlay of 15N-1H correlation spectra at 500 MHz for F508del NBD1 ΔRIΔRE (black; background) and the H620Q variant, F508del H620Q NBD1 ΔRIΔRE (red; foreground). Arrows indicate a subset of peaks that shift in the mutant. Asterisks show a separate subset of peaks that shift in the H620Q variant but do not shift upon addition of CFFT-001. B, close-up of boxed area of A. C, same overlay as in B, with a third layer showing the addition of compound to the H620Q variant (green; foreground). D, side chain of His-620 interacting with the side chain of Phe-640. E, ribbon diagram of WT NBD1 ΔRIΔRE (2PZE) with unassigned residues in cyan, and assigned residues that do not shift upon H620Q mutation are shown in light gray. The N atoms for residues that show chemical shift changes upon mutation are shown as spheres colored with a linear gradient from light pink to red, where light pink corresponds to the smallest shifts (beginning at 7 Hz; see supplemental Fig. S3) and red to the largest shifts. The N atoms for residues whose chemical shift changes upon mutation yet cannot be identified with certainty are shown as magenta spheres. His-620 and Phe-508 are shown as green and orange spheres, respectively.
Dual Corrector/Potentiator Activity of CFFT-001
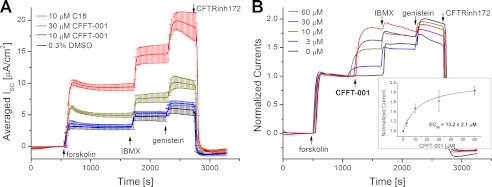
To probe the conformational effects of binding of small molecule CFTR modulators, we focused on the dual corrector/potentiator N-cyclohexyl-4-(6-methyl-3-pyridinyl) pyrimidine-2-amine (CFFT-001, supplemental Fig. S2) that was originally synthesized in the CFTR modulator drug discovery program of EPIX Pharmaceuticals, Inc. funded by the Cystic Fibrosis Foundation Therapeutics (Bethesda, MD). The electrophysiological data in Fig. 3 illustrate the dual acting properties of CFFT-001. After CFTR was maximally activated by subsequent additions of 10 μm forskolin, 100 μm IBMX, and 20 μm genistein, cells incubated for 24 h with 30 μm CFFT-001 showed a robust peak current increase of 68% compared with control (Fig. 3A), pointing to an increase in membrane-localized channels and illustrating the corrector activity of the compound.
FIGURE 3.
The dual-acting corrector-potentiator activity of CFFT-001. A, FRT cells stably expressing CFTR F508del were incubated for 24 h at 37 °C with either 0.3% DMSO, 10 or 30 μm CFFT-001, or 10 μm C18 before short circuit currents were recorded in Ussing chambers at 27 °C. The traces are base line-corrected averages of three individual recordings, except for the C18 positive control (n = 2). Error bars indicate the S.D. of the mean trace value. After a 20-min equilibration period and base-line acquisition, CFTR was activated maximally by additions of 10 μm forskolin, 100 μm IBMX, and 20 μm genistein prior to inhibition by 20 μm CFTR inhibitor 172 (CFTRinh172). Current increases due to CFFT-001 incubation are significant with p = 0.0059 (unpaired t test). B, current traces are averages of short circuit traces from cells that were incubated for 24 h with DMSO. Currents were normalized to the forskolin-elicited current (n = 3). After the forskolin-induced current stabilized, CFFT-001 was added to final concentrations of 0, 3, 10, 30, and 60 μm. Differences in the peak currents after all agonist additions, including 100 μm IBMX and 20 μm genistein, are not statistically significant. The inset shows the normalized current increases plotted against the corresponding dose of CFFT-001. Error bars are the S.D. of the mean current increases. The data were fitted with a Hill function (nH = 1, R2 = 0.996), which yielded an EC50 concentration of 13.2 ± 2.1 μm.
After activation of CFTR with 10 μm forskolin, acute addition of CFFT-001 to 0, 3, 10, 30, or 60 μm final concentrations in the half-chamber bathing the apical membrane of the FRT cells leads to a clear dose-dependent current increase (Fig. 3B), as expected for a CFTR potentiator. Consistent with the concept of an absolute biophysical limit for the maximal open probability, the additional current increases due to subsequent additions of 100 μm IBMX and 20 μm genistein have the inverse rank order of the CFFT-001-mediated effect: the IBMX/genistein response is largest without any acute addition of CFFT-001 and smallest for the highest dose of the compound. It should be noted that the >60% increase in inhibitor-sensitive current over the DMSO control condition observed after 24 h of incubation with 30 μm CFFT-001 (Fig. 3A) is not seen after acute addition of the compound (Fig. 3B), clearly distinguishing the corrector and potentiator activities of CFFT-001 in these assays.
CFFT-001 Compound Reduces Helicity of H8 and H9 in F508del NBD1 ΔRIΔRE
To determine whether the dual corrector/potentiator CFFT-001 compound binds to F508del NBD1 ΔRIΔRE and to monitor any conformational changes that may result, we recorded NMR spectra following several titration points of CFFT-001. Spectra were obtained after addition of each of several aliquots of CFFT-001 (1:1, 2:1, and 3:1, compound:protein) to generate a series of spectra representing changes as a function of apparent compound concentration up to 750:250 μm of CFFT-001:NBD1. Due to the relatively low aqueous solubility of CFFT-001 (∼30 μm), we have not saturated the complex, and our stated compound concentrations are likely to be significantly over-estimated, consistent with expectations from the 13 μm EC50 for potentiation. Titrations beyond ∼3:1 (apparent compound:protein concentrations) requiring >750 μm apparent compound concentrations were not performed due to precipitation of CFFT-001 in the aqueous NMR buffer (only including up to 1.5% DMSO to minimize protein destabilization due to this organic solvent).
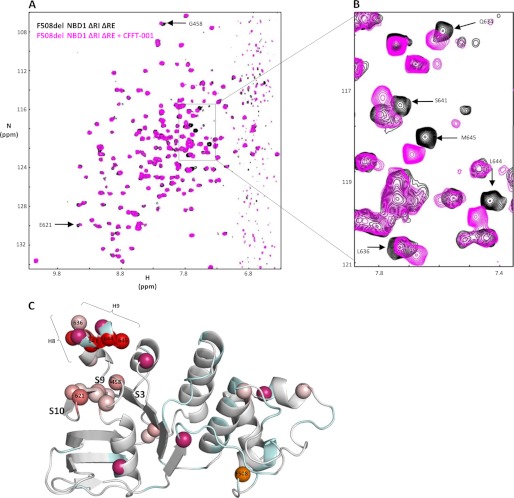
Fig. 4, A and B, shows a spectral overlay for F508del NBD1 ΔRIΔRE in the apo state and after the final CFFT-001 titration point, with arrows pointing to a subset of peaks that shift. Comparison of this set of peaks with a subset of those resulting from the H620Q mutation (compare Fig. 2 with Fig. 4) shows significant similarities, including both the identity of perturbed resonances and the direction of the chemical shift changes. These similarities are consistent with a common effect of the H620Q substitution and CFFT-001 on H8 and H9, although additional peak shifts reflecting other conformational changes in the mutant are present as well (Fig. 2A). To account for small shifts caused by the compound solvent DMSO, chemical shifts in the presence of DMSO at appropriate concentrations were subtracted from chemical shift changes due to CFFT-001. Mapping of the resulting chemical shift changes (Fig. 4C) onto a ribbon diagram of the structure of WT NBD1 ΔRIΔRE clearly shows that the greatest peak shifts map to residues on helices H8 and H9. Similar to the H620Q variant, residues on both surfaces of these helices are affected by CFFT-001, as opposed to one surface of the helix as one might expect for a direct drug interaction with the helix. The linearity of the size of peak shifts as a function of added CFFT-001 indicates equilibrium between two states that are in fast exchange on the NMR timescale, with addition of the compound shifting the equilibrium to one of the states. The direction (toward the center of the amide proton chemical shift range at ∼8.0 ppm) of most of the amide proton chemical shift changes is consistent with a conformational equilibrium shift toward coil, shifting the underlying H8/H9 helix-coil equilibrium further toward disordered states (supplemental Fig. S4B). There are also smaller chemical shift changes in residues 458 and 462 (located in the loop between S3 and helix H1) and β-strands S9 and S10, a region contacted by helices H8 and H9.
FIGURE 4.
CFFT-001 compound reduces helicity of H8 and H9 in F508del NBD1 ΔRIΔRE. A, overlay of 15N-1H correlation spectra at 500 MHz for F508del NBD1 ΔRIΔRE in the absence (black; background) and presence of the final titration point (3:1) of CFFT-001 (magenta). CFFT-001 was added in 250, 500, and 750 μm apparent concentrations to a 250 μm sample of F508del NBD1 ΔRIΔRE. B, close-up of boxed area of A. Arrows in A and B indicate peaks that shift upon addition of compound. Compare Fig. 2, A and B, with Fig. 4, A and B. C, ribbon diagram of WT NBD1 ΔRIΔRE as described in Fig. 2 with the color gradient representing chemical shift changes due to compound addition.
Importantly, the size of the chemical shift change is not directly correlated with proximity to the compound. Rather, the size of the shift is related to changes in the immediate chemical environment of each amide proton-nitrogen pair due to either direct binding or indirect structural and dynamic effects. H8 and H9 form an unlikely interaction surface for CFFT-001, with their polar, charged, and irregular surfaces. The compound is hydrophobic and contains planar aromatic rings, suggesting that a flatter surface composed of hydrophobic residues in S3, S9, and S10 could constitute a more suitable site for drug binding. We propose that CFFT-001 binds to a hydrophobic surface formed by residues of these strands lying below H8 and H9, thereby disrupting the interaction between the sheet and the helices and reducing their helicity. The small shifts in the hydrophobic surface residues are attributable to the small fraction bound, the relatively minor conformational adjustments to these strands upon compound binding, and the expectation of poorer sensitivity of the observed resonances of the backbone atoms to compound binding than unobserved resonances of side chain atoms likely in direct contact with the compound. The larger shifts observed for H8 and H9 are attributable to the more dramatic backbone conformational changes upon ligand binding.
In addition to the F508del NBD1 ΔRIΔRE protein, we have also titrated the compound into WT NBD1 ΔRIΔRE and saw nearly identical peak shifts (supplemental Fig. S5) and a similar reduction of helicity. Interestingly, titration of the compound into the F508del H620Q NBD1 ΔRIΔRE leads to resonances of H9 shifting more toward coil (Fig. 2C, arrows). Thus, the compound further pushes the conformational equilibrium shift already present in the H620Q variant, in a qualitatively additive fashion, and the mutation does not inhibit compound binding.
Effect of Deletion of H9 on Binding of CFFT-001
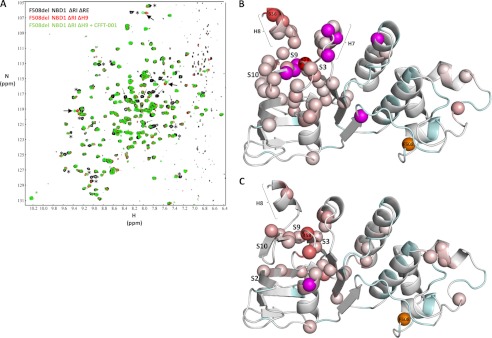
Although the most significant peak shifts observed upon addition of compound map to residues in the C-terminal helices, we hypothesize that CFFT-001 has a direct interaction with residues of S3, S9, and S10 that results in shifting the H8/H9 conformational equilibrium. To address this hypothesis and to test whether removal of these helices would allow greater accessibility for the compound to the surface of the sheet, we attempted to probe binding to NBD1 with both H8 and H9 deleted. The protein, however, was unstable, and sufficient yields and purity could not be attained. Notably, we were able to express and purify NBD1 in which residues 637–646 comprising H9 were deleted (Fig. 5A), consistent with the lack of density or conformational heterogeneity for residues of H9 in various crystal structures of NBD1 (supplemental Fig. S4D) (40, 60, 61) and our NMR data showing fraying of H9 (supplemental Fig. S4C). Comparison of spectra of F508del NBD1 ΔRIΔRE with F508del NBD1 ΔRIΔH9 (Fig. 5A) shows that deletion of residues 637–646 leads to significant chemical shift changes in addition to absence of peaks of deleted residues. Chemical shift changes can be noted in H7, H8, S3, S9, S10, and elsewhere in the protein (Fig. 5B). The largest shifts were observed for Gly-458 (in the loop between S3 and H1), confirming the loss of binding interaction to H9, and, as expected, Arg-636, at the C terminus before the truncation. We titrated CFFT-001 (1:1, 2:1, and 3:1, apparent compound:protein concentrations) into F508del NBD1 ΔRIΔH9 spectra and observed that the largest shift, again, was for Gly-458 (Fig. 5, A and C). This highlights the sensitivity of S3, S9, and S10 to compound binding, providing evidence for direct binding of the compound on the surface of this portion of the NBD1 core. Comparison of Figs. 5C and 4C shows a more extensive effect of compound binding (including S2 and the intersubdomain region) and more significant peak shifts in constructs lacking H9, suggestive of higher affinity binding. This is expected as moving H9 away to facilitate accessibility to the actual binding site on the surface of strands S3, S9, and S10 requires energy, so deleting it should lead to more favorable binding energy. These data also indicate that NBD1 retains the same basic fold in the absence of H9 and support our model of compound binding to the β-strands below the C-terminal helices and not to H9 itself (Fig. 5C).
FIGURE 5.
Effect of deletion of helix H9 on CFFT-001 binding. A, overlay of 15N-1H correlation spectra at 500 MHz for 250 μm F508del NBD1 ΔRIΔRE (387–646, Δ405–436) (black; background) and 250 μm F508del NBD1 ΔRI ΔH9 (387–636, Δ405–436) with (green, foreground) and without (red, middle ground) CFFT-001 (750 μm apparent concentration). Small arrows indicate peaks that are lost as a result of the deletion, whereas asterisks mark peaks that are affected by the deletion. Large arrows indicate peaks that shift upon compound addition. B and C, ribbon diagrams of WT NBD1 as described in Fig. 2 with the color gradients for N atoms representing chemical shift changes upon deletion of H9 (B) and compound addition to F508del NBD1 ΔRIΔH9 (C).
Simulated Docking of CFFT-001 onto NBD1
To address whether the region around S3/S9/S10 and H8/H9 contains a suitable binding site for CFFT-001, we carried out docking simulations. Examination of the crystal structure of F508del NBD1 ΔRIΔRE (Protein Data Bank code 2PZF) did not reveal any compound binding sites in this region. Because NMR data suggest that the binding does not occur to the ground state described in the crystal structure, we utilized REMD simulations starting from the structure of the ATP-bound NBD1 to sample higher energy states. In accordance with NMR data, we identified conformations in which H8 and H9 showed substantial fraying of helical structure as well as large distances between His-620 in S9 and Phe-640 in H9. We performed a site search procedure (41) on several of these conformations, leading to the identification of potential compound binding sites in this region that were not present in the ground state. These potential binding sites were subjected to docking simulations using the flexible docking procedure in Discovery Studio (50) and the induced fit protocol in Schrodinger (52, 53, 62). Docking resulted in multiple binding modes to the surface of S3/S9/S10 (Fig. 6). Interestingly, H9 contributed to the binding in many cases but not usually through specific H-bonds or π-π interactions with the ligand.
FIGURE 6.
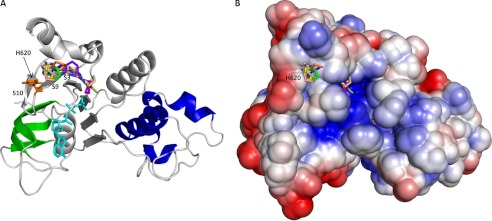
Model of interaction of CFFT-001 with NBD1. Multiple binding modes for CFFT-001 interaction with F508del NBD1 ΔRIΔRE. A, ribbon diagram of WT NBD1 as in Fig. 1 with the compounds and ATP shown as stick models. Compounds in each binding mode are represented by different colors, whereas ATP and the side chain of His-620 are shown in cyan and orange, respectively. B, electrostatic surface representation of NBD1 (red, negative potential; blue, positive potential; white, hydrophobic) showing the same compounds as described in A.
Because CFFT-001 also binds to NBD1 in the absence of H9, we repeated the docking simulations after removing residues comprising H9 (637–646) from the previously chosen REMD trajectory conformers. Once more, multiple binding modes were obtained in which the ligand comes into close contact with the hydrophobic S3/S9/S10 surface (supplemental Fig. S6). A few specific interactions with binding site residues (e.g. π-cation interactions with Lys-464) were observed in some cases. Similar results were obtained upon docking CFFT-001 to chain B of Protein Data Bank code 2PFZ, which lacks H9. In all of these binding modes, interaction of CFFT-001 with the surface of strands S3, S9, and S10 is not expected to be significantly perturbed by mutation of H620Q, which is adjacent to but not directly at the proposed binding surface (Fig. 6 and supplemental Fig. S6). Note also that ATP did not contact the compound in any of the REMD or docking simulations.
Direct Interaction between CFFT-001 and Mutations of NBD1 That Decrease Thermal Stability
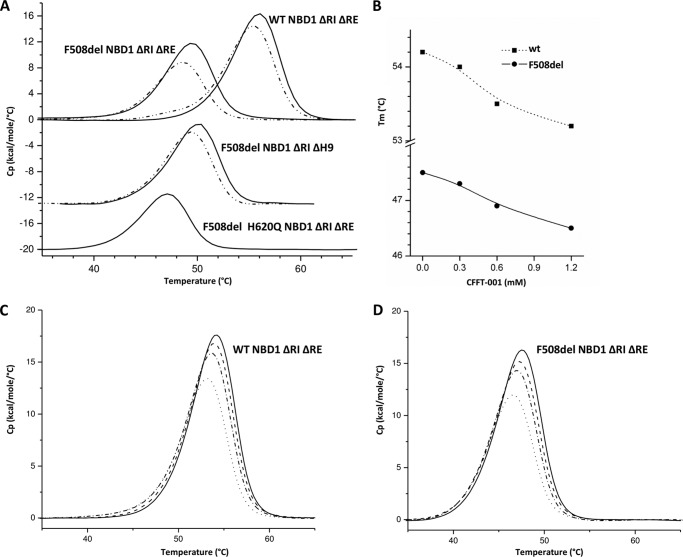
Having examined conformational changes within NBD1 as a result of mutation and compound addition using NMR approaches, we used differential scanning calorimetry (DSC) to probe these perturbations. The data in Fig. 7A illustrate that both the H620Q variant and the deletion of H9 reduce the thermal stability of NBD1. The midpoint of thermal denaturation, Tm, is reduced by ∼1–2 degrees in each, comparing F508del NBD1 ΔRIΔRE (Tm in 2 mm ATP = 49.4 °C and 5 mm ATP = 51.0 °C) to either F508del H620Q NBD1 ΔRIΔRE (bottom curve, Tm in 2 mm ATP = 47.1 °C) or F508del NBD1 ΔRIΔH9 (middle curve, Tm in 5 mm ATP = 50.4 °C). Furthermore, direct CFFT-001 binding to NBD1 is indicated by the consistent reduction in Tm of ∼1 degree when the compound is incubated at ∼1 mm (apparent concentration) with the various NBD1 constructs, for F508del NBD1 ΔRIΔRE from 49.4 to 48.6 °C, at 2 mm ATP, and for F508del NBD1 ΔRIΔH9 from 50.4 to 49.4 °C, at 5 mm ATP. In addition, there is a clear concentration-dependent loss of thermal stability (Fig. 7, B–D). Note that although ATP concentration changes the absolute Tm values, it is not the Tm values but the changes in Tm upon mutation or CFFT-001 binding (at constant ATP concentration) that are important. Given that ATP is already saturated by 2 mm and there are no contacts between ATP and His-620, helix H9, or the compound, the changes in Tm values should not be significantly affected by the ATP concentration.
FIGURE 7.
Differential scanning calorimetry of NBD1. A, DSC traces for WT and F508del NBD1 ΔRIΔRE (upper curves) in the absence (solid lines) and presence (dashed lines) of CFFT-001; buffer includes 2 mm ATP. DSC traces for F508del NBD1 ΔRIΔH9 (middle curves) in the absence (solid line) and presence (dashed line) of CFFT-001; buffer includes 5 mm ATP. DSC traces for F508del H620Q NBD1 ΔRIΔRE (lower curve); buffer includes 2 mm ATP. B, concentration dependence of Tm for WT and F508del NBD1 ΔRIΔRE, in the presence of 1 mm ATP. Concentration-dependent DSC traces of data shown in B for WT NBD1 ΔRIΔRE (C) and F508del NBD1 ΔRIΔRE (D). Solid lines show the NBD1 alone; addition of CFFT-001 at respective concentrations of 0.3 mm (dashed lines), 0.6 mm (filled squares on a dashed line) and 1.2 mm (filled squares on a solid line).
Importantly, the negative Tm shift observed upon binding of CFFT-001 is diagnostic of the compound interacting and stabilizing a conformation of NBD1 that is less thermostable, compatible with binding to a state in which the C-terminal helices are released from the NBD1 core. The data are consistent with both mutation and compound binding facilitating a shift in the underlying conformational equilibrium that leads to the preferential population of the state having the C-terminal helices released from the core. These data also indicate that deletions of Phe-508 and H9 do not eliminate the CFFT-001 binding site because the Tm values of NBD1 containing F508del or both mutations are equally affected by CFFT-001. Overall, the DSC data are in agreement with the effects of deletion of H9, an H620Q variant, and addition of CFFT-001 on NBD1 inferred from NMR data for these constructs.
CFTR-specific Sequence Conservation of Strands S9 and S10
CFTR differs structurally from other members of the ABC C family by the addition of the RI and R region phosphoregulatory segments that have been proposed to interact with the core of NBD1 in a phosphorylation-dependent fashion (31, 63). We hypothesized that these interactions require a binding site that is unique to CFTR. We expected to find CFTR-specific conservation of surface residues in the NBD1 of CFTR that differ significantly from their counterparts in other ABC C family members and that would point to RI and R region-specific functional surfaces on NBD1 involved in regulation of channel activity and/or misfolding. We further hypothesized that these surfaces could potentially be linked to the conformational changes within NBD1 identified by our mutational and compound binding studies. As such, an analysis of sequence conservation was undertaken, with a specific focus on separating conservation common to ABC transporters from conservation that is unique to CFTR.
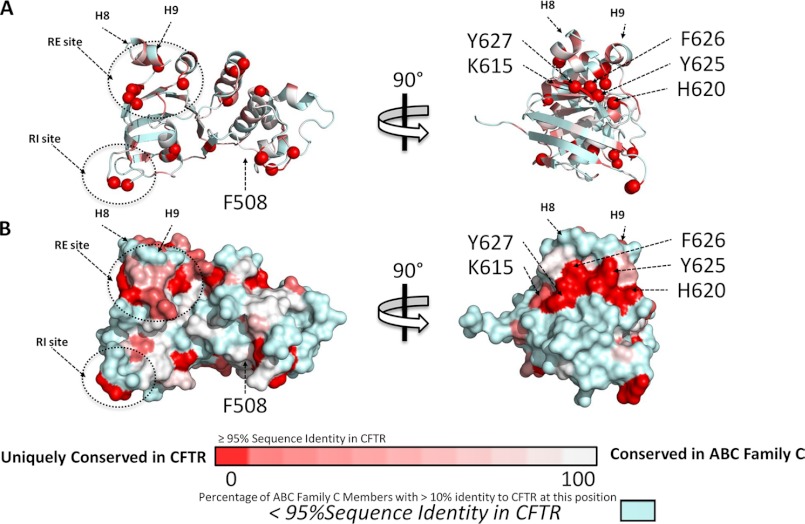
We prepared a sequence alignment comparing NBD1s from CFTR to sequences from each of the 11 other ABC C family members, using all 12 human sequences to identify homologs while filtering by annotation to ensure that the 12 members were kept distinct. From this, we determined the degree to which conserved CFTR residues (>95% conserved) in NBD1 match those found in other ABC C family members (see supplemental Table S1 for sequence alignment). Fig. 8 depicts the location of highly conserved residues in NBD1, colored according to the number of ABC C family members that have >10% sequence identity to CFTR at that position, with highly conserved residues that are unique to CFTR shown in red. Of these uniquely conserved residues, the largest solvent exposed cluster occurs in β-strands S9 and S10, with His-620, Tyr-625, Phe-626, and Tyr-627 having no matches to other ABC C family members, and Lys-615 only having a match to one. Significantly, S10 contains three unique hydrophobic aromatics in CFTR, but in all other ABC C family members, S10 has instead at least one strongly polar residue, primarily Glu, Arg, Lys, and Gln. This region of high conservation, namely S9 and S10, coincides with the hydrophobic region we have described here as the site of CFFT-001 binding immediately below helices H8 and H9 and is also the location of residues shown to affect channel activity, including His-620 (56). This high degree of CFTR-specific conservation supports the observation that this region plays a role in conformational changes related to CFTR function and/or processing.
FIGURE 8.
Sequence conservation unique to CFTR. Sequence profiles were determined for each of the 12 ABC subfamily C members, including CFTR. Conservation values were mapped onto the structure of WT CFTR NBD1 (Protein Data Bank 2PZE). Residues with <95% conservation in CFTR (by sequence identity) are colored in light blue. Residues with >95% conservation are colored in a gradient from red to white, with the residues that are unique to CFTR in red, and the residues that are present (>10% sequence identity) in other subfamily C members colored from dark pink (one matching ABC) to white (11 matching ABCs). A, ribbon diagrams in two orientations, with the N atoms of unique residues shown as spheres. B, solvent accessible surfaces for the same two orientations.
DISCUSSION
Molecular level knowledge on the conformational changes within NBD1 that are relevant to channel activity and folding energetics of CFTR is currently limited. Mutations, deletions and pharmacological effects are most often measured in the context of a full length CFTR molecule and usually in whole cells where many proteins are involved. NMR has enabled us to probe these effects at a residue-specific level and demonstrate that the H620Q substitution associated with higher channel open probability and a dual corrector/potentiator compound give rise to similar conformational changes within NBD1. These results, as well as their synergy with other dynamic and energetic data on NBD1 (29, 31, 32, 64), provide evidence for conformational properties of NBD1 that likely are involved in regulation of channel gating and in folding that may be tested in the future by studies on full-length CFTR.
The significance of the DSC results for the H620Q and H9 deletion can be better appreciated in the context of our current understanding of the NBD1 thermal unfolding pathway derived from a comprehensive analysis of previous DSC data on WT and F508del NBD1 (32, 64). Deletion of F508 leads to a Tm reduction of 6–7 °C (Fig. 7) resulting from the combined effects of a lower thermodynamic stability of the native state (ΔG) and a faster rate for the formation of an aggregation-prone intermediate. The H620Q and ΔH9 mutations also may reduce the thermodynamic stability of NBD1, or accelerate the rate of aggregation, or both. It is reasonable to expect that either structural modification would reduce the Tm of the protein because dynamic, but productive, interactions with the remainder of NBD1 have been disrupted or eliminated.
Mass action effects provide a clear, albeit unexpected, interpretation for the concentration-dependent decrease in the NBD1 Tm upon CFFT-001 binding. Typically, ligand binding to the native receptor results in an apparent increase in stability and Tm for receptor unfolding. However, here the Tm decreases, indicating that CFFT-001 binds preferentially to a conformation that is populated to a lower extent in its absence. Any such conformational equilibrium shift toward a state having reduced structural contacts gives rise to a reduction in Tm, based on thermodynamic principles, although compound binding makes this the more stable state. NMR data point to the release and loss of helicity for H8 and H9 upon CFFT-001 binding, yielding a conformation that would qualify as the non-native state to which CFFT-001 binds.
The H620Q variant, originally identified in CF patients demonstrating pancreatic insufficiency, has previously been shown to increase the Po of single CFTR channels, indicating an effect on the gating properties of CFTR (56–58). Both the H620Q variant and the CFFT-001 compound cause a shift of helices H8 and H9 from a preexisting helix-coil conformational equilibrium toward the coil state. These results suggest that perturbations affecting this conformational equilibrium coincide with conditions that promote channel opening and/or impede channel closing. Because the H620Q variant displays additional peak shifts relative to those observed for compound binding, there are certainly other consequences of the mutation. The Becq group (56) has demonstrated that the region surrounding H620 is critical for trafficking and gating. Although H620Q does not affect maturation/processing, an H620P mutation does, highlighting the importance of this position and the nature of the residue in it.
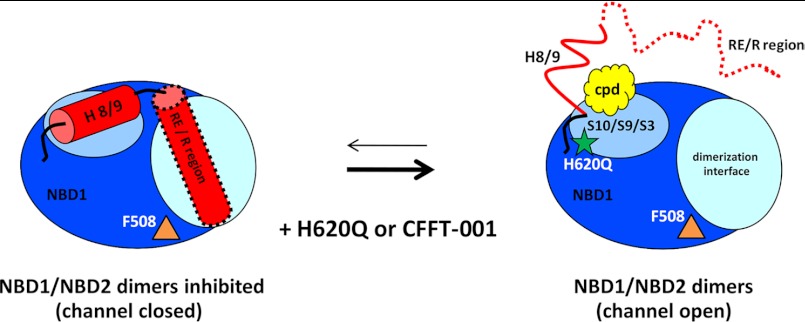
Previous models for the interaction of the RE/R region with NBD1 suggest that the RE is dynamic but that it can interact with the proposed NBD1/NBD2 heterodimerization interface on the core of NBD1 (4, 31, 61, 63). Work from our laboratory on murine NBD1 containing the RE (31) as well as isolated full-length R region with murine (63) and human6 NBD1 shows that the RE, which consists of the first 30 residues of the R region, transiently interacts with the surface of the NBD1 core in helical conformations. The RE contains two PKA phosphorylation sites, at 660 and 670. One effect of PKA-mediated phosphorylation of the RE or R region is to reduce this interaction and the helical structure6 (4, 31), shifting the conformational ensemble and potentially facilitating NBD heterodimerization and enhancing channel activity. Our current data on NBD1 lacking the RE, which is a mimic for the extreme of this conformational equilibrium in which the RE is always off of the surface, suggest that H9 may be thought of as the first part of the RE and R region with a conformational equilibrium between helical states bound to the core of NBD1 and coil states not interacting with the NBD1 core. This is consistent with the significant conformational heterogeneity of H9 in NBD1 crystal structures (supplemental Fig. S4D) and with previous definitions of the C-terminal NBD1 boundary (19, 40, 65). Thus, we hypothesize that conformational changes, as elicited by H620Q or CFFT-001, result in a shift in an underlying conformational equilibrium of regulatory interactions that are normally involved in gating. This shift involves release of H8 and H9 from the surface of S3, S9, and S10 that helps displace the RE/R region from the NBD1 dimerization interface (Fig. 9). This should promote NBD dimerization and lead to an enhanced open probability, increased channel activity for H620Q mutant channels and the potentiating effect of CFFT-001. It is possible that H620Q and CFFT-001 act to force an “unnatural” gating; however, the likelihood of this is low, considering that the portion of NBD1 affected appears to be a regulatory hot spot based on our sequence analysis and the observed effects of mutations here (56). Note that although phosphorylation is expected to have an overlapping effect to compound binding in similarly disrupting the R region:NBD1 interface, R region phosphorylation has numerous other effects including enhancement of R region interactions with other parts of CFTR (9) and modulations of R region binding with other binding partners.6 It is also possible that CFFT-001 may bind more strongly to full-length CFTR in the cell where many other factors contribute toward shifting this conformational equilibrium, including phosphorylation by kinases other than PKA (66) as well as multiple binding partners for the R region that compete with NBD1 for interaction.6 Such a scenario is consistent with our observation that PKA phosphorylation of CFTR is required to observe potentiator activity for CFFT-001 in cells.
FIGURE 9.
Simplified schematic model for functional dynamics within NBD1. NBD1 (dark blue) is shown as residing in two populations in a dynamic equilibrium. When channels are closed, NBD1/NBD2 heterodimers are inhibited due to the steric hindrance of the RE/R region interacting with NBD1. Addition of compound or the H620Q mutation shifts this equilibrium by reducing H9 helicity and contacts with the NBD1, subsequently leading to release of the RE/R region from the dimerization interface, relieving the inhibition and facilitating NBD1/NBD2 heterodimerization and channel opening.
The CFFT-001 compound may contribute to the corrector effect by binding to the core of NBD1 and stabilizing the folded state of the core NBD1 (not including H8 and H9) or by promoting NBD dimerization that could stabilize the folding of full-length CFTR. As shown by DSC, CFFT-001 does not stabilize the folded state of an isolated NBD1, including H8 and H9, but it may stabilize the NBD1 core within full-length CFTR. Binding of CFFT-001 to the uniquely conserved S9 and S10 site is consistent with CFTR-specific potentiating and/or correcting effects. Mutations of residues in these C-terminal strands of NBD1, specifically G622D and G628R, have been demonstrated to perturb the pharmacological effects of dual “MPB (benzo(c)quinolizinium)” compounds, characterized by their ability to both activate CFTR and rescue defective trafficking (56). This suggests that the C-terminal part of NBD1 impacts both gating and trafficking and plays a role in the mechanism of action for these dual acting compounds. It may also be that the mechanisms of action for the CFFT-001 corrector and/or potentiator activities occur at separate sites on the CFTR molecule and involve other domains of CFTR or proteins, making them impossible to dissect using an isolated NBD1.
The identification of S9 and S10 as uniquely conserved and as the site of potentiating mutations and potentiator/corrector compound binding may suggest that this region is an allosteric regulator of channel function or folding. With an understanding of allostery based on modulation of the energy landscape (67), NBD1 can be seen as a sensitive energetically malleable domain, with conformational sampling poised to be affected by a variety of perturbations such as compound binding (29). Thus, although the helix-coil transition for H8 and H9 may be qualitatively correlated with potentiation under some conditions, it is not necessarily quantitatively correlated with potentiation under all conditions, because the set of conformations accessible under different conditions (i.e. phosphorylated or not, nucleotide-bound state, etc.) may not be the same. A similar argument can be made for an underlying allosteric mechanism of suppression by the “3M” (G550E/R553M/R555K) mutations (17) which can improve CFTR processing in the absence of F508; even though they do not directly address the structural changes at the Phe-508 site, these mutations apparently change the equilibrium distribution of conformations accessed by NBD1 to be more similar to that of wild-type. The large number of substitutions in NBD1 that can suppress the F508del mutation supports such a general allosteric view of NBD1 with the structural changes we observe in the H620Q variant and upon CFFT-001 binding being one part of the NBD1 conformational equilibria. The conformational dynamics involving H8/H9 and the start of the R region highlighted here may facilitate other changes in CFTR that are not encompassed by the highly simplified model presented in Fig. 9, including modulation of the dynamic R region interaction hub,6 and allosteric effects within NBD1 on ATP binding and hydrolysis or NBD/intracellular domain coupling. These may be predicted to have potentiator and corrector consequences.
We have demonstrated that both a mutation and a compound elicit similar conformational changes within NBD1 that shift an underlying equilibrium involving helical structure in H8/H9 and their interaction with the uniquely conserved CFTR-specific region of β-strands S3/S9/S10. Because the H620Q mutation and CFFT-001 are both associated with an increase in the open probability of CFTR channels and because H8/H9 lead directly into the RE/R region within the context of full-length CFTR, we have hypothesized that this conformational shift at H8/H9 releases the R region from the NBD1/NBD2 dimerization interface, allowing heterodimers to form and thereby enhancing channel open probability and possibly processing. The uniquely conserved β-strands S9/S10 may thus be a key CFTR-specific lynchpin for integrating phosphoregulatory signals from the R region to NBD1 making it an attractive site for CFTR-specific therapeutics. This is consistent with previous evidence (56) pointing to β-strands S9 and S10 as a potential target for the design of more potent and selective CFTR modulators. These studies provide a foundation for further detailed analysis of the effects of mutations and other compounds on underlying conformational changes within NBD1 to provide insights into dynamic processes required for proper CFTR function and the rescue of these processes in the diseased state.
Supplementary Material
Acknowledgments
We thank Drs. Ranjith Muhandiram and Lewis E. Kay for assistance with NMR experiments and Dr. Phil Thomas for stimulating discussions. Julia Barette, David Lam and Leah Smith are acknowledged for preliminary and preparative work in studies of CFTR modulators.
This work was supported by Cystic Fibrosis Foundation Therapeutics Grants FORMAN05XX0 (to J. D. F.-K.), BROUIL08XX0 and BROUIL07XX0 (to C. G. B.), and SENDER09XX0 (to H. S.) and Cystic Fibrosis Canada (J. D. F.-K.).

This article contains supplemental Figs. S1–S6 and Table S1.
P. A. Chong and J. D. Forman-Kay, manuscript in preparation.
Z. Bozoky and J. D. Forman-Kay, manuscript in preparation.
- CF
- cystic fibrosis
- CFTR
- CF transmembrane conductance regulator
- NBD
- nucleotide binding domain
- DSC
- differential scanning calorimetry
- RE
- regulatory extension
- RI
- regulatory insertion
- MSD
- membrane-spanning domain
- DMSO
- dimethyl sulfoxide
- REMD
- replica exchange molecular dynamics
- IBMX
- isobutylmethylxanthine
- FRT
- Fischer rat thyroid
- PDB
- Protein Data Bank.
REFERENCES
- 1. Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. (1989) Identification of the cystic fibrosis gene: Genetic analysis. Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 2. Rommens J. M., Iannuzzi M. C., Kerem B., Drumm M. L., Melmer G., Dean M., Rozmahel R., Cole J. L., Kennedy D., Hidaka N. (1989) Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 245, 1059–1065 [DOI] [PubMed] [Google Scholar]
- 3. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 4. Lewis H. A., Buchanan S. G., Burley S. K., Conners K., Dickey M., Dorwart M., Fowler R., Gao X., Guggino W. B., Hendrickson W. A., Hunt J. F., Kearins M. C., Lorimer D., Maloney P. C., Post K. W., Rajashankar K. R., Rutter M. E., Sauder J. M., Shriver S., Thibodeau P. H., Thomas P. J., Zhang M., Zhao X., Emtage S. (2004) Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 23, 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ostedgaard L. S., Baldursson O., Welsh M. J. (2001) Regulation of the cystic fibrosis transmembrane conductance regulator Cl− channel by its R domain. J. Biol. Chem. 276, 7689–7692 [DOI] [PubMed] [Google Scholar]
- 6. Chappe V., Hinkson D. A., Howell L. D., Evagelidis A., Liao J., Chang X. B., Riordan J. R., Hanrahan J. W. (2004) Stimulatory and inhibitory protein kinase C consensus sequences regulate the cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A. 101, 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hallows K. R., Raghuram V., Kemp B. E., Witters L. A., Foskett J. K. (2000) Inhibition of cystic fibrosis transmembrane conductance regulator by novel interaction with the metabolic sensor AMP-activated protein kinase. J. Clin. Invest. 105, 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chappe V., Irvine T., Liao J., Evagelidis A., Hanrahan J. W. (2005) Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J. 24, 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mense M., Vergani P., White D. M., Altberg G., Nairn A. C., Gadsby D. C. (2006) In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 25, 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mornon J. P., Lehn P., Callebaut I. (2009) Molecular models of the open and closed states of the whole human CFTR protein. Cell Mol. Life Sci. 66, 3469–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hwang T. C., Sheppard D. N. (2009) Gating of the CFTR Cl− channel by ATP-driven nucleotide-binding domain dimerization. J. Physiol. 587, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bobadilla J. L., Macek M., Jr., Fine J. P., Farrell P. M. (2002) Cystic fibrosis: A worldwide analysis of CFTR mutations–correlation with incidence data and application to screening. Hum. Mutat. 19, 575–606 [DOI] [PubMed] [Google Scholar]
- 14. Welsh M. J., Smith A. E. (1993) Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 73, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 15. Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., Welsh M. J. (1992) Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 16. Sato S., Ward C. L., Krouse M. E., Wine J. J., Kopito R. R. (1996) Glycerol reverses the misfolding phenotype of the most common cystic fibrosis mutation. J. Biol. Chem. 271, 635–638 [DOI] [PubMed] [Google Scholar]
- 17. Teem J. L., Berger H. A., Ostedgaard L. S., Rich D. P., Tsui L. C., Welsh M. J. (1993) Identification of revertants for the cystic fibrosis ΔF508 mutation using STE6-CFTR chimeras in yeast. Cell 73, 335–346 [DOI] [PubMed] [Google Scholar]
- 18. Pissarra L. S., Farinha C. M., Xu Z., Schmidt A., Thibodeau P. H., Cai Z., Thomas P. J., Sheppard D. N., Amaral M. D. (2008) Solubilizing mutations used to crystallize one CFTR domain attenuate the trafficking and channel defects caused by the major cystic fibrosis mutation. Chem. Biol. 15, 62–69 [DOI] [PubMed] [Google Scholar]
- 19. Aleksandrov A. A., Kota P., Aleksandrov L. A., He L., Jensen T., Cui L., Gentzsch M., Dokholyan N. V., Riordan J. R. (2010) Regulatory insertion removal restores maturation, stability and function of ΔF508 CFTR. J. Mol. Biol. 401, 194–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dalemans W., Barbry P., Champigny G., Jallat S., Dott K., Dreyer D., Crystal R. G., Pavirani A., Lecocq J. P., Lazdunski M. (1991) Altered chloride ion channel kinetics associated with the ΔF508 cystic fibrosis mutation. Nature 354, 526–528 [DOI] [PubMed] [Google Scholar]
- 21. Zhang F., Kartner N., Lukacs G. L. (1998) Limited proteolysis as a probe for arrested conformational maturation of ΔF508 CFTR. Nat. Struct. Biol. 5, 180–183 [DOI] [PubMed] [Google Scholar]
- 22. Van Goor F., Hadida S., Grootenhuis P. D., Burton B., Stack J. H., Straley K. S., Decker C. J., Miller M., McCartney J., Olson E. R., Wine J. J., Frizzell R. A., Ashlock M., Negulescu P. A. (2011) Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. U.S.A. 108, 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clancy J. P., Rowe S. M., Accurso F. J., Aitken M. L., Amin R. S., Ashlock M. A., Ballmann M., Boyle M. P., Bronsveld I., Campbell P. W., De Boeck K., Donaldson S. H., Dorkin H. L., Dunitz J. M., Durie P. R., Jain M., Leonard A., McCoy K. S., Moss R. B., Pilewski J. M., Rosenbluth D. B., Rubenstein R. C., Schechter M. S., Botfield M., Ordoñez C. L., Spencer-Green G. T., Vernillet L., Wisseh S., Yen K., Konstan M. W. (2012) Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 67, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Accurso F. J., Rowe S. M., Clancy J. P., Boyle M. P., Dunitz J. M., Durie P. R., Sagel S. D., Hornick D. B., Konstan M. W., Donaldson S. H., Moss R. B., Pilewski J. M., Rubenstein R. C., Uluer A. Z., Aitken M. L., Freedman S. D., Rose L. M., Mayer-Hamblett N., Dong Q., Zha J., Stone A. J., Olson E. R., Ordoñez C. L., Campbell P. W., Ashlock M. A., Ramsey B. W. (2010) Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N. Engl. J. Med. 363, 1991–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Goor F., Hadida S., Grootenhuis P. D., Burton B., Cao D., Neuberger T., Turnbull A., Singh A., Joubran J., Hazlewood A., Zhou J., McCartney J., Arumugam V., Decker C., Yang J., Young C., Olson E. R., Wine J. J., Frizzell R. A., Ashlock M., Negulescu P. (2009) Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. U.S.A. 106, 18825–18830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Goor F., Yu H., Burton B., Huang T., Hoffman B., Negulescu P. A. (2011) VX-770 potentiation of CFTR forms with channel gating defects in vitro. Pediatr. Pulmonol. 34, 215 [Google Scholar]
- 27. Yu H., Burton B., Huang C. J., Worley J., Cao D., Johnson J. P., Jr., Urrutia A., Joubran J., Seepersaud S., Sussky K., Hoffman B. J., Van Goor F. (2012) Ivacaftor potentiation of multiple CFTR channels with gating mutations. J. Cyst. Fibros. 11, 237–245 [DOI] [PubMed] [Google Scholar]
- 28. Lukacs G. L., Verkman A. S. (2012) CFTR: Folding, misfolding, and correcting the ΔF508 conformational defect. Trends Mol. Med. 18, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chong P. A., Kota P., Dokholyan N. V., Forman-Kay J. D. (2012) Dynamics intrinsic to CFTR function and stability in Cystic Fibrosis: Molecular Basis, Physiological Changes, and Therapeutic Strategies (John Riordan R. B., Quinton P., eds) Cold Spring Harbor Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanelis V., Chong P. A., Forman-Kay J. D. (2011) NMR spectroscopy to study the dynamics and interactions of CFTR. Methods Mol. Biol. 741, 377–403 [DOI] [PubMed] [Google Scholar]
- 31. Kanelis V., Hudson R. P., Thibodeau P. H., Thomas P. J., Forman-Kay J. D. (2010) NMR evidence for differential phosphorylation-dependent interactions in WT and ΔF508 CFTR. EMBO J. 29, 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Protasevich I., Yang Z., Wang C., Atwell S., Zhao X., Emtage S., Wetmore D., Hunt J. F., Brouillette C. G. (2010) Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 19, 1917–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 34. Johnson B. A., Blevins R. A. (1994) NMR view: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 35. Goddard T. D., Kneller D. G. (2008) Sparky 3, version 3, University of California, San Francisco [Google Scholar]
- 36. Yang D., Kay L. E. (1999) Improved 1HN-detected triple resonance TROSY-based experiments. J. Biomol. NMR 13, 3–10 [DOI] [PubMed] [Google Scholar]
- 37. Salzmann M., Pervushin K., Wider G., Senn H., Wüthrich K. (1998) TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rule G. S., Hitchens T. K. (2006) Fundamentals of Protein NMR Spectroscopy, Springer, Utrecht University, Utrecht, The Netherlands [Google Scholar]
- 39. Pedemonte N., Lukacs G. L., Du K., Caci E., Zegarra-Moran O., Galietta L. J., Verkman A. S. (2005) Small-molecule correctors of defective ΔF508-CFTR cellular processing identified by high-throughput screening. J. Clin. Invest. 115, 2564–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Atwell S., Brouillette C. G., Conners K., Emtage S., Gheyi T., Guggino W. B., Hendle J., Hunt J. F., Lewis H. A., Lu F., Protasevich II, Rodgers L. A., Romero R., Wasserman S. R., Weber P. C., Wetmore D., Zhang F. F., Zhao X. (2010) Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng. Des. Sel. 23, 375–384 [DOI] [PubMed] [Google Scholar]
- 41. Accelrys (2005–2009) Discovery Studio Modeling Environment, 2.5 Ed., Accelrys Software, Inc., San Diego [Google Scholar]
- 42. Berendsen H. J., van der Spoel D., van Drunen R. (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91, 43–56 [Google Scholar]
- 43. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 44. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side chain torsion potentials for the Amber ff99SB protein force field. Prot. Struct. Funct. Bioinformat. 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patriksson A., van der Spoel D. (2008) A temperature predictor for parallel tempering simulations. Phys. Chem. Chem. Phys. 10, 2073–2077 [DOI] [PubMed] [Google Scholar]
- 46. Hockney R. W., Goel S. P., Eastwood J. W. (1974) Quiet high-resolution computer models of a plasma. J. Comput. Phys. 14, 148–158 [Google Scholar]
- 47. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: An W log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 48. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 49. Hess B., Bekker H., Berendsen H., Fraaije J. (1997) LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 50. Koska J., Spassov V. Z., Maynard A. J., Yan L., Austin N., Flook P. K., Venkatachalam C. M. (2008) Fully automated molecular mechanics based induced fit protein-ligand docking method. J. Chem. Inf. Model. 48, 1965–1973 [DOI] [PubMed] [Google Scholar]
- 51. Schrodinger, LLC. (2010) Maestro, version 9.1, Schrodinger, New York [Google Scholar]
- 52. Sharma S., Ding F., Dokholyan N. V. (2007) Multiscale modeling of nucleosome dynamics. Biophys. J. 92, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schrodinger, LLC (2010) Schrodinger Suite 2010 Induced Fit Docking protocol, Glide, version 5.6, New York, NY [Google Scholar]
- 54. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Edgar R. C. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Billet A., Melin P., Jollivet M., Mornon J. P., Callebaut I., Becq F. (2010) C terminus of nucleotide binding domain 1 contains critical features for cystic fibrosis transmembrane conductance regulator trafficking and activation. J. Biol. Chem. 285, 22132–22140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei L., Vankeerberghen A., Cuppens H., Droogmans G., Cassiman J. J., Nilius B. (1998) Phosphorylation site-independent single R-domain mutations affect CFTR channel activity. FEBS Lett. 439, 121–126 [DOI] [PubMed] [Google Scholar]
- 58. Vankeerberghen A., Wei L., Teng H., Jaspers M., Cassiman J. J., Nilius B., Cuppens H. (1998) Characterization of mutations located in exon 18 of the CFTR gene. FEBS Lett. 437, 1–4 [DOI] [PubMed] [Google Scholar]
- 59. Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D. (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lewis H. A., Wang C., Zhao X., Hamuro Y., Conners K., Kearins M. C., Lu F., Sauder J. M., Molnar K. S., Coales S. J., Maloney P. C., Guggino W. B., Wetmore D. R., Weber P. C., Hunt J. F. (2010) Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J. Mol. Biol. 396, 406–430 [DOI] [PubMed] [Google Scholar]
- 61. Lewis H. A., Zhao X., Wang C., Sauder J. M., Rooney I., Noland B. W., Lorimer D., Kearins M. C., Conners K., Condon B., Maloney P. C., Guggino W. B., Hunt J. F., Emtage S. (2005) Impact of the ΔF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 280, 1346–1353 [DOI] [PubMed] [Google Scholar]
- 62. Sherman W., Beard H. S., Farid R. (2006) Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 67, 83–84 [DOI] [PubMed] [Google Scholar]
- 63. Baker J. M., Hudson R. P., Kanelis V., Choy W. Y., Thibodeau P. H., Thomas P. J., Forman-Kay J. D. (2007) CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 14, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang C., Protasevich I., Yang Z., Seehausen D., Skalak T., Zhao X., Atwell S., Spencer Emtage J., Wetmore D. R., Brouillette C. G., Hunt J. F. (2010) Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 19, 1932–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Csanády L., Chan K. W., Nairn A. C., Gadsby D. C. (2005) Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain. J. Gen. Physiol. 125, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Seavilleklein G., Amer N., Evagelidis A., Chappe F., Irvine T., Hanrahan J. W., Chappe V. (2008) PKC phosphorylation modulates PKA-dependent binding of the R domain to other domains of CFTR. Am. J. Physiol. Cell Physiol. 295, C1366–1375 [DOI] [PubMed] [Google Scholar]
- 67. Wrabl J. O., Gu J., Liu T., Schrank T. P., Whitten S. T., Hilser V. J. (2011) The role of protein conformational fluctuations in allostery, function, and evolution. Biophys. Chem. 159, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chang P., Ghosh S. (December 29, 2010) Patent WO 2010/151747 A1
- 69. Hadida R. S., Van Goor F., Miller M., McCartney J., Arumugam V. (February 22, 2007) Patent WO 2007/021982 A2
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.