Background: Functional relationships between the microRNA and cellular hypoxia response pathways are unknown.
Results: Dicer is down-regulated in chronic hypoxia; this mechanism maintains the induction of hypoxia-inducible factor-α subunits and hypoxia-responsive genes.
Conclusion: Loss of Dicer-dependent microRNA regulation is important for maintaining the concerted cellular response to hypoxia.
Significance: Altogether, we provide a newer perspective into the post-transcriptional pathways that regulate the cellular hypoxic response.
Keywords: Endothelial Cell, Endothelium, Hypoxia, Hypoxia-inducible Factor (HIF), MicroRNA, Molecular Biology, Molecular Cell Biology, Dicer
Abstract
The processes by which cells sense and respond to ambient oxygen concentration are fundamental to cell survival and function, and they commonly target gene regulatory events. To date, however, little is known about the link between the microRNA pathway and hypoxia signaling. Here, we show in vitro and in vivo that chronic hypoxia impairs Dicer (DICER1) expression and activity, resulting in global consequences on microRNA biogenesis. We show that von Hippel-Lindau-dependent down-regulation of Dicer is key to the expression and function of hypoxia-inducible factor α (HIF-α) subunits. Specifically, we show that EPAS1/HIF-2α is regulated by the Dicer-dependent microRNA miR-185, which is down-regulated by hypoxia. Full expression of hypoxia-responsive/HIF target genes in chronic hypoxia (e.g. VEGFA, FLT1/VEGFR1, KDR/VEGFR2, BNIP3L, and SLC2A1/GLUT1), the function of which is to regulate various adaptive responses to compromised oxygen availability, is also dependent on hypoxia-mediated down-regulation of Dicer function and changes in post-transcriptional gene regulation. Therefore, functional deficiency of Dicer in chronic hypoxia is relevant to both HIF-α isoforms and hypoxia-responsive/HIF target genes, especially in the vascular endothelium. These findings have relevance to emerging therapies given that we show that the efficacy of RNA interference under chronic hypoxia, but not normal oxygen availability, is Dicer-dependent. Collectively, these findings show that the down-regulation of Dicer under chronic hypoxia is an adaptive mechanism that serves to maintain the cellular hypoxic response through HIF-α- and microRNA-dependent mechanisms, thereby providing an essential mechanistic insight into the oxygen-dependent microRNA regulatory pathway.
Introduction
Cellular adaptation to hypoxia is governed by the HIF3 family of heterodimeric transcription factors (1–3). Aryl hydrocarbon receptor nuclear translocator (ARNT/HIF-1β) is constitutively expressed and stable, whereas the HIF-α (that is HIF-1α or HIF-2α) subunit is regulated by an oxygen-dependent degradation process and targeted for ubiquitin-mediated destruction under normoxia. Hypoxia has long been associated with alterations to blood vessel function. For example, hypoxia elicits dramatic changes in the physiology (4) and gene expression in the vascular endothelium (5–7). These gene expression changes are dependent in major part on HIF isoforms and result in alterations to the endothelial phenotype that can ultimately result in endothelial activation and dysfunction.
It has been known for some time that the post-transcriptional regulation of hypoxia-responsive/HIF target genes, especially mRNA stabilization, is a key component of an integrated hypoxic response, especially in the vascular endothelium. Recent studies have determined a link between hypoxia and the regulation of microRNAs, although the global mechanism(s) remains largely unknown (8). Dicer is a key endoribonuclease that processes precursor microRNAs (pre-microRNAs) into mature microRNAs and cleaves double-stranded RNAs into small interfering RNAs (9). Dicer also aids in the incorporation of microRNAs into the RNA-induced silencing complex (RISC) (10), which silences gene expression via changes in target mRNA stability and/or translation, thereby establishing Dicer as a critical regulator of post-transcriptional gene silencing. Increases and decreases in Dicer expression have broad functional effects on cellular phenotype (11). Given that hypoxia and microRNAs both regulate translation and mRNA stability (12, 13), it is surprising that so little is known about the contribution of changes in oxygen bioavailability, or sensing, to microRNA biogenesis and more importantly the functional consequences of such regulation.
In light of the potential for hypoxia to regulate mRNAs post-transcriptionally and considering that HIF-α isoforms and many hypoxia-responsive/HIF target genes are regulated by microRNAs, it was of interest to determine whether global changes in microRNA biogenesis could play a functional role in chronic hypoxia. We report here that the microRNA pathway is functionally integrated with the cellular hypoxia response pathway, especially in the vascular endothelium. We show in vitro in human cell types and in vivo in the mouse that chronic hypoxia impairs Dicer expression and activity, resulting in global consequences on microRNA biogenesis. VHL-dependent and HIF-independent down-regulation of Dicer is key to the sustained expression and function of HIF-α subunits in chronic hypoxia. Full expression of hypoxia-responsive/HIF target genes in chronic hypoxia (e.g. VEGFA, VEGFR1, VEGFR2, BNIP3L, and GLUT1), the function of which is to regulate various adaptive responses to compromised oxygen availability, is also dependent on the hypoxia-mediated down-regulation of Dicer function and changes in post-transcriptional gene regulation. These findings add important conceptual and mechanistic insight into the interaction between two very important cellular pathways, namely the microRNA pathway and the cellular hypoxia response pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
Human umbilical vein endothelial cells (HUVEC) isolated from multiple independent donors were cultured as described previously (7). Early passage HUVEC (passage 3–5) were used in these studies. Human dermal microvascular endothelial cells (Lonza) were cultured in EGM-2MV (Lonza). Human aortic smooth muscle cells (ScienCell) were cultured in smooth muscle cell medium (ScienCell). HepG2, RCC4, 786-O, and UMRC2 cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Hyclone). 786-O clones stably expressing pRetroSUPER-empty or pRetroSUPER-HIF2α shRNA were described previously (14). Cells were grown at 37 °C in 5% CO2 in a humidified Steri-Cycle incubator (ThermoForma, Model 370). Total cellular protein and RNA were extracted using the mirVana PARIS kit (Ambion) according to the manufacturer's recommendations. Nuclear/cytoplasmic partitioning was performed as described previously (7). For MG-132 treatment, confluent HUVEC were treated with MG-132 (EMD Chemicals) at a final concentration of 10 μm or DMSO after which cells were subjected to 0 or 24 h of hypoxia.
Hypoxia Treatment
Cells were subjected to 1% O2 in a temperature- and humidity-controlled incubator within a sealed anaerobic system (ThermoForma, Model 1025). A hypoxic environment (1% O2) was achieved and maintained using a high purity anaerobic gas mixture (5% CO2, 10% H2, 85% N2; Linde). Desferrioxamine treatment was performed as described previously (7).
Quantitative RT-PCR
First strand cDNA synthesis and qRT-PCR were performed as described previously (7). TaqMan human microRNA assays and a TaqMan microRNA reverse transcription kit (Applied Biosystems) were used for microRNA measurements according to the manufacturer's recommendations. Results were normalized to 18 S rRNA levels. mRNA and 18 S rRNA relative -fold changes were calculated using either absolute quantification with plasmid standard curves or the comparative Ct method, which was used for precursor microRNAs and microRNAs. A predetermined amount of in vitro synthesized, capped, and polyadenylated luciferase mRNA was added to each sample immediately before RNA extraction and measured by qRT-PCR to control for efficiencies of RNA extraction and first strand cDNA synthesis. Primer sequences are shown in supplemental Table S3.
Immunoblot
Total cellular protein was size-fractionated on NuPAGE Novex 4–12% Bis-Tris or 3–8% Tris acetate gels (Invitrogen) using the XCell SureLock Mini-Cell (Invitrogen) and transferred onto 0.45-μm nitrocellulose membranes using the XCell II Blot Module (Invitrogen) according to the manufacturer's recommendations. The following primary antibodies were used: anti-Drosha (ab12286, Abcam), anti-DGCR8 (N-19) (sc-48473, Santa Cruz Biotechnology), anti-Exportin-5 (ab31351, Abcam), anti-Dicer (13D6) (ab14601, Abcam), anti-TRBP (ab42018, Abcam), anti-Argonaute 1 (Ago1) (NB100–2817, Novus Biologicals), anti-Ago2 (ab57113, Abcam), anti-HIF-1α (AF1935, R&D Systems or 610958, BD Pharmingen), anti-HIF-2α (NB100–122, Novus Biologicals), anti-GLUT1 (ab652, Abcam), anti-BNIP3L (N0399, Sigma-Aldrich), anti-VEGFR1 (Y103) (ab32152, Abcam), anti-VEGFR2 (C-1158) (sc-504, Santa Cruz Biotechnology), anti-HA (12CA5) (11583816001, Roche Applied Science); anti-VHL (Ig32) (556347, BD Pharmingen), anti-LMNA (346) (sc-7293, Santa Cruz Biotechnology), and anti-Vinculin (V9264, Sigma-Aldrich). The following secondary antibodies were used: horseradish peroxidase (HRP)-conjugated rabbit anti-mouse IgG (heavy and light) (ab6728, Abcam), HRP-conjugated goat anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology), and HRP-conjugated donkey anti-goat IgG (sc-2020, Santa Cruz Biotechnology). Signal quantification was performed using NIH ImageJ and normalized to loading control (LMNA).
Global MicroRNA Profiling in Hypoxia
Microarray analysis of global microRNA expression in HUVEC was performed by Exiqon using miRCURY LNA Arrays (v10.0, Exiqon) annotated according to miRBase v11. qRT-PCR measurements of global mouse kidney microRNA expression were performed by Applied Biological Materials, covering 726 mature mouse microRNAs annotated according to miRBase v16.
Manipulation of Dicer and MicroRNA Expression
Dicer knockdowns were performed using Oligofectamine (Invitrogen) as described previously (7) using Dicer-specific siRNAs versus scrambled non-silencing siRNA described previously (15). LMNA knockdown was performed using siGENOME LMNA control siRNA (Dharmacon) to control for nonspecific RNA interference (RNAi) effects. Cells were harvested 72 h after transfection. For Dicer overexpression, 70–80% confluent HUVEC grown on 100-mm dishes were transiently transfected with either 5 μg of Dicer plasmid (Hannon laboratory) or pcDNA3 (Invitrogen) together with 3 μg of pMACs KK.II plasmid (Miltenyi Biotec) using Effectene (Qiagen) according to the manufacturer's recommendations. 48–72 h after plasmid transfection, cells were immunomagnetically purified using the MACSelect KK transfected cell selection kit (Miltenyi Biotec) according to the manufacturer's recommendations. Purified cells were grown on 12-well plates for an additional 24 h before they were subjected to 0 or 24 h of hypoxia. For microRNA overexpression, microRNA mimics (Dharmacon) were transfected at a final concentration of 40 nm.
HIF-α/VEGFA 3′-UTR Luciferase Assay
70–80% confluent HUVEC grown on 60-mm dishes were transiently transfected with either 2 μg of pLuc-Ctrl (described previously in Ref. 16) or pLuc-Empty-3′-UTR (SwitchGear Genomics) encoding firefly luciferase (GL3/luc+) versus chimeric luciferase reporter-human HIF-1α/HIF-2α/VEGFA 3′-UTR, i.e. pLuc-HIF1A/HIF2A/VEGFA-3′-UTR (pLuc-HIF1A/HIF2A-3′-UTR, SwitchGear Genomics) together with 0.4 μg of pRL-SV40 (Promega) (i.e. control for transfection efficiency) and either 2 μg of Dicer plasmid or pcDNA3 (Invitrogen). 24 h after plasmid transfection, cells were subjected to 0 or 24 h of hypoxia after which luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's recommendations.
For miR-185 overexpression assays, microRNA target binding sites in pLuc-HIF2A-3′-UTR were mutated using standard recombinant DNA techniques and replaced with sequences that do not contain any known mRNA regulatory elements. 70–80% confluent HUVEC grown on 60-mm dishes were transiently transfected with 2 μg of luciferase reporter constructs, either 2 μg of Dicer plasmid or pcDNA3, and 0.4 μg of pRL-SV40. MicroRNA mimics (Dharmacon) were transfected at a final concentration of 40 nm 24 h after plasmid transfection. 24 h after microRNA mimic transfection, cells were subjected to 0 or 24 h of hypoxia after which luciferase activities were measured as described above.
Dicer-dependent and Dicer-independent RNAi Assays
70–80% confluent HUVEC grown on 60-mm dishes were transiently transfected with 4 μg of pLuc-Ctrl and 0.4 μg of pRL-SV40. The following RNAi methods were compared. 24 h after plasmid transfection, cells were transfected with one of the following: siRNA: Silencer Firefly Luciferase (GL3) siRNA (Ambion) or siGENOME Non-Targeting siRNA Number 3 (Dharmacon) at a final concentration of 50 nm; Dicer substrate siRNA (DsiRNA): Dicector Fluc-S1 DS Positive Control or Dicector DS Scrambled Neg (Integrated DNA Technologies) at a final concentration of 20 nm; and shRNA: 3.2 μg of psiRNA-LucGL3 with 0.8 μg of psiRNA-LacZ (Invivogen) or 4 μg of psiRNA-LacZ. Dicer-specific siRNAs were transfected at a final concentration of 50 nm. For Dicer overexpression experiments, cells were transfected with 2 μg each of pLuc-Ctrl and Dicer plasmid or pcDNA3,1.6 μg of psiRNA-LucGL3, and 2.4 μg of psiRNA-LacZ. 16 h after siRNA/shRNA/DsiRNA transfection, cells were subjected to 0, 4, or 24 h of hypoxia after which luciferase activities were measured as described above.
In Vitro Assay for RISC Activity
In vitro mRNA cleavage experiments were performed using cytoplasmic extracts as described previously (17).
RNA Polymerase II Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as described previously (7). ChIP primers are available upon request.
mRNA Half-life Determination
Confluent HUVEC were subjected to 4 h of normoxia or hypoxia after which actinomycin D (BioShop Canada) was added at a final concentration of 2 μg/ml. Cells were then subjected to normoxia or hypoxia during which total cellular RNA was extracted at 0, 2, 4, 6, and 24 h.
Global Protein Synthesis Measurements
[3H]Leucine (PerkinElmer Life Sciences) was added to the growth medium of confluent HUVEC grown on 12-well plates at a final concentration of 2 μCi/well for the last hour of normoxia or hypoxia after which cells were washed with ice-cold PBS, and the incorporated radioactivity was precipitated with 15% TCA for 20 min. Cells were then washed with DNase/RNase-free distilled water and solubilized with 0.1 m NaOH and 0.1% SDS. Radioactivity from each well was counted in a liquid scintillation counter. Each experiment was performed in duplicate. As a complementary approach, [35S]methionine (PerkinElmer Life Sciences) was added to the growth medium of confluent HUVEC grown on 12-well plates at a final concentration of 10 mCi/well for the last 0, 20, 40, or 60 min of normoxia or hypoxia after which cells were washed with ice-cold PBS, incubated in 10% TCA for 20 min, washed three times with 100% ethanol, and dried at 45 °C. The precipitates were dissolved in 0.3 n NaOH for 20 min and neutralized with 0.3 n HCl. Radioactivity in the resultant mixture was then measured using a liquid scintillation counter. The rate of [35S]methionine incorporation was determined by calculating the slope of the radioactivity-incubation time plot.
Dicer-specific Protein Synthesis and Half-life Measurements
Dicer-specific [35S]methionine incorporation experiments were performed as described previously (18) using an anti-Dicer (13D6) antibody (ab14601, Abcam) (1:50 dilution).
Animals and Hypoxia Treatment
C57BL/6J mice were obtained from The Jackson Laboratory. Mice between 8 and 12 weeks of age were subjected to either atmospheric O2 or 8% O2 for 24 h after which they were sacrificed and organs were harvested.
Clear Cell Renal Cell Carcinoma (CCRCC) Microarray Analysis
Microarray analysis of 105 primary CCRCC samples and 12 non-diseased kidney samples was performed as described previously (19) (Gene Expression Omnibus accession number GSE14762). Institutional review board approval was obtained from each participating institution, and informed consent was obtained from all participants.
Study Approval
All animal studies were performed in accordance with the guidelines of the Canadian Council on Animal Care and were approved by the University of Toronto Animal Care Committee.
Statistical Analysis
All data sets represent the mean ± S.E. of at least three independent experiments unless otherwise stated. Statistical analysis was performed using a two-tailed t test or analysis of variance where appropriate. A two-tailed Welch's t test was used to analyze comparisons from the CCRCC microarray data.
RESULTS
Global Effects of Chronic Hypoxia on MicroRNA Expression in Human Endothelial Cells
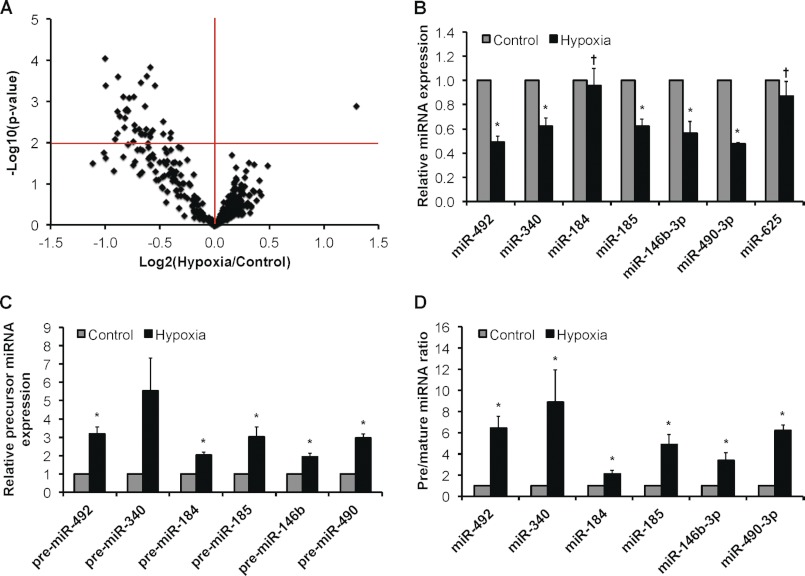
We performed a microRNA microarray analysis on HUVEC subjected to either acute (4-h) or chronic (24-h) hypoxia (1% O2). Results revealed that 360 microRNAs exhibited detectable expression above background in HUVEC, and we were able to confirm the majority of microRNAs reported previously by others as the most highly expressed in endothelial cells under basal conditions (data not shown). Significantly, volcano plot analysis of the microarray results revealed 36 microRNAs that were differentially regulated by chronic hypoxia at p < 0.01 compared with normoxia/control conditions (Fig. 1A and supplemental Table S1). Importantly, we observed a common pattern in which chronic hypoxia resulted in the down-regulation of 35 of the 36 differentially regulated microRNAs (supplemental Table S1). The expression levels of seven such hypoxia-regulated microRNAs were validated using qRT-PCR (Fig. 1B) along with seven other internal controls (supplemental Fig. S1A). Notably, we confirmed the up-regulation of miR-210, a hypoxia-inducible microRNA (8, 20). Genomic loci encoding the 68 microRNAs (obtained from miRBase) indicated no apparent clustering of polycistronic primary microRNA transcripts (pri-microRNAs) (data not shown). MicroRNAs mature through three intermediates: a pri-microRNA, a pre-microRNA, and a microRNA-microRNA* duplex (9). Interestingly, we observed the accumulation of precursor (i.e. pri/pre-microRNAs) species under chronic hypoxia (Fig. 1C). This accumulation of precursor species extends to microRNAs that were not significantly regulated by hypoxia (supplemental Fig. S1B). This suggested that microRNA biogenesis is impaired under chronic hypoxia and is best illustrated by defining the ratio of precursor to mature microRNA species (Fig. 1D and supplemental Fig. S1C).
FIGURE 1.
Global effect of chronic hypoxia on microRNA expression in endothelial cells. A, volcano plot of 360 detectable microRNAs in 24-h hypoxic versus control HUVEC as measured by microarray profiling (n = 3). Expression levels of representative down-regulated microRNAs (B) and corresponding precursor microRNAs (pri/pre-microRNAs) (C) and the ratio of precursor microRNAs to their mature counterparts in normoxic versus 24-h hypoxic HUVEC (D) are shown. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with normoxia and 4-h hypoxia, respectively.
Following specific knockdown of Dicer in HUVEC under normoxia (supplemental Fig. S2A), we observed a decrease in mature microRNA levels (supplemental Fig. S2B) with a corresponding accumulation of the precursor species (supplemental Fig. S2, C and D). Comparable profiles of microRNA versus precursor microRNA expression following Dicer knockdown and chronic hypoxia and broad class effects on a significant number of microRNAs suggested the hypothesis that Dicer function is deficient in endothelial cells under chronic hypoxia.
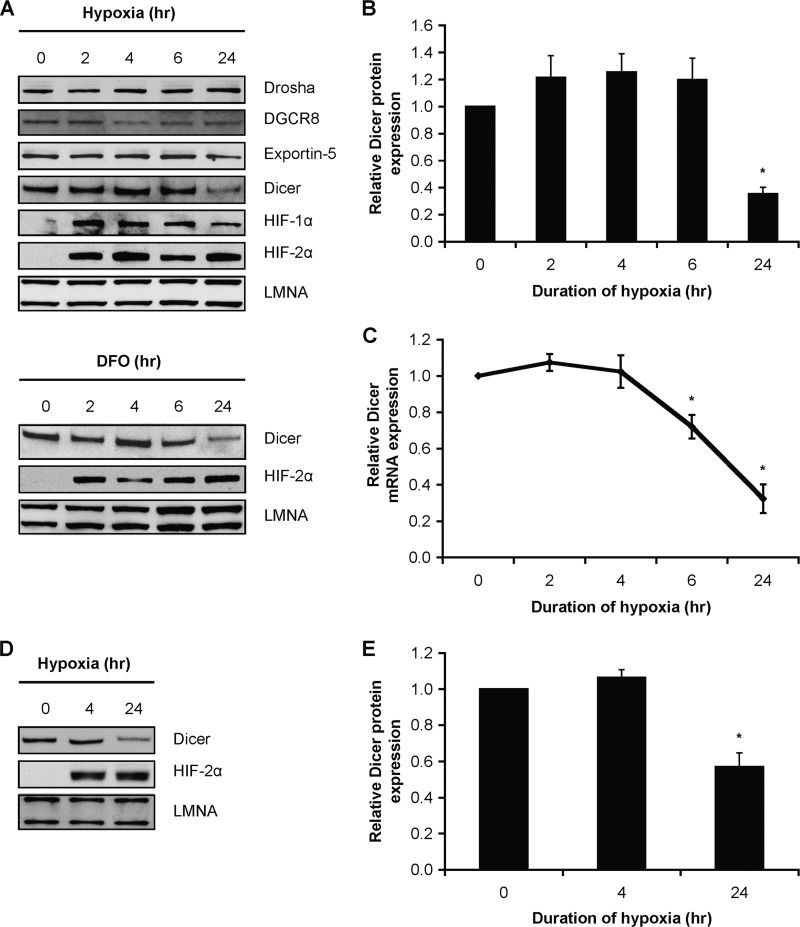
Therefore, we examined the expression of Dicer and other microRNA biogenesis/effector components (9) in hypoxic HUVEC. We observed a significant decrease in Dicer protein and mRNA expression in HUVEC under chronic hypoxia (Fig. 2, A–C) as well as in HUVEC treated with the hypoxia mimetic desferrioxamine (Fig. 2A). We confirmed this decrease in Dicer expression in a variety of primary human cell types, e.g. human microvascular endothelial cells (Fig. 2, D and E) and human aortic smooth muscle cells (data not shown) as well as cancer cell lines (i.e. HepG2; data not shown). Results indicated that Drosha expression remained unchanged, and DiGeorge syndrome critical region gene 8 (DGCR8) and Exportin-5 expression were slightly reduced (Fig. 2A and supplemental Fig. S3, A–C). Although chronic hypoxia led to a fall in Exportin-5 protein expression in HUVEC (supplemental Fig. S3C), we found that precursor microRNA species accumulated in the cytoplasm (supplemental Fig. S3D). This suggests that even though Exportin-5 can be rate-limiting in certain settings (21) we failed to define a major functional delay in the nuclear/cytoplasmic transport of pre-microRNAs, which is a known function of Exportin-5. Thus, taken together, these findings strongly support our hypothesis that the accumulation of precursor microRNAs is attributable mainly to a deficiency in Dicer, the key microRNA processing enzyme in the cytoplasm, rather than impairment of nuclear processing or nuclear-cytoplasmic export of microRNAs.
FIGURE 2.
Effect of chronic hypoxia on Dicer expression. A, representative immunoblots of hypoxic (upper panel) and desferrioxamine (DFO)-treated (lower panel) HUVEC. B, quantification of Dicer immunoblots in A. Data represent mean ± S.E. (error bars) (n = 3). C, Dicer mRNA levels in hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 4). D, representative immunoblots of hypoxic human dermal microvascular endothelial cells. E, quantification of Dicer immunoblots in D. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with 0 h.
Dicer-dependent Regulation of HIF-α Isoforms and Hypoxia-responsive Genes in Chronic Hypoxia
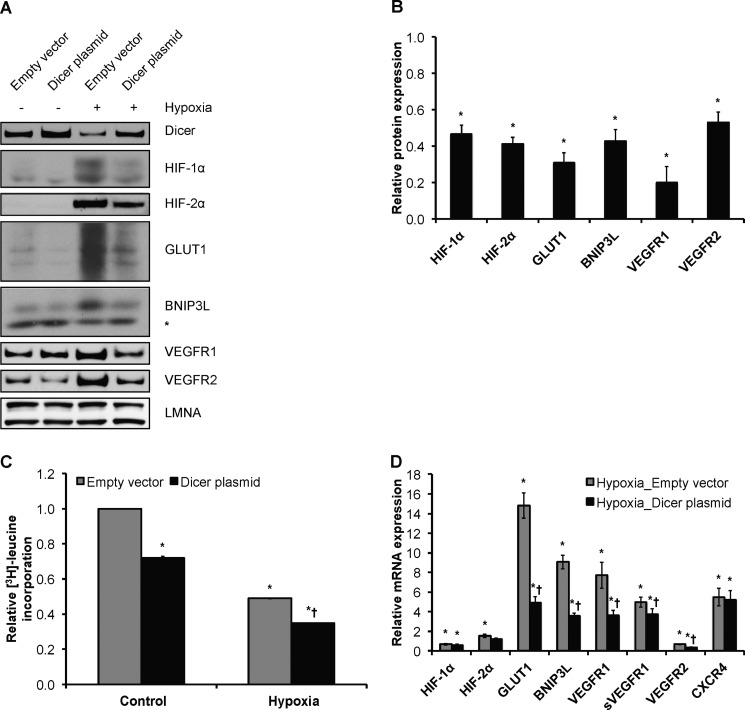
To determine the biological consequences of Dicer down-regulation on the ability of cells to sense and respond to ambient oxygen concentration, we measured the effects of forced Dicer overexpression on HIF-α isoforms and several well known hypoxia-responsive genes that promote various cellular processes, including anaerobic metabolism, cell survival, and angiogenesis such as GLUT1, BNIP3L, VEGFA, and its receptors VEGFR1 and VEGFR2 (Figs. 3–5). As in most hypoxia-responsive cells, exposure to hypoxia led to robust increases in steady-state protein levels of HIF-1α and, as known for endothelial cells, HIF-2α (Figs. 2A and 3A). Importantly, results indicated that Dicer overexpression in HUVEC resulted in the significant attenuation of endogenous HIF-1α and HIF-2α protein expression under chronic hypoxia (Fig. 3, A and B). Notably, the same effect was observed to occur for the various hypoxia-responsive/HIF target genes tested whereby the hypoxic induction of GLUT1, BNIP3L, VEGFR1, and VEGFR2 protein expression (22–25) was significantly attenuated following forced Dicer overexpression in hypoxic HUVEC (Fig. 3, A and B). Of note, hypoxia has profound effects on global translational rates (12). Although a variety of mechanisms are operative in hypoxia (26), and microRNAs are known to basally target up to 60% of coding genes (27), the relative contribution of microRNAs to the global translational repression during hypoxia has yet to be defined. Measurements of global protein synthesis with [3H]leucine incorporation indicated that Dicer overexpression decreased global protein synthesis in both normoxic and chronically hypoxic HUVEC (Fig. 3C). This is consistent with the classic role of microRNAs in global translational inhibition and underscores the relevance of functional Dicer deficiency in chronic hypoxia.
FIGURE 3.
Dicer-dependent down-regulation of HIF-α isoforms and hypoxia-responsive genes in chronic hypoxia. A, representative immunoblots of control versus 24-h hypoxic Dicer-overexpressing and control vector-transfected HUVEC. * denotes a nonspecific band. B, quantifications of HIF-1α, HIF-2α, GLUT1, BNIP3L, VEGFR1, and VEGFR2 immunoblots in A. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with hypoxia-empty vector. C, relative global protein synthesis, i.e. [3H]leucine incorporation, in control versus 24-h hypoxic Dicer-overexpressing and control vector-transfected HUVEC. A representative experiment is shown. Data represent mean ± S.E. (error bars) of duplicate measurements. * and † denote p < 0.05 compared with control-Empty vector and hypoxia-empty vector, respectively. D, mRNA levels of HIF-2α, GLUT1, BNIP3L, total VEGFR1, soluble VEGFR1 (sVEGFR1), VEGFR2, HIF-1α, and CXCR4 in control versus 24-h hypoxic Dicer-overexpressing and control vector-transfected HUVEC. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with control-empty vector and hypoxia-empty vector, respectively.
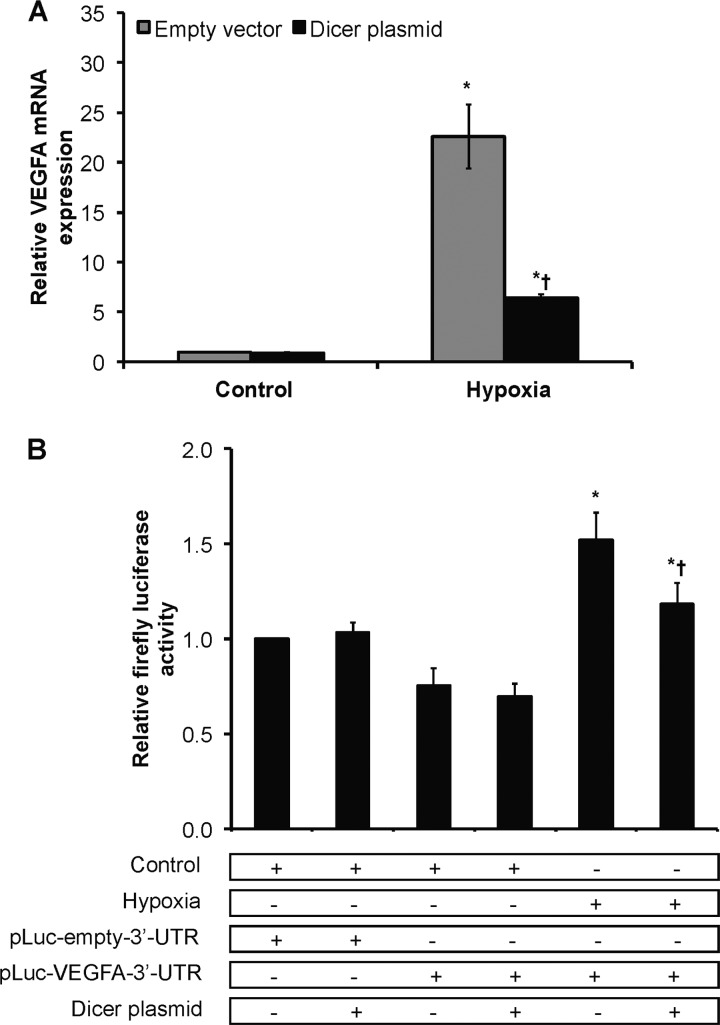
FIGURE 4.
Dicer-dependent, HIF-independent down-regulation of VEGFA in chronic hypoxia. A, VEGFA mRNA levels in control versus 24-h hypoxic Dicer-overexpressing and control vector-transfected HUVEC. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with control-empty vector and hypoxia-empty vector, respectively. B, relative firefly luciferase activity. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with control and hypoxic non-over-expressing HUVEC, respectively, with pLuc-VEGFA-3′-UTR.
FIGURE 5.
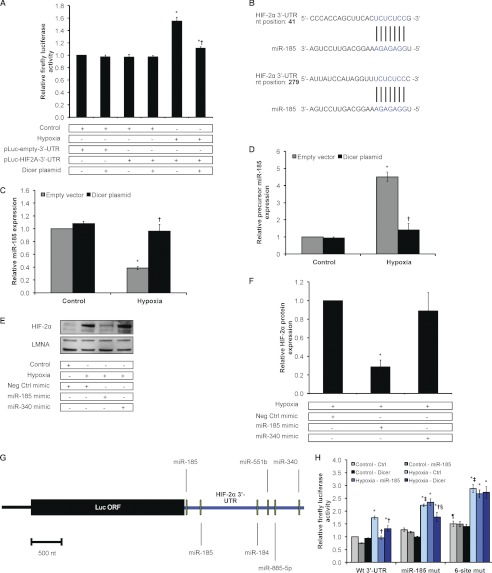
Loss of HIF-2α regulation by the Dicer-dependent miR-185 in chronic hypoxia. A, relative firefly luciferase activity. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with control and 24-h hypoxic non-over-expressing HUVEC, respectively, with pLuc-HIF2A-3′-UTR. B, miR-185 binding sites in HIF-2α 3′-UTR predicted by TargetScan and miRanda algorithms. Levels of miR-185 (C) and precursor miR-185 (D) in control versus 24-h hypoxic Dicer-overexpressing and control vector-transfected HUVEC are shown. Data represent mean ± S.E. (error bars) (n = 3). * and † denote p < 0.05 compared with control-empty vector and hypoxia-empty vector, respectively. E, representative immunoblots of control versus 24-h hypoxic HUVEC transfected with microRNA mimics. F, quantification of HIF-2α immunoblots in E. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with control-negative control (Neg Ctrl) mimic. G, schematic of chimeric luciferase-HIF-2α 3′-UTR reporter construct with the indicated binding sites for the top five hypoxia-regulated microRNAs predicted to target HIF-2α. nt, nucleotides; Luc, luciferase. H, relative firefly luciferase activity. Data represent mean ± S.E. (error bars) (n = 3). *, †, ‡, and § denote p < 0.05 compared with the corresponding control-control (Ctrl), hypoxia-control, hypoxia-control-WT 3′-UTR, and hypoxia-miR-185, respectively. ¶ denotes p < 0.05 compared with control-control of WT 3′-UTR. Wt 3′-UTR, wild-type HIF-2α 3′-UTR; miR-185 mut, wild-type HIF-2α 3′-UTR with mutations in both miR-185 binding sites; 6-site mut, wild-type HIF-2α 3′-UTR with mutations in binding sites of all five microRNAs.
Taken together, the data presented so far indicate that the loss of Dicer activity is functionally relevant to the maintenance of HIF-α expression levels and its activity as a transcription factor under chronic hypoxia. Indeed, we observed that the hypoxic/HIF induction of HIF-2α, GLUT1, BNIP3L, VEGFA, VEGFR1, and soluble VEGFR1 mRNA levels (23–25, 28) in chronic hypoxia was significantly blunted by Dicer overexpression (Figs. 3D and 4A). Similarly, the hypoxia-mediated decrease in VEGFR2 mRNA expression (23) was further accentuated by Dicer overexpression (Fig. 3D). Notably, Dicer overexpression did not affect the hypoxia-mediated changes in mRNA levels of HIF-1α or CXCR4 (Fig. 3D) (29, 30). Importantly, two recent independent studies that examined the kinetics of microRNA-mediated translational inhibition and mRNA degradation have found that mRNA degradation follows translational inhibition (31, 32). Thus, this could explain why we observed a Dicer/microRNA-mediated decrease in HIF-1α at the protein (Fig. 3, A and B) but not mRNA level (Fig. 3D). The same likely holds true for CXCR4, which has been shown to be a target for microRNAs (Fig. 3D) (33).
Two pathways may be relevant here. First, the data presented above indicate that functional loss of Dicer activity in chronic hypoxia leads to increased HIF transcriptional activity due in part to hypoxia-induced loss of microRNA repression of HIF-α expression. However, because a number of hypoxia-induced genes such as VEGFA (34) and VEGFR2 (22) are known to be profoundly regulated at the post-transcriptional level by hypoxia, especially chronic hypoxia, this raises the second possibility that loss of microRNA-mediated repression may be mechanistically relevant to the induction of specific hypoxia-responsive genes. To test this latter model, we investigated VEGFA as a prototypic gene and created chimeric luciferase reporter constructs representing the 3′-untranslated regions of VEGFA located downstream of the open reading frame of luciferase and transfected HUVEC under normoxia versus chronic hypoxia. The activity of a chimeric luciferase-VEGFA 3′-UTR construct was increased under chronic hypoxia compared with control luciferase as reported by others (34) (Fig. 4B). Importantly, Dicer overexpression significantly attenuated this induction. This indicates that microRNA-mediated mRNA degradation/translational inhibition is involved in the regulation of hypoxia-inducible genes such as VEGFA, which is a known target for microRNAs (35).
Our finding that a broad number of microRNAs are affected by functional deficiency of Dicer in chronic hypoxia is significant given that most studies performed to date have stressed the single microRNA-single target mRNA paradigm. Importantly, in silico analyses identified 36 (i.e. 53%) hypoxia-regulated microRNAs that can putatively target nine prototypical hypoxia-regulated genes such as HIF-α subunits, VEGFA, VEGFR1, VEGFR2, GLUT1, and BNIP3L among others (data not shown). This indicates that a large proportion of hypoxia-regulated microRNAs can potentially regulate genes involved in the cellular response to hypoxia. Consistent with the observation that a number of microRNAs target specific mRNAs, the HIF-1α isoform has been shown recently to be negatively regulated directly by a number of microRNAs at the post-transcriptional level such as miR-20b, miR-199a, the miR-17–92 cluster, and miR-519c (36). In our microarray analysis, miR-519c and miR-92a were differentially regulated by hypoxia at p < 0.05. In addition, a recent study has identified miR-424 as a hypoxia-inducible microRNA (supplemental Fig. S1, A–C) that indirectly stabilizes HIF-1α by targeting and destabilizing CUL2, which is involved in HIF-1α degradation (37). Furthermore, the constitutively expressed HIF-1β subunit has also been shown to be a direct target of miR-107 (38).
However, in contrast to HIF-1α and HIF-1β, no studies have been performed to the authors' knowledge that examine the targeting of HIF-2α by microRNAs, especially under hypoxia. Thus, to investigate Dicer-mediated microRNA regulation of HIF-2α, we first transfected chimeric luciferase reporter constructs representing the 3′-untranslated regions of HIF-2α located downstream of the open reading frame of luciferase into HUVEC under normoxia versus chronic hypoxia. Similar experiments were performed using luciferase HIF-1α 3′-UTR constructs for comparison. Importantly, hypoxia led to increased and decreased reporter activity for the luciferase HIF-2α and HIF-1α constructs, respectively (Fig. 5A and supplemental Fig. S4A), which is consistent with the reported differential effects of hypoxia on mRNA levels for HIF-2α and HIF-1α in HUVEC (Fig. 3D) (29) as well as other cell types (39). Importantly, Dicer overexpression in the setting of hypoxia significantly blunted the luciferase activity of chimeric HIF-2α and HIF-1α 3′-UTR reporters (Fig. 5A and supplemental Fig. S4A). These results strongly suggested that HIF-2α is subject to Dicer-mediated microRNA regulation.
Loss of HIF-2α Regulation by the Dicer-dependent miR-185 in Chronic Hypoxia
Given the above evidence, we performed an in silico analysis on our hypoxia-regulated microRNAs to determine the top candidate microRNAs that can target HIF-2α. The criteria used included microRNA conservation, the number of target sites in HIF-2α, target site conservation, identification by at least TargetScan and miRanda prediction algorithms, and expression levels as measured by microarray and qRT-PCR. MiR-185 is a highly conserved microRNA species that is predicted to have two target binding sites in the human HIF-2α 3′-UTR (Fig. 5B). Notably, we observed that the down-regulation of miR-185 under chronic hypoxia is Dicer-dependent: mature miR-185 levels fell under chronic hypoxia (Fig. 1B), whereas its precursor species accumulated (Fig. 1, C and D). Importantly, we found that Dicer overexpression under hypoxic conditions rescued mature miR-185 levels (Fig. 5C) and restored precursor levels to normoxic levels (Fig. 5D). Thus, the hypoxic regulation of miR-185 is Dicer-dependent. Importantly, overexpression of miR-185 in hypoxic HUVEC resulted in the reduction of endogenous HIF-2α protein (Fig. 5, E and F) and mRNA levels (supplemental Fig. S4B). In contrast, although miR-340 was predicted to target HIF-2α, it failed to modify HIF-2α expression alone. This underscores the importance of direct experimentation versus prediction algorithms. We utilized firefly luciferase-HIF-2α 3′-UTR chimeric reporters that contained either wild-type HIF-2α 3′-UTR, wild-type 3′-UTR with mutations in both miR-185 binding sites, or wild-type 3′-UTR with multiple microRNA binding site mutations (Fig. 5, G and H, and supplemental Fig. S4, C–H). Notably, overexpression of both miR-185 and Dicer were able to attenuate the hypoxic induction of wild-type construct expression (Fig. 5H). In contrast, for the miR-185 luciferase-HIF-2α mutant construct, Dicer, but not miR-185 overexpression, was able to attenuate the hypoxic induction of construct expression (Fig. 5H). Lastly, expression of the six-site mutant construct was not affected by either miR-185 or Dicer overexpression, suggesting that additional Dicer- and/or microRNA-independent pathways may also be operative in HIF-2α post-transcriptional regulation (Fig. 5H).
Overall, these results indicate that miR-185 targets HIF-2α. In chronic hypoxia, the decrease in miR-185 expression maintains HIF-2α induction. Significantly, this is the first description that the hypoxia-induced attenuation of Dicer function is critically important in maintaining the expression of HIF-α subunits via the regulation of HIF-α-targeting microRNAs. Ours is also the first study to identify the direct regulation of HIF-2α by a specific microRNA, i.e. miR-185. This is especially valuable because there is a growing consensus that HIF-2α has unique roles in hypoxia signaling that are distinct from HIF-1α, especially in endothelial cells (40) and cancers such as VHL-associated renal cell carcinoma (41).
Efficacy of Dicer-dependent Versus Dicer-independent RNA Interference in Chronic Hypoxia
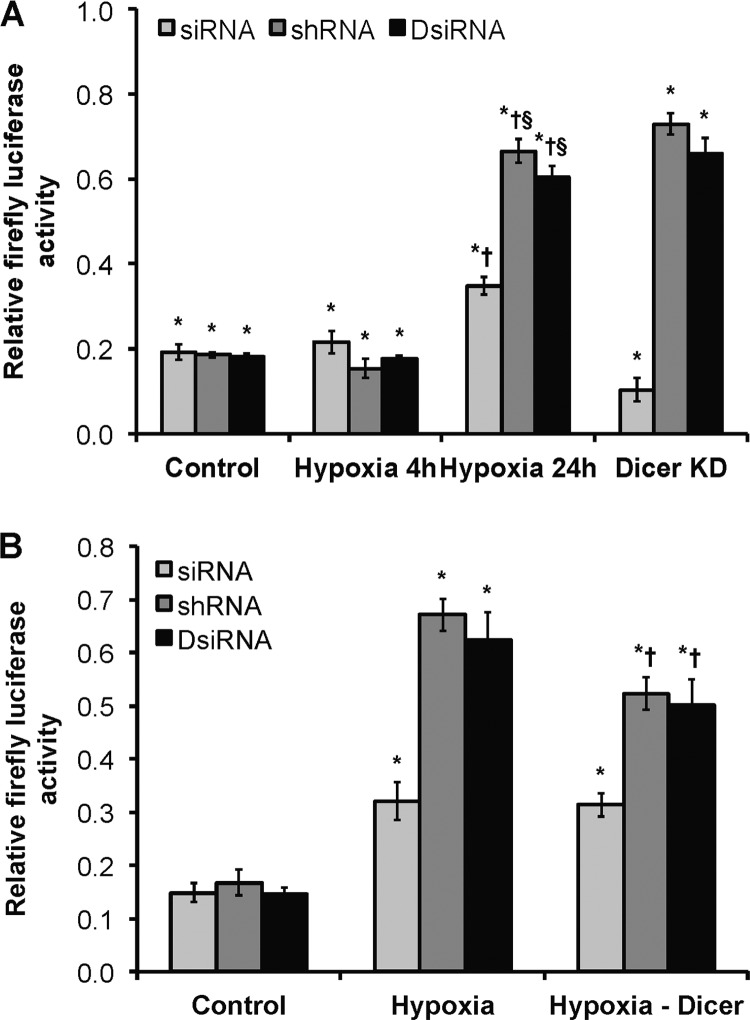
Next, we examined the effect of hypoxic Dicer down-regulation on different RNAi knockdown methods. This concept is relevant to the application of RNAi-based therapeutics in disease, especially in the setting of cancer and cellular hypoxia. We designed a luciferase assay whereby the specific knockdown of a firefly luciferase reporter in HUVEC was performed using a Dicer-independent method (i.e. siRNA) versus Dicer-dependent knockdown methods (i.e. shRNA or DsiRNA) (42) (Fig. 6A). Results indicated that all three approaches were effective in knocking down luciferase expression under normoxia and acute hypoxia. However, the efficiencies of Dicer-dependent knockdown approaches were markedly reduced under chronic hypoxia compared with normoxia. Similar results were obtained in normoxic cells when Dicer had been knocked down with siRNA (Fig. 6A). Importantly, the observed phenotype was significantly rescued when Dicer was overexpressed in chronically hypoxic HUVEC (Fig. 6B). Similar results were obtained when the experiments were performed in HepG2 cells (data not shown). Taken together, these results strongly indicate that functional Dicer activity is reduced under chronic hypoxia. These findings have implications for RNA interference therapies and suggest that impairment of global pre-microRNA processing in disease settings may differentially affect exogenous Dicer-dependent and -independent approaches.
FIGURE 6.
Effect of chronic hypoxia on Dicer-dependent versus Dicer-independent RNA interference. A, relative firefly luciferase activity. * and † denote p < 0.05 compared with corresponding non-silencing control and control, respectively. § denotes p < 0.05 compared with 24-h hypoxia-siRNA. Dicer KD, Dicer knockdown in control HUVEC. B, relative firefly luciferase activity. * and † denote p < 0.05 compared with corresponding control and hypoxia, respectively. Data represent mean ± S.E. (error bars) (n = 3).
Effects of Chronic Hypoxia on RISC Activity and Argonaute Protein Expression
Although Dicer is a functional component of the RISC (10), the ∼2-fold decrease in efficiency of the siRNA/Dicer-independent knockdown and the significant but incomplete rescue following Dicer overexpression under chronic hypoxia suggested that microRNA activity is also affected by additional factors in chronic hypoxia. Data presented here indicate that Ago1, Ago2, and TRBP protein and mRNA levels decreased in chronic hypoxia at least in the vascular endothelium (supplemental Fig. S5, A–G). Contrasting results have been described in vascular smooth muscle cells (43). A recent report suggests that prolyl 4-hydroxylation regulates Ago2 stability (44), although the physiologic role of oxygen dependence of this post-translational modification is unclear. Therefore, we performed additional functional assays in hypoxic HUVEC. We chose to assay RISC activity under conditions where the mature microRNA (i.e. let-7a) being functionally assessed did not change in extracts of hypoxic versus normoxic HUVEC (supplemental Fig. S1A). Correspondingly, we observed a ∼56% decrease in let-7a-dependent RISC activity in the setting of chronic hypoxia (supplemental Fig. S5H). These results suggest that hypoxia down-regulates RISC expression and function at least in the vascular endothelium.
Effects of Chronic Hypoxia on Dicer mRNA and Protein
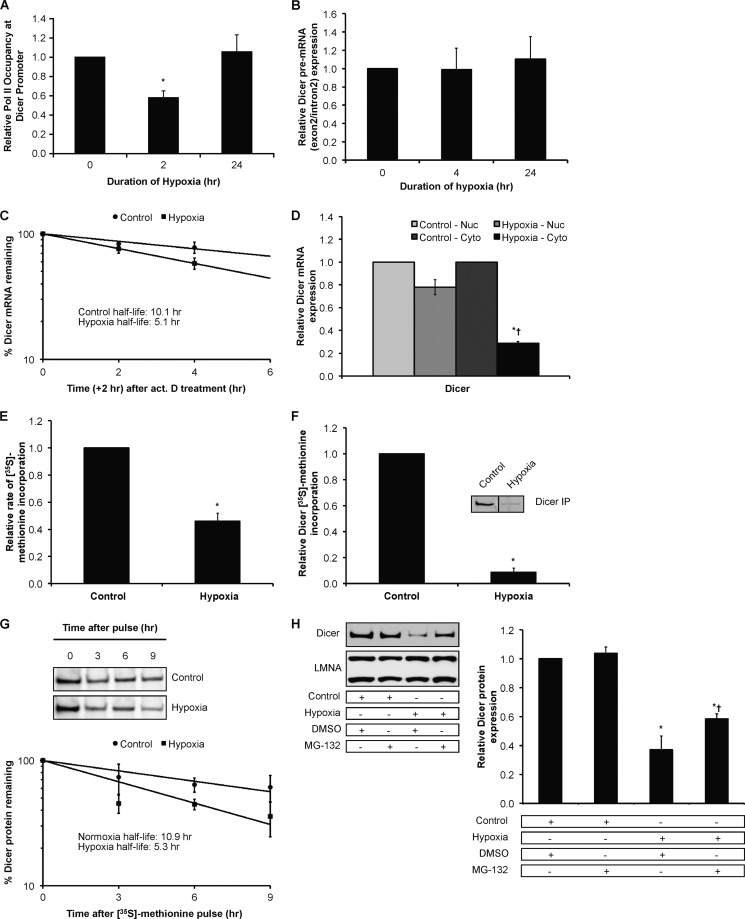
To determine the mechanisms underlying the decrease in Dicer mRNA under hypoxia, we performed RNA polymerase II ChIP experiments and precursor mRNA (pre-mRNA) measurements to ascertain whether Dicer is regulated at the transcriptional level by hypoxia. Results indicated that polymerase II occupancy at the major Dicer promoter in HUVEC (Fig. 7A) as well as downstream genomic regions shared in common between all Dicer mRNA variants (data not shown) decreased with acute (2-h) but not chronic (24-h) hypoxia. Measurements of Dicer pre-mRNA, which reflects transcription, indicated that at 4 and 24 h the fall in Dicer transcription was no longer evident (Fig. 7B). Importantly, mRNA half-life experiments performed using actinomycin D showed that Dicer mRNA half-life was decreased under hypoxia (∼5.1 h) versus normoxia (∼10.1 h) (Fig. 7C).
FIGURE 7.
Effect of chronic hypoxia on Dicer mRNA and protein. A, relative RNA polymerase II (Pol II) occupancy at the Dicer proximal promoter in hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with 0 h. B, Dicer pre-mRNA measurements in hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 4). C, Dicer mRNA half-life measurements in control versus hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 3). D, Dicer mRNA levels in nuclear (Nuc) versus cytoplasmic (Cyto) fractions of control versus 24-h hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 3). * and † denote statistical significance (p < 0.05) compared with hypoxia-nuclear and control-cytoplasmic, respectively. Relative rate of global protein synthesis (E) and relative Dicer protein synthesis (F) in normoxic versus 24-h hypoxic HUVEC measured using [35S]methionine incorporation. A representative gel is shown for F. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with normoxia. G, Dicer protein half-life measurements in control versus hypoxic HUVEC. Data represent mean ± S.E. (error bars) (n = 3). A representative gel from each condition is shown. H, representative immunoblots of control versus 24-h hypoxic HUVEC treated with MG-132 (left panel). Quantification of Dicer immunoblots is shown in the right panel. Data represent mean ± S.E. (error bars) (n = 3). * and † denote statistical significance (p < 0.05) compared with control-DMSO and hypoxia-DMSO, respectively. IP, immunoprecipitation.
Additionally, Exportin-5 protein expression fell in chronic hypoxia (Fig. 2A and supplemental Fig. S3C). A recent study reported that Exportin-5 is a post-transcriptional regulator of Dicer expression that can mediate the nuclear-cytoplasmic export of Dicer mRNA (45). Therefore, we wanted to assess whether the decrease in Exportin-5 expression in chronic hypoxia plays a role in the hypoxic down-regulation of steady-state Dicer mRNA. To do so, we measured Dicer mRNA levels in nuclear versus cytoplasmic fractions of HUVEC. The results indicated no preferential nuclear accumulation of Dicer mRNA in chronic hypoxia (Fig. 7D) and that the down-regulation of Dicer mRNA occurred predominantly in the cytoplasm. These results suggest that changes in nuclear-cytoplasmic export due to decreases in Exportin-5 expression in chronic hypoxia do not play a significant role in the down-regulation of Dicer mRNA. Taken together, these results indicate that Dicer mRNA levels decrease in hypoxic HUVEC primarily due to post-transcriptional mechanisms in the cytoplasm.
We and others have provided evidence for translational and post-translational regulation of Dicer (46, 47). For example, alternative promoters and exonic splicing variants within human Dicer mRNA transcripts, especially within the 5′-UTR, have profound effects on translational efficiency (46). 5′-Rapid amplification of cDNA ends indicated that the major mRNA species in normoxic and hypoxic HUVEC are inefficient substrates for the translational machinery at least under basal conditions (data not shown). Indeed, measurement of global protein synthesis rates and Dicer-specific protein synthesis with [35S]methionine pulse experiments indicated that although hypoxia decreased the rate of global protein synthesis by ∼50% (12) (Fig. 7E) Dicer protein synthesis decreased to ∼10% of the normoxic rate under chronic hypoxia (Fig. 7F). Furthermore, Dicer-specific [35S]methionine pulse-chase experiments indicated that Dicer protein half-life was decreased under hypoxia (∼5.3 h) versus normoxia (∼10.9 h) (Fig. 7G). The proteasomal inhibitor MG-132 blunted the decrease in Dicer protein (Fig. 7H), suggesting that the decreased stability/enhanced degradation of Dicer protein under hypoxic conditions is dependent in part on a functional proteasome. RISC components TRBP and Ago1/2 are known to contribute to Dicer stability (10). Their decreased expression in hypoxia may also be functionally relevant to decreases in Dicer expression. Therefore, these results together indicate that decreased mRNA levels, decreased mRNA translation, and impaired protein stability all contribute to decreases in Dicer expression under chronic hypoxia.
Effects of Chronic Hypoxia on Dicer Expression and Function in Mouse Organs
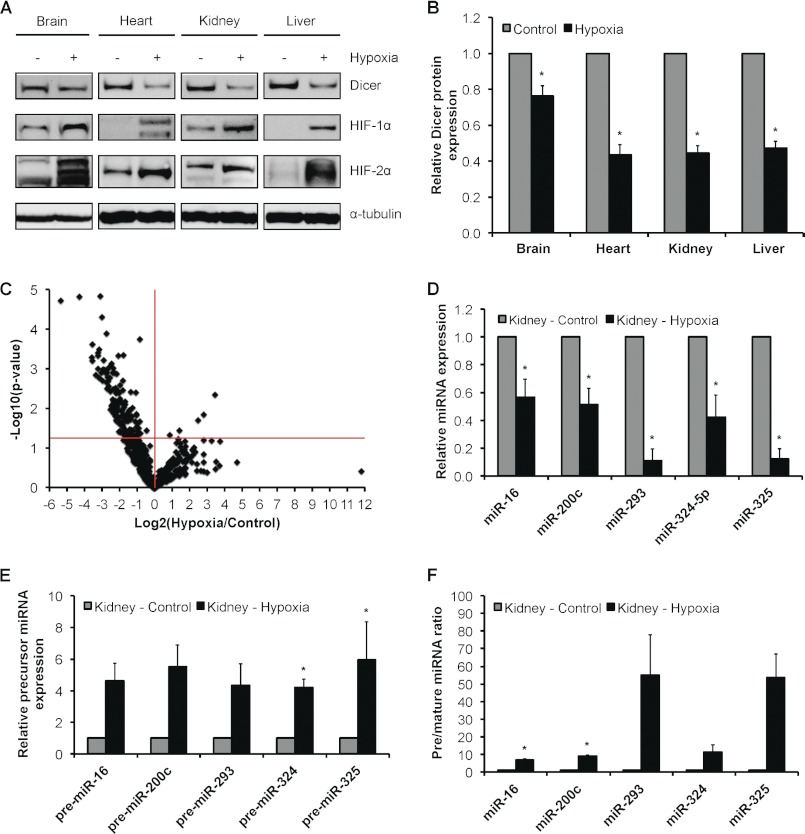
Dicer is key for survival. It has been shown that Dicer knock-out is embryonic lethal (48). In addition, severe morbidity and mortality have been reported for multiple cell type/tissue-specific Dicer knock-outs (49–54). Thus, it is especially challenging to use these models to examine the in vivo relationship between hypoxia and Dicer/microRNA biology. In order to extend and establish the in vivo relevance of our major findings, we first measured Dicer expression in normoxic versus chronically hypoxic (8% O2 for 24 h) mouse organs (Fig. 8, A and B). Consistent with our in vitro observations, results indicated that Dicer protein expression is significantly reduced under chronic hypoxia in multiple organs, including brain, heart, kidney, and liver. We then asked whether this down-regulation of Dicer has any biological impact on microRNA expression and processing. Thus, we used qRT-PCR to obtain a profile of microRNA expression in normoxic versus chronically hypoxic mouse kidneys. In total, 726 mature mouse microRNA species were measured, and volcano plot analysis indicated that 148 species were regulated by hypoxia at p < 0.05 (Fig. 8C and supplemental Table S2). Importantly, 143 (i.e. 97%) of these hypoxia-regulated microRNAs were down-regulated, whereas five were up-regulated. This observation is significant because it bears a striking resemblance to the trend we identified in vitro whereby 88% of hypoxia-regulated microRNAs were down-regulated under chronic hypoxia. Interestingly, in silico analysis showed that a significant number, i.e. 62 (42%), of the hypoxia-regulated mouse kidney microRNAs are predicted to target nine prototypic hypoxia-regulated genes (data not shown), five of which are shown to represent down-regulated microRNAs (Fig. 8D). Together with our in vitro screen, these results strongly suggest that a large proportion of hypoxia-regulated microRNAs can regulate genes involved in the cellular response to hypoxia in a tissue/cell type-dependent manner. Next, to show that the decrease in mature microRNA expression is Dicer-dependent, we measured the precursor levels of five representative microRNA species that fall in hypoxic mouse kidneys and expressed the ratios of pre/mature levels (Fig. 8, E and F). As in our in vitro studies, we observed the accumulation of precursors even for microRNAs for which mature expression did not change at steady state (supplemental Fig. 1D). Thus, we recapitulated in vivo the down-regulation of Dicer expression and function in chronic hypoxia that we observed in vitro.
FIGURE 8.
Effect of chronic hypoxia on Dicer expression and function in mouse organs. A, representative immunoblots of control versus 24-h hypoxic mouse brain, heart, kidney, and liver. B, quantification of Dicer immunoblots in A. Data represent mean ± S.E. (error bars) (n = 5/6). * denotes p < 0.05 compared with normoxia. C, volcano plot of 582 detectable microRNAs in 24-h hypoxic versus control mouse kidney as measured by qRT-PCR profiling (n = 4). Expression levels of representative down-regulated microRNAs (D) and corresponding precursor microRNAs (E) and the ratio of precursor microRNAs to their mature counterparts (F) are shown. Data represent mean ± S.E. (error bars) (n = 4). * denotes statistical significance (p < 0.05) compared with kidney-control.
Down-regulation of Dicer in Chronic Hypoxia Is VHL Protein-dependent
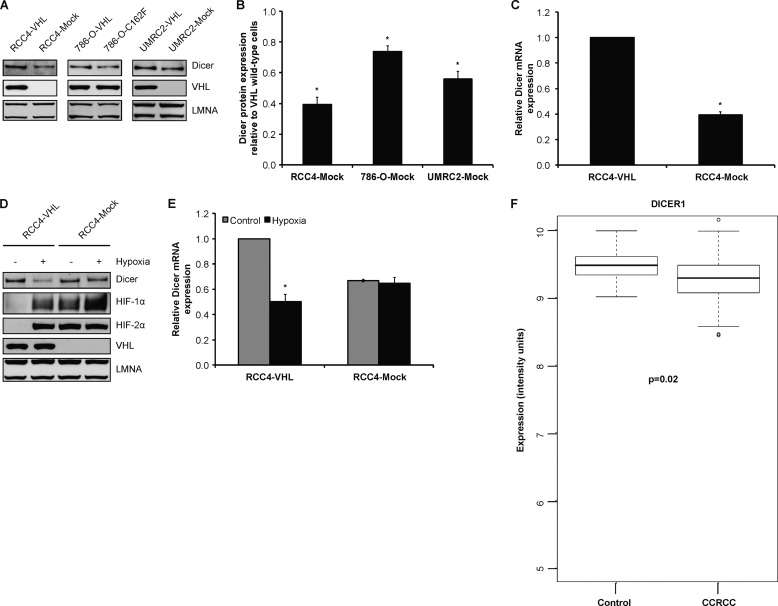
The VHL protein regulates gene transcription by ubiquitylating HIF-α subunits and targeting them for proteasomal degradation in an oxygen-dependent manner (55). Specifically, VHL functions as a substrate recognition component of an E3 ubiquitin ligase, ECV (Elongin B/C-Cullin 2-VHL), which directly binds oxygen-dependent prolyl hydroxylated HIF-α for ubiquitylation and subsequent destruction via the 26 S proteasome (55). Consequently, cells that are deficient in VHL functional activity constitutively express hypoxia-inducible genes. Functional loss of VHL results in the hereditary cancer predisposition of VHL disease and underlies the vast majority of sporadic CCRCC, the predominant form of kidney cancer. Common to most cancer-causing VHL mutations is a failure in down-regulating HIF-α. Therefore, we sought to define the relationship between VHL status in CCRCC and Dicer expression both in vitro and in vivo. Measurement of Dicer expression in three different VHL-deficient CCRCC cell lines (e.g. RCC4, 786-O, and UMRC2) that had been stably reconstituted with either wild-type or mutant VHL showed that Dicer protein levels were lower in VHL-deficient (RCC4 and UMRC2) or mutant (786-O-C162F mutant) cells relative to their wild-type VHL-expressing counterparts (Fig. 9, A and B). Lower Dicer mRNA levels were also observed in RCC4 VHL-deficient versus VHL wild-type cells (Fig. 9C). Moreover, measurement of Dicer expression in normoxic versus chronically hypoxic RCC4 showed that chronic hypoxia led to decreased Dicer protein and mRNA levels in VHL wild-type but not VHL-deficient RCC4 cells (Fig. 9, D and E). To understand the in vivo implications of this work, we performed a microarray analysis of 105 CCRCC tumor samples versus 12 non-diseased renal cortex controls as we have described previously (19) and observed that Dicer mRNA is significantly lower in CCRCC tumors (by ∼20%) (Fig. 9F). Taken together, these findings indicate that Dicer expression is at least in part dependent on VHL.
FIGURE 9.
Down-regulation of Dicer in chronic hypoxia is VHL-dependent. A, representative immunoblots of VHL-deficient RCC4, 786-O, and UMRC2 cells reconstituted with wild-type or mutant VHL. B, quantification of Dicer immunoblots in RCC4, 786-O, and UMRC2. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with the corresponding wild-type VHL-reconstituted condition. C, Dicer mRNA levels in RCC4-VHL and RCC4-Mock cells. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with RCC4-VHL. Representative immunoblots (D) and Dicer mRNA levels (E) in control versus 24-h hypoxic RCC4-VHL and RCC4-Mock cells are shown. Data represent mean ± S.E. (error bars) (n = 3). * denotes p < 0.05 compared with corresponding control. F, microarray analysis of Dicer mRNA levels in CCRCC tumors (n = 105) versus non-diseased renal cortex controls (n = 12, p = 0.02).
Down-regulation of Dicer mRNA Expression in Chronic Hypoxia Is Not HIF-dependent
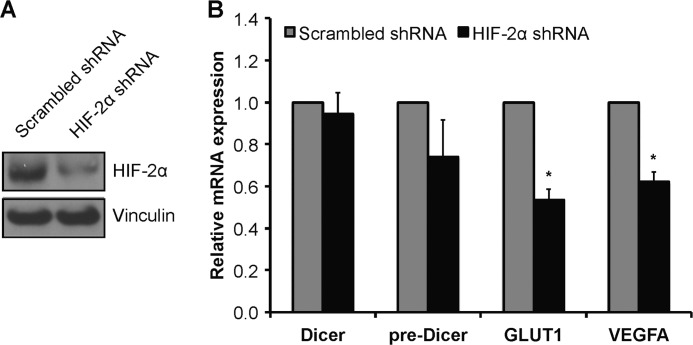
Given that VHL is a critical regulator of HIF-α, we wanted to test whether the down-regulation of Dicer in hypoxia is HIF-dependent. In its classical role as a transcription factor, HIF could bind directly to the Dicer promoter to mediate down-regulation of Dicer transcription as we and others have shown to occur for several genes, including CDH1/E-cadherin and RABEP1/Rabaptin-5 (56–60). Thus, we performed an extensive in silico analysis, including the use of the transcriptional regulatory element database (TRED) (61, 62), to determine whether hypoxia response elements exist in the Dicer promoter, i.e. ±1500 bp of the transcription start site, for the most commonly expressed Dicer mRNA variant in human, mouse, and rat (46). Results indicated that there are no hypoxia response elements in sequences upstream or downstream of the Dicer transcription start site, especially in the proximal promoter (data not shown). However, given that functional hypoxia response elements can operate at large distances (e.g. human EPO gene (63)), we directly assessed HIF dependence. We assessed Dicer expression in VHL- and HIF-1α-deficient, HIF-2α-expressing 786-O clones that had been stably transfected with either an HIF-2α-targeting shRNA or a scrambled shRNA (Fig. 10A) (7, 14). Notably, HIF-2α is the predominant HIF-α isoform in endothelial cells, and it is also the only HIF-α isoform expressed in 786-O cells. Results indicated that Dicer mRNA and pre-mRNA (Fig. 10B) expression levels were not affected by cellular HIF-α status. In contrast, GLUT1 and VEGFA mRNA expression was blunted in the HIF-2α knockdown cells (Fig. 10B). Taken together, these findings indicate that the down-regulation of Dicer mRNA in hypoxia is not HIF-dependent. In addition to HIF-mediated effects, VHL can also influence gene expression via post-transcription mechanisms involving RNA-binding proteins. Examples include HuR, which regulates VEGFA mRNA stability (64), and heterogeneous nuclear ribonucleoprotein A2, which regulates GLUT1 expression (65), both in a VHL-dependent manner. Additionally, HIF can down-regulate gene expression via non-transcriptional mechanisms (66). For example, HIF has been shown to mediate the cytoplasmic proteasomal degradation of Foxp3 (67). Future studies will be required to elucidate the mechanism(s) underlying the VHL dependence of Dicer expression.
FIGURE 10.
Down-regulation of Dicer mRNA in chronic hypoxia is not HIF-dependent. Representative immunoblots (A) and mRNA levels (B) of Dicer mRNA, Dicer pre-mRNA, GLUT1, and VEGFA in 786-O clones that have been stably transfected with either a HIF-2α-targeting shRNA or a scrambled shRNA are shown. Data represent mean ± S.E. (error bars) (n = 5). * denotes statistical significance (p < 0.05) compared with scrambled shRNA.
DISCUSSION
Cellular hypoxia regulates gene expression through transcriptional and post-transcriptional mechanisms. Post-transcriptional regulation of hypoxia-responsive/HIF target genes, especially mRNA stabilization, is a key component of an integrated hypoxic response. We have found for the first time that the microRNA pathway is functionally integrated with the cellular hypoxia response pathway, especially in the vascular endothelium. Specifically, chronic hypoxia leads to the down-regulation of Dicer function, which has profound effects on the microRNA pathway both in vitro and in vivo. Especially relevant are the generalized decrease in expression of mature microRNAs and the accumulation of precursor microRNA species in chronic hypoxia. Importantly, the hypoxic down-regulation of Dicer serves as a key mechanism that aids in the maintenance of HIF-α induction in chronic hypoxia as well as the induction of numerous key hypoxia-responsive/HIF target genes. Dicer expression has been observed to be decreased in hypoxic pulmonary artery smooth muscle cells (43) as well as rat lungs (68). However, the biological significance of such a decrease has not been appreciated until the current work. Here, we show that the loss of microRNA-mediated repression is an important mechanism that is operative in the cellular response to hypoxia. We found that the decrease in Dicer function in the hypoxic vascular endothelium is mediated at multiple levels. Hypoxia led to a transient decrease in Dicer gene transcription. Hypoxia also led to decreases in Dicer mRNA stability and in Dicer protein stability. A functional proteasome was necessary for decreases in Dicer protein expression. We also provide evidence that Dicer is regulated in hypoxia via VHL-dependent processes. Moreover, the decrease in Dicer transcription is HIF-independent at least in VHL-deficient CCRCC cells.
Evidence presented here indicates that even though distinct mRNA species may be differentially up- or down-regulated in hypoxia the loss of Dicer-mediated microRNA repression is nonetheless functionally relevant and important to a broad variety of hypoxia-responsive genes. For instance, stabilization of VEGFA mRNA contributes to the observed increases in VEGFA mRNA expression in hypoxia, especially after the initial HIF-dependent transcriptional activation. As an example, hypoxia leads to increases in VEGFA mRNA and translational efficiency in chronically hypoxic HUVEC (7). We report here that the hypoxic induction of VEGFA mRNA is blunted if Dicer is overexpressed in hypoxic endothelial cells. Moreover, chimeric RNAs that represent heterologous reporter proteins ligated to the human VEGFA 3′-UTR exhibit increased expression in hypoxic HUVEC. This increase, which is dependent on VEGFA sequences, is blunted when Dicer is overexpressed. As with VEGFA, we observed that the hypoxic induction of GLUT1, BNIP3L, and VEGFR1 mRNA and protein is significantly impaired when Dicer is overexpressed in hypoxic endothelial cells.
On the other hand, as reported previously by others, VEGFR2 mRNA decreases whereas VEGFR2 protein increases in hypoxic endothelial cells (22, 23). Interestingly, Dicer overexpression in chronic hypoxia leads to further decreases in both VEGFR2 protein and especially mRNA. We observed the same effect with HIF-1α mRNA (29, 39) and luciferase-HIF-1α-3′-UTR construct expression in chronic hypoxia, which are further decreased when Dicer is overexpressed. Thus, we take these data to indicate that even though distinct mRNA species may be differentially up- or down-regulated in hypoxia the loss of Dicer-mediated microRNA repression is nonetheless functionally relevant and important to a broad variety of hypoxia-responsive genes.
We found that the loss of microRNA function is relevant for the post-transcriptional induction and/or maintenance of both HIF-α isoforms, especially HIF-2α. HIF-2α is directly targeted by Dicer-dependent microRNAs. Regulation of the human HIF-2α 3′-UTR by miR-185 has not been reported previously. Significantly, we show not only that HIF-2α is a direct target of miR-185 in blood vessels in normoxia but also that the processing of miR-185 is impaired in chronic hypoxia, which prevents it from suppressing HIF-2α function. miR-185 was down-regulated in HUVEC in chronic hypoxia, whereas miR-185 precursors accumulated. Chimeric RNAs that represent heterologous reporter proteins ligated to the human HIF-2α 3′-UTR were increased in expression in hypoxic HUVEC. Mutation of the miR-185 target sites in the 3′-UTR of HIF-2α blunted induction of the reporter. Forced overexpression of Dicer or miR-185 in HUVEC under chronic hypoxia blunted HIF-2α mRNA, protein, and chimeric 3′-UTR reporter induction. Our work underscores that predicted 3′-UTR binding sites for microRNAs need to be validated. For example, although miR-340 fell in hypoxia and the precursor accumulated, mutation of the target site or re-expression of miR-340 did not affect HIF-2α expression.
The broad pattern effect of chronic hypoxia on an important number of mature microRNAs and the accumulation of precursor microRNA species is a new finding. Importantly, the accumulation of precursors even occurs for mature microRNAs that did not exhibit a significant change in expression. This observation suggests that Dicer is functionally deficient in chronic hypoxia. Except for microRNAs that are known to be transcriptionally up-regulated by hypoxia (e.g. miR-210) (8, 20), it is a challenge to explain why all mature microRNAs do not fall with chronic hypoxia. One potential contributing factor is the existence of Dicer-independent microRNA biogenesis pathways (69–71). Importantly, future studies will be required to better understand the relationship between steady-state microRNA expression and target mRNA abundance, especially in disease settings such as hypoxia. The inherent stability of mature microRNAs under normal conditions (9) may be relevant here. Interestingly, a recent kinetic analysis indicated that the rate of microRNA decay can be accelerated through target regulation in a manner that is dependent on mRNA target concentration (72). This newer view would argue that those microRNAs that target hypoxia-inducible mRNAs (e.g. HIF-2α and VEGFA) will be selectively destabilized in hypoxia due to increased mRNA target concentration (72, 73). We hypothesize that hypoxic increases in hypoxia-inducible/HIF target gene expression driven initially by enhanced transcriptional HIF activity in hypoxia contribute to the preferential down-regulation of unique microRNA species such as those observed in this study, which then sets up a reinforcing post-transcriptional stabilization of these same hypoxia-inducible/HIF target genes.
Notably, we observed that there was no significant overlap between the hypoxia-regulated microRNAs detected in vitro versus those detected in vivo. In addition to cross-species differences in microRNA profiles, this discrepancy is likely accounted for by the dynamic and integrative effects of several factors. First, in vivo organs like the kidney consist of multiple cell types (e.g. podocytes, a variety of tubular epithelial cell types, mesangial cells, and a small contribution of endothelial cells, etc.) as compared with a single cell type (e.g. HUVEC) studied in vitro, and different cell types often exhibit unique basal microRNA profiles (74). In addition, basal exposure levels of oxygen are very different in the two settings. HUVEC cultured in vitro are exposed to a pO2 of 160 mm Hg, whereas the pO2 in in vivo tissues is much lower (75). Different cell types in an organ may respond differently to changes in pO2. Moreover, unlike in vitro hypoxia, hypoxia in vivo is associated with important concurrent physiological changes, e.g. blood flow, oxidant stress injury, etc., all of which may regulate microRNA expression. Thus, the interplay between multiple factors and their combined net effect in the in vivo setting of hypoxia likely account for the differences observed between in vitro and in vivo hypoxia. However, it is important to remember that even given these differences our major finding that Dicer expression and activity are down-regulated in hypoxia was observed both in vitro and in vivo. Additionally, a significant proportion of hypoxia-regulated microRNAs were predicted to target hypoxia-inducible genes both in vitro (53%) and in vivo (42%). Thus, given that a single mRNA can be targeted by multiple microRNAs and that a single microRNA can target multiple mRNAs (76), this observation supports our model that the initial transcriptional up-regulation of hypoxia-inducible/HIF target genes contributes to the preferential down-regulation of both common and unique tissue/cell type-dependent microRNA species, which then sets up a reinforcing post-transcriptional stabilization of these same hypoxia-inducible genes.
Given our findings in this current study, it would be interesting to test the hypothesis that other cellular stresses/stimuli also lead to the down-regulation of Dicer and that it is functionally relevant to the biology of that particular stress/stimulus. Indeed, it has been reported that Dicer expression is down-regulated by multiple stresses, including reactive oxygen species, phorbol esters, Ras oncogene, and IFN-α (77). Importantly, however, the biological significance of Dicer down-regulation in these cases has not been investigated. Thus, the functional relevance of such changes in Dicer expression, especially in the context of the biology of those stresses, needs to be examined in future studies. As in the current work, assessments of the ratios of precursor to mature microRNA species may be helpful in these studies.
Notably, most microRNA studies performed to date have focused on the “single microRNA-single target” paradigm, including in hypoxia, with preferences toward up-regulated microRNA species. For example, miR-210 (8) and miR-424 (37) are both hypoxia-inducible microRNAs that play important roles in hypoxia biology, especially miR-424, which is involved in the indirect regulation of HIF-α isoforms (37). Importantly, these studies provide important mechanistic insight into hypoxia biology. However, in this current study, we have shown for the first time that the microRNA pathway is functionally integrated with the cellular hypoxia response pathway on a higher lever that goes beyond the single microRNA-single target paradigm. Specifically, Dicer, the key cytoplasmic microRNA processing enzyme, is down-regulated in chronic hypoxia. This reduction in expression results in impaired Dicer function, which we argue participates in the adaptive mechanisms to maintain the concerted cellular response to hypoxia. Specifically, this is achieved via both loss of microRNA regulation of HIF-α isoforms and hypoxia-responsive/HIF target genes and reduced transcriptional induction of HIF target genes as a result of decreased HIF expression/activity. Consequently, the hypoxia-mediated down-regulation of Dicer acts to maintain the induction of hypoxia-responsive genes through both transcriptional (HIF activity) and post-transcriptional (i.e. microRNA regulation) mechanisms. Importantly, this represents a novel, previously uncharacterized facet of the regulation of the cellular hypoxic response by microRNAs.
The findings reported here are relevant to microRNA function in cancer. Dicer status has prognostic implications in a number of human malignancies (78, 79). A general down-regulation of microRNAs in tumors compared with normal tissues has been observed (74), and recent studies have focused on Dicer as a haploinsufficient tumor suppressor gene where deletion of a single copy of Dicer in murine tumors led to reduced survival compared with controls (79). This is consistent with reports of impaired shRNA-mediated gene silencing in ovarian cancer cell lines with low Dicer expression (78) and impaired microRNA processing in murine tumors containing only a single copy of the Dicer gene (79). Significantly, the data we present here indicate that cellular hypoxia is also relevant to decreased Dicer expression and function in tumors. In this regard, a characteristic feature of solid tumors is the presence of cells at very low oxygen tensions; these hypoxic cells confer an angiogenic phenotype and tumor resistance to radiotherapy and chemotherapy and are selected for a more malignant phenotype (60). Recent studies have revealed that miR-103/107 and let-7 can target Dicer (80, 81). These microRNAs are functionally integrated into cellular pathways involved in cancer development. Thus, it is of interest that even though members of the miR-103/107 and let-7 families are present in HUVEC they do not significantly decrease in hypoxia. Our findings that chronic hypoxia impairs Dicer function are important in cancer given that they provide a new mechanism for the general down-regulation of microRNAs in tumors. Finally, we report here that functional loss of Dicer activity is VHL-dependent. However, the down-regulation of Dicer mRNA expression is not HIF-dependent. Loss of VHL function is key mechanistically to the majority of familial and sporadic cases of CCRCC. Moreover, 62% of renal cell tumors have hemizygous loss of the DICER1 locus (82). Conceptually, therefore, loss of Dicer function in CCRCC could involve three distinct pathways, namely deletion/mutations of the Dicer gene, loss of VHL-dependent regulation of Dicer, and intratumoral hypoxia. The relative contribution of each or all of these pathways to prognosis and therapy of patients with CCRCC warrants further study.
Taken together, these findings represent a newer perspective into the regulation of the cellular hypoxic response. Specifically, we are the first to show that loss of Dicer-dependent microRNA regulation is important for the concerted cellular response to hypoxia. Importantly, our findings suggest that the hypoxic down-regulation of Dicer plays a significant role in maintaining the cellular response to hypoxia by preserving the expression of HIF-α via microRNA regulation, including the direct regulation of HIF-2α by the Dicer-dependent miR-185 as we have shown here as well as the expression of other essential hypoxia-responsive genes via HIF-α and microRNA regulation. Additionally, the down-regulation of Dicer by chronic hypoxia also serves as a potential explanation for the global microRNA down-regulation observed in tumors versus normal tissue (74). The finding that Dicer is functionally deficient in chronic hypoxia is highly relevant to gene therapy approaches that are Dicer-dependent versus Dicer-independent as we have shown in this study. These findings provide significant new insight for hypoxia biology and the development of RNAi-based therapeutics.
Supplementary Material
Acknowledgment
We thank Dr. Gregory Hannon from the Howard Hughes Medical Institute at Cold Spring Harbor Laboratory for providing us with the full-length human Dicer coding sequence plasmid construct.
This work was supported in part by Heart and Stroke Foundation Grant T-6777 (to P. A. M.) and Canadian Institutes of Health Research Grant CIHR MOP77718 (to M. O.).

This article contains supplemental Figs. S1–S5 and Tables S1–S3.
- HIF
- hypoxia-inducible factor
- VHL
- von Hippel-Lindau
- VEGFR
- VEGF receptor
- RISC
- RNA-induced silencing complex
- HUVEC
- human umbilical vein endothelial cells
- qRT-PCR
- quantitative RT-PCR
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CCRCC
- clear cell renal cell carcinoma
- pre-microRNA
- precursor microRNA
- pri-microRNA
- primary microRNA transcript
- DsiRNA
- Dicer substrate siRNA
- Ago
- argonaute
- pre-mRNA
- precursor mRNA
- TRBP
- TAR (HIV-1) RNA binding protein 2
- LMNA
- lamin A/C
- DGCR8
- DiGeorge syndrome critical region gene 8.
REFERENCES
- 1. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2011) Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365, 537–547 [DOI] [PubMed] [Google Scholar]
- 3. Semenza G. L. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho J. J., Man H. S., Marsden P. A. (2012) Nitric oxide signaling in hypoxia. J. Mol. Med. 90, 217–231 [DOI] [PubMed] [Google Scholar]
- 5. McQuillan L. P., Leung G. K., Marsden P. A., Kostyk S. K., Kourembanas S. (1994) Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am. J. Physiol. Heart Circ. Physiol. 267, H1921–H1927 [DOI] [PubMed] [Google Scholar]
- 6. Faller D. V. (1999) Endothelial cell responses to hypoxic stress. Clin. Exp. Pharmacol. Physiol. 26, 74–84 [DOI] [PubMed] [Google Scholar]
- 7. Fish J. E., Matouk C. C., Yeboah E., Bevan S. C., Khan M., Patil K., Ohh M., Marsden P. A. (2007) Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J. Biol. Chem. 282, 15652–15666 [DOI] [PubMed] [Google Scholar]
- 8. Fasanaro P., D'Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M. C., Martelli F. (2008) MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 10. Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merritt W. M., Bar-Eli M., Sood A. K. (2010) The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 70, 2571–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koritzinsky M., Wouters B. G. (2007) Hypoxia and regulation of messenger RNA translation. Methods Enzymol. 435, 247–273 [DOI] [PubMed] [Google Scholar]
- 13. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G., Jr. (2003) Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suárez Y., Fernández-Hernando C., Pober J. S., Sessa W. C. (2007) Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 100, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 16. Mawji I. A., Robb G. B., Tai S. C., Marsden P. A. (2004) Role of the 3′-untranslated region of human endothelin-1 in vascular endothelial cells. Contribution to transcript lability and the cellular heat shock response. J. Biol. Chem. 279, 8655–8667 [DOI] [PubMed] [Google Scholar]
- 17. Robb G. B., Brown K. M., Khurana J., Rana T. M. (2005) Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 12, 133–137 [DOI] [PubMed] [Google Scholar]
- 18. He J. Z., Ho J. J., Gingerich S., Courtman D. W., Marsden P. A., Ward M. E. (2010) Enhanced translation of heme oxygenase-2 preserves human endothelial cell viability during hypoxia. J. Biol. Chem. 285, 9452–9461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koeman J. M., Russell R. C., Tan M. H., Petillo D., Westphal M., Koelzer K., Metcalf J. L., Zhang Z., Matsuda D., Dykema K. J., Houseman H. L., Kort E. J., Furge L. L., Kahnoski R. J., Richard S., Vieillefond A., Swiatek P. J., Teh B. T., Ohh M., Furge K. A. (2008) Somatic pairing of chromosome 19 in renal oncocytoma is associated with deregulated EGLN2-mediated [corrected] oxygen-sensing response. PLoS Genet. 4, e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fasanaro P., Greco S., Lorenzi M., Pescatori M., Brioschi M., Kulshreshtha R., Banfi C., Stubbs A., Calin G. A., Ivan M., Capogrossi M. C., Martelli F. (2009) An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 284, 35134–35143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grimm D., Streetz K. L., Jopling C. L., Storm T. A., Pandey K., Davis C. R., Marion P., Salazar F., Kay M. A. (2006) Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 [DOI] [PubMed] [Google Scholar]
- 22. Waltenberger J., Mayr U., Pentz S., Hombach V. (1996) Functional upregulation of the vascular endothelial growth factor receptor KDR by hypoxia. Circulation 94, 1647–1654 [DOI] [PubMed] [Google Scholar]
- 23. Gerber H. P., Condorelli F., Park J., Ferrara N. (1997) Differential transcriptional regulation of the two vascular endothelial growth factor receptor genes. Flt-1, but not Flk-1/KDR, is up-regulated by hypoxia. J. Biol. Chem. 272, 23659–23667 [DOI] [PubMed] [Google Scholar]
- 24. Takeda N., Maemura K., Imai Y., Harada T., Kawanami D., Nojiri T., Manabe I., Nagai R. (2004) Endothelial PAS domain protein 1 gene promotes angiogenesis through the transactivation of both vascular endothelial growth factor and its receptor, Flt-1. Circ. Res. 95, 146–153 [DOI] [PubMed] [Google Scholar]
- 25. Abaci H. E., Truitt R., Luong E., Drazer G., Gerecht S. (2010) Adaptation to oxygen deprivation in cultures of human pluripotent stem cells, endothelial progenitor cells, and umbilical vein endothelial cells. Am. J. Physiol. Cell Physiol. 298, C1527–C1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ward M. E., Toporsian M., Scott J. A., Teoh H., Govindaraju V., Quan A., Wener A. D., Wang G., Bevan S. C., Newton D. C., Marsden P. A. (2005) Hypoxia induces a functionally significant and translationally efficient neuronal NO synthase mRNA variant. J. Clin. Investig. 115, 3128–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Purpura K. A., George S. H., Dang S. M., Choi K., Nagy A., Zandstra P. W. (2008) Soluble Flt-1 regulates Flk-1 activation to control hematopoietic and endothelial development in an oxygen-responsive manner. Stem Cells 26, 2832–2842 [DOI] [PubMed] [Google Scholar]
- 29. Chamboredon S., Ciais D., Desroches-Castan A., Savi P., Bono F., Feige J. J., Cherradi N. (2011) Hypoxia-inducible factor-1α mRNA: a new target for destabilization by tristetraprolin in endothelial cells. Mol. Biol. Cell 22, 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schioppa T., Uranchimeg B., Saccani A., Biswas S. K., Doni A., Rapisarda A., Bernasconi S., Saccani S., Nebuloni M., Vago L., Mantovani A., Melillo G., Sica A. (2003) Regulation of the chemokine receptor CXCR4 by hypoxia. J. Exp. Med. 198, 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bazzini A. A., Lee M. T., Giraldez A. J. (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Djuranovic S., Nahvi A., Green R. (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tano N., Kim H. W., Ashraf M. (2011) microRNA-150 regulates mobilization and migration of bone marrow-derived mononuclear cells by targeting Cxcr4. PLoS One 6, e23114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vumbaca F., Phoenix K. N., Rodriguez-Pinto D., Han D. K., Claffey K. P. (2008) Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell. Biol. 28, 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hua Z., Lv Q., Ye W., Wong C. K., Cai G., Gu D., Ji Y., Zhao C., Wang J., Yang B. B., Zhang Y. (2006) MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cha S. T., Chen P. S., Johansson G., Chu C. Y., Wang M. Y., Jeng Y. M., Yu S. L., Chen J. S., Chang K. J., Jee S. H., Tan C. T., Lin M. T., Kuo M. L. (2010) MicroRNA-519c suppresses hypoxia-inducible factor-1α expression and tumor angiogenesis. Cancer Res. 70, 2675–2685 [DOI] [PubMed] [Google Scholar]
- 37. Ghosh G., Subramanian I. V., Adhikari N., Zhang X., Joshi H. P., Basi D., Chandrashekhar Y. S., Hall J. L., Roy S., Zeng Y., Ramakrishnan S. (2010) Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J. Clin. Investig. 120, 4141–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamakuchi M., Lotterman C. D., Bao C., Hruban R. H., Karim B., Mendell J. T., Huso D., Lowenstein C. J. (2010) p53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 6334–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uchida T., Rossignol F., Matthay M. A., Mounier R., Couette S., Clottes E., Clerici C. (2004) Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: implication of natural antisense HIF-1α. J. Biol. Chem. 279, 14871–14878 [DOI] [PubMed] [Google Scholar]
- 40. Skuli N., Simon M. C. (2009) HIF-1α versus HIF-2α in endothelial cells and vascular functions: is there a master in angiogenesis regulation? Cell Cycle 8, 3252–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raval R. R., Lau K. W., Tran M. G., Sowter H. M., Mandriota S. J., Li J. L., Pugh C. W., Maxwell P. H., Harris A. L., Ratcliffe P. J. (2005) Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25, 5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim D. H., Behlke M. A., Rose S. D., Chang M. S., Choi S., Rossi J. J. (2005) Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 [DOI] [PubMed] [Google Scholar]
- 43. Wu C., So J., Davis-Dusenbery B. N., Qi H. H., Bloch D. B., Shi Y., Lagna G., Hata A. (2011) Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 31, 4760–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qi H. H., Ongusaha P. P., Myllyharju J., Cheng D., Pakkanen O., Shi Y., Lee S. W., Peng J., Shi Y. (2008) Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455, 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bennasser Y., Chable-Bessia C., Triboulet R., Gibbings D., Gwizdek C., Dargemont C., Kremer E. J., Voinnet O., Benkirane M. (2011) Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat. Struct. Mol. Biol. 18, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh S., Bevan S. C., Patil K., Newton D. C., Marsden P. A. (2005) Extensive variation in the 5′-UTR of Dicer mRNAs influences translational efficiency. Biochem. Biophys. Res. Commun. 335, 643–650 [DOI] [PubMed] [Google Scholar]
- 47. Matskevich A. A., Moelling K. (2008) Stimuli-dependent cleavage of Dicer during apoptosis. Biochem. J. 412, 527–534 [DOI] [PubMed] [Google Scholar]
- 48. Bernstein E., Kim S. Y., Carmell M. A., Murchison E. P., Alcorn H., Li M. Z., Mills A. A., Elledge S. J., Anderson K. V., Hannon G. J. (2003) Dicer is essential for mouse development. Nat. Genet. 35, 215–217 [DOI] [PubMed] [Google Scholar]
- 49. O'Rourke J. R., Georges S. A., Seay H. R., Tapscott S. J., McManus M. T., Goldhamer D. J., Swanson M. S., Harfe B. D. (2007) Essential role for Dicer during skeletal muscle development. Dev. Biol. 311, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schaefer A., O'Carroll D., Tan C. L., Hillman D., Sugimori M., Llinas R., Greengard P. (2007) Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 204, 1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen J. F., Murchison E. P., Tang R., Callis T. E., Tatsuguchi M., Deng Z., Rojas M., Hammond S. M., Schneider M. D., Selzman C. H., Meissner G., Patterson C., Hannon G. J., Wang D. Z. (2008) Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. U.S.A. 105, 2111–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harvey S. J., Jarad G., Cunningham J., Goldberg S., Schermer B., Harfe B. D., McManus M. T., Benzing T., Miner J. H. (2008) Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J. Am. Soc. Nephrol. 19, 2150–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ho J. J., Marsden P. A. (2008) Dicer cuts the kidney. J. Am. Soc. Nephrol. 19, 2043–2046 [DOI] [PubMed] [Google Scholar]
- 54. Singh M. K., Lu M. M., Massera D., Epstein J. A. (2011) MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J. Biol. Chem. 286, 41036–41045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaelin W. G., Jr. (2008) The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 [DOI] [PubMed] [Google Scholar]
- 56. Mazure N. M., Chauvet C., Bois-Joyeux B., Bernard M. A., Nacer-Chérif H., Danan J. L. (2002) Repression of α-fetoprotein gene expression under hypoxic conditions in human hepatoma cells: characterization of a negative hypoxia response element that mediates opposite effects of hypoxia inducible factor-1 and c-Myc. Cancer Res. 62, 1158–1165 [PubMed] [Google Scholar]
- 57. Chen K. F., Lai Y. Y., Sun H. S., Tsai S. J. (2005) Transcriptional repression of human cad gene by hypoxia inducible factor-1α. Nucleic Acids Res. 33, 5190–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eltzschig H. K., Abdulla P., Hoffman E., Hamilton K. E., Daniels D., Schönfeld C., Löffler M., Reyes G., Duszenko M., Karhausen J., Robinson A., Westerman K. A., Coe I. R., Colgan S. P. (2005) HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J. Exp. Med. 202, 1493–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Evans A. J., Russell R. C., Roche O., Burry T. N., Fish J. E., Chow V. W., Kim W. Y., Saravanan A., Maynard M. A., Gervais M. L., Sufan R. I., Roberts A. M., Wilson L. A., Betten M., Vandewalle C., Berx G., Marsden P. A., Irwin M. S., Teh B. T., Jewett M. A., Ohh M. (2007) VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol. Cell. Biol. 27, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Y., Roche O., Yan M. S., Finak G., Evans A. J., Metcalf J. L., Hast B. E., Hanna S. C., Wondergem B., Furge K. A., Irwin M. S., Kim W. Y., Teh B. T., Grinstein S., Park M., Marsden P. A., Ohh M. (2009) Regulation of endocytosis via the oxygen-sensing pathway. Nat. Med. 15, 319–324 [DOI] [PubMed] [Google Scholar]
- 61. Zhao F., Xuan Z., Liu L., Zhang M. Q. (2005) TRED: a transcriptional regulatory element database and a platform for in silico gene regulation studies. Nucleic Acids Res. 33, D103–D107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang C., Xuan Z., Zhao F., Zhang M. Q. (2007) TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 35, D137–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Semenza G. L., Wang G. L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Datta K., Mondal S., Sinha S., Li J., Wang E., Knebelmann B., Karumanchi S. A., Mukhopadhyay D. (2005) Role of elongin-binding domain of von Hippel Lindau gene product on HuR-mediated VPF/VEGF mRNA stability in renal cell carcinoma. Oncogene 24, 7850–7858 [DOI] [PubMed] [Google Scholar]
- 65. Pioli P. A., Rigby W. F. (2001) The von Hippel-Lindau protein interacts with heteronuclear ribonucleoprotein a2 and regulates its expression. J. Biol. Chem. 276, 40346–40352 [DOI] [PubMed] [Google Scholar]
- 66. Greer S. N., Metcalf J. L., Wang Y., Ohh M. (2012) The updated biology of hypoxia-inducible factor. EMBO J. 31, 2448–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dang E. V., Barbi J., Yang H. Y., Jinasena D., Yu H., Zheng Y., Bordman Z., Fu J., Kim Y., Yen H. R., Luo W., Zeller K., Shimoda L., Topalian S. L., Semenza G. L., Dang C. V., Pardoll D. M., Pan F. (2011) Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell 146, 772–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Caruso P., MacLean M. R., Khanin R., McClure J., Soon E., Southgate M., MacDonald R. A., Greig J. A., Robertson K. E., Masson R., Denby L., Dempsie Y., Long L., Morrell N. W., Baker A. H. (2010) Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler. Thromb. Vasc. Biol. 30, 716–723 [DOI] [PubMed] [Google Scholar]
- 69. Cheloufi S., Dos Santos C. O., Chong M. M., Hannon G. J. (2010) A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang J. S., Maurin T., Robine N., Rasmussen K. D., Jeffrey K. L., Chandwani R., Papapetrou E. P., Sadelain M., O'Carroll D., Lai E. C. (2010) Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Havens M. A., Reich A. A., Duelli D. M., Hastings M. L. (2012) Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 40, 4626–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Baccarini A., Chauhan H., Gardner T. J., Jayaprakash A. D., Sachidanandam R., Brown B. D. (2011) Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 21, 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chatterjee S., Grosshans H. (2009) Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461, 546–549 [DOI] [PubMed] [Google Scholar]
- 74. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 75. Brahimi-Horn M. C., Pouysségur J. (2007) Oxygen, a source of life and stress. FEBS Lett. 581, 3582–3591 [DOI] [PubMed] [Google Scholar]
- 76. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 77. Wiesen J. L., Tomasi T. B. (2009) Dicer is regulated by cellular stresses and interferons. Mol. Immunol. 46, 1222–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Merritt W. M., Lin Y. G., Han L. Y., Kamat A. A., Spannuth W. A., Schmandt R., Urbauer D., Pennacchio L. A., Cheng J. F., Nick A. M., Deavers M. T., Mourad-Zeidan A., Wang H., Mueller P., Lenburg M. E., Gray J. W., Mok S., Birrer M. J., Lopez-Berestein G., Coleman R. L., Bar-Eli M., Sood A. K. (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 359, 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kumar M. S., Pester R. E., Chen C. Y., Lane K., Chin C., Lu J., Kirsch D. G., Golub T. R., Jacks T. (2009) Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23, 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jakymiw A., Patel R. S., Deming N., Bhattacharyya I., Shah P., Lamont R. J., Stewart C. M., Cohen D. M., Chan E. K. (2010) Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer 49, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martello G., Rosato A., Ferrari F., Manfrin A., Cordenonsi M., Dupont S., Enzo E., Guzzardo V., Rondina M., Spruce T., Parenti A. R., Daidone M. G., Bicciato S., Piccolo S. (2010) A microRNA targeting dicer for metastasis control. Cell 141, 1195–1207 [DOI] [PubMed] [Google Scholar]
- 82. Forbes S. A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J. W., Futreal P. A., Stratton M. R. (2008) The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. Chapter 10, Unit 10.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.