Background: Targeted CDK4/6 inhibition is a novel therapeutic strategy undergoing PhaseI/II clinical trials for the treatment of solid tumors.
Results: CDK4/6 inhibition antagonizes the cytotoxic mechanism(s) of traditional chemotherapies and alters DNA repair processes.
Conclusion: CDK4/6 inhibition attenuates the cellular response to cytotoxic chemotherapies.
Significance: Understanding of cell cycle and transcriptional effects of CDK4/6 inhibition is critical for clinical utilization.
Keywords: Cancer Biology, CDK (Cyclin-dependent Kinase), Cell Cycle, Checkpoint Control, DNA Damage, DNA Damage Response, E2F Transcription Factor, Retinoblastoma (Rb)
Abstract
The RB/E2F axis represents a critical node of cell signaling that integrates a diverse array of signaling pathways. Recent evidence has suggested a role for E2F-mediated gene transcription in DNA damage response and repair, as well as apoptosis signaling. Herein, we investigated how repression of E2F activity via CDK4/6 inhibition and RB activation impacts the response of triple negative breast cancer (TNBC) to frequently used therapeutic agents. In combination with taxanes and anthracyclines CDK4/6 inhibition and consequent cell cycle arrest prevented the induction of DNA damage and associated cell death in an RB-dependent manner; thereby demonstrating antagonism between the cytostatic influence of the CDK-inhibitor and cytotoxic agents. As many of these effects were secondary to cell cycle arrest, γ-irradiation (IR) was utilized to examine effects of CDK4/6 inhibition on direct DNA damage. Although E2F controls a number of genes involved in DNA repair (e.g. Rad51), CDK4/6 inhibition did not alter the overall rate of DNA repair, rather it significantly shifted the burden of this repair from homologous recombination (HR) to non-homologous end joining (NHEJ). Together, these data indicate that CDK4/6 inhibition can antagonize cytotoxic therapeutic strategies and increases utilization of error-prone DNA repair mechanisms that could contribute to disease progression.
Introduction
The retinoblastoma (RB)3 tumor suppressor is a critical regulator of numerous cellular events involved in cell cycle progression, replication, DNA repair, apoptosis, and mitosis (1, 2). In the clinical setting, RB-status is emerging as a key determinant of cancer progression, recurrence, and therapeutic response. For example, in breast cancers, elevated expression of a recently characterized RB-loss signature is associated with poor overall disease outcome, but improved response to chemotherapy and longer relapse-free survival among ER-negative subpopulations (3).
RB functions as a transcriptional repressor of the E2F family of transcription factors, and when active results in a prominent G1 cell cycle arrest. Activation of RB is regulated by its phosphorylation state, which is directly controlled by cellular cyclin/cyclin-dependent kinase (CDK) complexes. Phosphorylation of the RB protein by the CDK4/6 holoenzyme is essential for entry into the cell cycle from G1-phase. Recently, PD0332991, a small molecule inhibitor of D-type cyclin/CDK 4/6 activity, has been developed that demonstrates specific activity in models that retain functional RB protein (4–8). Such specificity provides the opportunity to disconnect the cell cycle machinery from pro-growth signaling by specifically preventing the initial phosphorylation event of RB protein. As such, the ability of PD0332991 to inhibit E2F-mediated gene transcription in an RB-positive setting has been experimentally determined in multiple model systems (4–11). Responses among RB-proficient in vitro model systems display a potently cytostatic effect, and extended CDK4/6 inhibition can promote a senescence-like phenotype in specific settings (4). Human xenograft model systems of RB-positive breast, colon, prostate, ovarian, and glioblastoma have recapitulated the potently cytostatic effects of PD0332991 in vivo, and early results from Phase I clinical trials demonstrate that the side effects of PD0332991 are relatively well-tolerated (12, 13).
Currently, the clinical treatment for a wide array of human malignancies involves administration of genotoxic agents (e.g. taxanes and anthracyclines). Such agents are known to afford a degree of specificity by exploiting increased rates of cell proliferation present within the tumor. In the context of breast cancers, the aforementioned genotoxic agents, in addition to γ-irradiation (IR) are commonly used in the clinic. Such therapeutic agents rely on the direct or cell cycle-mediated induction of DNA damage to promote cellular apoptosis. Given the wide deployment of these agents in the treatment of breast cancers, and the pace with which pharmacological CDK4/6 inhibition (PD0332991) is proceeding toward the clinic, there is a need to determine how these agents, and the cellular processes that they control, will interact with each other in a combinatorial treatment scenario. In fact, PD0332991 is currently undergoing a phase I/II study of patients with advanced breast cancers in combination treatments with paclitaxel (NCT01320592). Thus, a critical question that must be answered is: Would clinical CDK4/6 inhibition be expected to work cooperatively with therapeutic agents that work most effectively in cycling cells?
Herein, we utilized a panel of triple negative breast cancer (TNBC) cell lines to examine the ability of CDK4/6 inhibition to modify the acute cellular response to S-phase (anthracycline) and mitosis-acting (taxane) chemotherapeutic agents, in addition to cell cycle independent IR-induced DNA damage. As TNBCs by definition lack the targets for anti-estrogen therapies and HER2 antagonists, yet still frequently retain functional RB protein, this breast cancer subgroup is an ideal candidate for both CDK4/6 inhibition and treatment with traditional genotoxic therapies (14). Clearly, a more complete picture of the consequence of CDK4/6 inhibition is necessary to thoroughly understand how it will be best employed in a clinical setting.
EXPERIMENTAL PROCEDURES
Cell Culture and Viral Infection
All cell lines were maintained in DMEM containing 10% fetal bovine serum, 100 units/ml penicillin/streptomycin, and 2 mm l-glutamine, and were cultured at 37 °C and 5% CO2. All cells were counted for experimental seeding using trypan blue exclusion. miRB and miNS-expressing retrovirus was produced and utilized as previously described (4).
Drug Treatments
All PD0332991 treatments were performed at a 500 nm concentration. DMSO was used as a vehicle control. In vitro pretreatment was accomplished by treating cells with PD0332991 24 h prior to genotoxic exposure. Concurrent treatments were performed by administering PD0332991 and chemotherapeutic agent simultaneously. In vitro doxorubicin and paclitaxel treatments were performed at 1 μm and 100 nm, respectively, while all IR treatments utilized a single 2 Gy dose. For acute cell cycle analyses, cells were harvested 24 h postgenotoxic exposure. Outgrowth (continuous PD) experiments were performed by plating cells at a density of 5 × 105 cells per plate and exposing them to corresponding combinations of PD0332991 and paclitaxel for 24 h. After 24 h, all drugs were washed from the plates and fresh PD0332991 was returned to PD0332991, Concurrent and PD0331991 pre-treat plates at a concentration of 500 nm, and replenished every 72 h for 7 days. Outgrowth (synchronized release) experiments were performed by plating 1 × 105 cells per plate and treating them with the corresponding drug combinations for 24 h. PD0332991 pretreatment plates were pretreated with PD0332991 for 24 h prior to paclitaxel administration. After 24 h in paclitaxel, all drugs were washed from the plate, and cells were allowed to recover for a period of 21 days or 100% confluency. At the indicated time points, cells were fixed and stained with a 1% crystal violet solution to visualize cell density and morphology.
Flow Cytometric Analysis
Cells were harvested and fixed in 70% EtOH overnight at 4 °C. Cells were labeled with BrdU for 1 h prior to harvest, and prepared for flow cytometry as previously described (15). Cell cycle analysis was performed using FlowJo 9.2.3 software (Ashland, OR). BrdU data is represented as a percentage of total population unless otherwise noted. Annexin V flow cytometry was performed using APC-conjugated Annexin V antibody (BD Pharmingen), following the manufacturer's recommended protocol. All experiments were performed in triplicate from a minimum of two independent experiments. Error bars represent standard deviation and *, p < 0.05.
Immunoblot Analysis
Cell lysates were resolved by SDS-PAGE and transferred to Immobilon-P membrane (Millipore) using standard methods. Proteins were detected using the following antibodies: Santa Cruz: Cyclin A (H432), MCM7 (141.2), Lamin B (M-20), Rad51 (H-92), Actin (I-19), CHK1 (G4) and FOXM1 (K-19). Cell Signaling: PARP (9542S) and p-CHK2 (T68) (2661P). Neomarkers: Cyclin D1 (Ab-3), DNA PKcs (Ab4), Ku (N3H10). Abcam: DNA PKcs S2056 (ab18192), DNA Ligase IV (ab26039). All primary antibodies were used at 1:1000 dilution and corresponding secondary antibodies at 1:5000.
Immunofluorescence
Cells were plated on coverslips and allowed to adhere overnight. At indicated time points, cells were then fixed in 3.7% formaldehyde and permeabilized in 0.4% Triton X-100 for 20–30 min at room temperature. Primary antibodies utilized include: Millipore: phospho-γ-H2AX (Ser-139) (05-636), phospho-Histone H3 (Ser-10) (06-570), and Santa Cruz Biotechnology: Rad51 (H-92). Image J software (version 1.45e) was utilized for foci measurement and image analysis. Primary antibodies were employed at 1:250 dilutions and secondary antibodies at 1:1000. All immunofluorescence was counter-stained with DAPI for visualization of nuclei.
DSB Repair Assays
Homologous recombination activity was quantified using a synthetic repair substrate (pDR-GFP) as previously described (16). Circular DR-GFP was digested using ISce1 enzyme. GFP activity was quantified via flow cytometry 72 h after substrate transfection using Lipofectamine 2000 as per manufacturers recommendations. Linear substrate was cotransfected alongside a DsRed expression construct to control for transfection efficiency. PD0332991 treated populations were pretreated with 500 nm PD0332991 24 h prior to transfection. Additionally, all populations were irradiated with 2 Gy IR exposure at time 0, prior to transfection. An equal volume of DMSO was used as a control. Data are presented as relative % GFP-positive (of 30,000 total events) normalized to the untreated (No PD0332991) condition.
NHEJ activity was measured using ISce1-linearized pEGFP-PEM1-AD2. Linearized plasmid was transfected using Lipofectamine 2000 immediately after 2 Gy IR exposure as per manufacturers recommendation alongside DsRed as a transfection control. This plasmid and assay has been previously described (17, 18). GFP activity was measured by flow cytometry 24 h post transfection under conditions identical to those described for HR assays. Data are presented as relative % GFP positive, normalized to untreated (No PD0332991) condition. All DNA repair constructs were kindly provided by Dr. Peter Stambrook (University of Cincinnati).
Xenografts and Immunohistochemistry
1 × 106 MDA-MB-231 breast cancer cells were injected s.c. into the flanks of 6–8 week-old athymic nude mice (Harlan Sprague-Dawley, Inc.). Cells were suspended in 150 μl of PBS and 50 μl Matrigel (BD Biosciences). Tumor growth was monitored using calipers. Once tumors reached 100–200 mm3 in size, mice were treated with doxorubicin via single interperitoneal injection (20 mg/kg dissolved in 0.9% saline), and were given PD0332991 (150 mg/kg in lactate buffer, pH4.0) or vehicle daily via oral gavage. Mice were euthanized 7 days post treatment initiation. At two hours before euthanization, mice were injected intraperitoneally with 150 mg/kg BrdUrd in 100 μl 0.9% saline. After euthanization, tumors were excised, fixed in neutral buffered formalin and paraffin embedded for sectioning. All animal experiments were conducted in accordance with the NIH Guide for Care and Use of Laboratory Animals and were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee. Immunohistochemical staining on xenograft sections utilized cyclin A (H432-Santa Cruz) and MCM7 (141.2-Santa Cruz) primary antibodies at a 1:250 concentration. Fluorescent staining for Rad51 was performed with H-92 primary antibody (Santa Cruz). Both methods were performed using previously described methods (15, 19). Image J software (version 1.45e) was utilized for image analysis.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism version 4.0c (GraphPad Prism Software, Inc.). Results were analyzed for statistical significance using a two- tailed Student's t-Test and standard deviation. For all analyses, p < 0.05 was considered significant.
RESULTS
CDK4/6 Inhibition Modifies the Response of RB-proficient Breast Cancer Cells to Anthracyclines
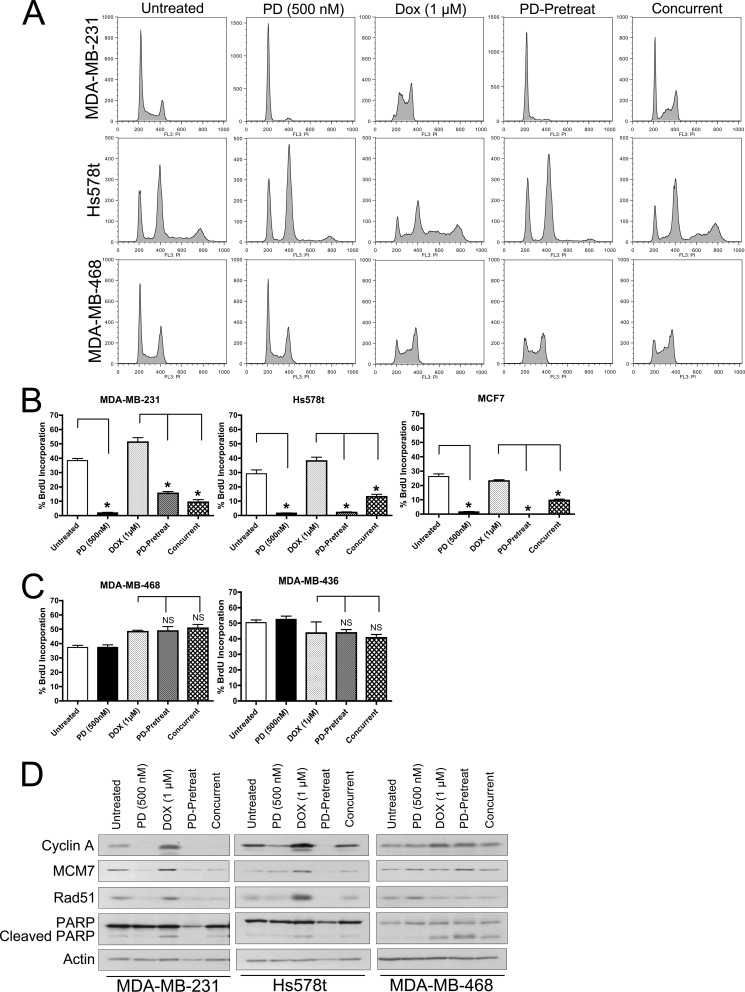
To determine the impact CDK4/6 inhibition on response to commonly used cytotoxic agents, PD0332991 was employed in combination with doxorubicin. In RB-proficient breast cancer cell lines (MDA-MB-231 and Hs578t), 24 h PD0332991 treatment resulted in a prominent G1 cell cycle arrest, recapitulating evidence observed in multiple other model systems (Fig. 1A) (4). This correlates to a prominently 2N population in MDA-MB-231 cells and prominent 2N and 4N populations in Hs578t cell populations, as these cells possess bimodal chromosome distribution. Importantly, in accordance with previously published studies, treatment with PD did not result in an increase in cell death as measured by Annexin V staining (supplemental Fig. S1). Treatment with 1 μm doxorubicin for 24 h resulted in an accumulation of S-phase cells, characteristic of previously described doxorubicin-mediated cell cycle arrest (20). Interestingly, 24 h pretreatment with PD0332991 prevents the accumulation of cells in S-phase upon exposure to doxorubicin and demonstrates a prominent G1 cell cycle arrest similar to that observed in PD0332991-only treated populations. Concurrent treatment with PD0332991 and doxorubicin results in a hybrid cell cycle distribution wherein the majority of cells are arrested in a 2N (or 4N for Hs578t) state, while a proportion of cells seemed to have escaped the negative cell cycle regulation by CDK4/6 inhibition. In comparison to the response of RB-positive cells to CDK4/6 inhibition, MDA-MB-468 cells (which possess a homozygous deletion of the RB1 gene) bypass the effects of CDK4/6 inhibition, as the corresponding cell cycle distribution is unchanged from the untreated state (Fig. 1A, bottom row). Furthermore, the cell cycle distribution afforded by doxorubicin treatment is not significantly altered in PD0332991-pretreated or concurrent treatment schedules. These data are consistent with the concept that CDK4/6 inhibition is unable to alter cell cycle progression in RB-null populations.
FIGURE 1.
CDK4/6 inhibition modifies the acute response of RB-positive breast cancer cells to anthracyclines. A, representative flow cytometric analysis of RB-positive (MDA-MB-231 and Hs578t) and RB-negative (MDA-MB-468) breast cancer cells treated with 500 nm PD0332991 for 24 h, 1 μm doxorubicin for 24 h, 24 h PD0332991 pretreatment followed by 24 h exposed to 1 μm doxorubicin (PD-Pretreat) or PD0332991/doxorubicin for 24 h (Concurrent). B, BrdU incorporation among ER-positive and ER-negative, RB-proficient breast cancer cell lines treated as described in A. Cells were pulse-labeled with BrdU for 1 h. Populations were then stained for BrdU and DNA content and subjected to FLOW cytometric analysis. Data are represented as percentage BrdU-positive of the total population. C, RB-deficient breast cancer cell lines treated as described in B. D, immunoblot analyses examining the ability of PD0332991 pretreatment to modify PARP cleavage and response of the RB/E2F axis in breast cancer cells treated with combinations of PD0332991 and doxorubicin as described in A. Actin served as a loading control.
To confirm these phenomena, quantification of BrdU incorporation among the various treatment schedules was performed (Fig. 1, B and C). As expected, RB-proficient cell lines demonstrate a dramatic reduction in BrdU incorporation upon exposure to PD0332991 (Fig. 1B, black bars), while rates of BrdU incorporation remain elevated in RB-positive populations exposed to doxorubicin, indicative of cell accumulation in S-phase (Fig. 1B, light gray bars). Importantly, pretreatment or concurrent treatment with PD0332991 results in significantly reduced levels of BrdU incorporation, which more closely resemble those observed among PD0332991-alone treatments. In comparison, RB-negative populations (MDA-MB-468 and MDA-MB-436) display no appreciable changes in BrdU incorporation in response to CDK4/6 inhibition in either single agent or combination treatment settings (Fig. 1C).
Biochemical analysis under all treatment conditions were performed to examine the status of critical RB/E2F pathway targets. As shown in Fig. 1D, among RB-positive populations (MDA-MB-231 and Hs578t), treatment with PD0332991 resulted in a decrease in RB-regulated target gene products essential for cell cycle progression (cyclin A), replication (MCM7), and DNA damage repair (Rad51). In contrast, CDK4/6 inhibition in RB-deficient MDA-MB-468 cells does not appreciably alter these targets. Downstream of the traditional DNA damage response induced by anthracyclines is the mechanism of poly ADP-ribose polymerase (PARP) cleavage, a pro-apoptotic signaling event. While basal levels of PARP cleavage remain unchanged in response to PD0332991 treatment alone, regardless of RB status, exposure to doxorubicin resulted in an increased abundance of cleaved PARP protein in both RB-proficient and -deficient populations. This event was inhibited by pretreatment with PD0332991 only in RB-positive populations, as MDA-MB-468 demonstrated PARP cleavage at levels indistinguishable from those treated with doxorubicin alone. Interestingly, concurrent treatment resulted in intermediate levels of PARP cleavage, again implying that the timing of PD0332991 dosing may be relevant to therapeutic outcome when used in combination with therapeutic agents requiring active S-phase.
CDK4/6 Inhibition Modifies the Response of RB-Proficient Breast Cancer Cells to Taxanes
While anthracyclines are a class of agents known to require active DNA replication for effective cytotoxicity and DNA damage induction, taxanes are a class of microtubule-stabilizing agents believed to function primarily through promoting mitotic catastrophe. As such, we sought to examine the influence of CDK4/6 inhibition on the cellular response to a frequently used chemotheraputic taxane, paclitaxel. As was previously observed in Fig. 1, 24 h PD0332991 treatment significantly reduces active cell cycle progression only in RB-proficient populations as measured by BrdU incorporation (Fig. 2A, left panel). While treatment with 100 nm paclitaxel alone resulted in an ∼50% decrease in BrdU incorporation as compared with untreated populations, both concurrent and pre-treatment with PD0332991 resulted in a more dramatic decrease in BrdU incorporation, indistinguishable from that observed with PD0332991 treatment alone. In comparison, the proliferative rate of RB-negative breast cancer cells is not significantly modified by CDK4/6 inhibition alone or in combination with paclitaxel treatment, regardless of scheduling (Fig. 2A, right panel).
FIGURE 2.
CDK4/6 inhibition modifies the response of RB-positive breast cancer cells to mitosis-acting taxanes. A, RB-proficient and RB-deficient breast cancer cell lines treated with 500 nm PD0332991 for 24 h, 100 nm paclitaxel for 24 h, 24 h PD0332991 pretreatment followed by 24 h exposure to 100 nm paclitaxel (PD-Pretreat) or PD0332991/paclitaxel for 24 h (Concurrent). Cells were pulse-labeled with BrdU for 1 h. Populations were then stained for BrdU and DNA content and subjected to FLOW cytometric analysis. Data are represented as percentage BrdU-positive of the total population. B, quantification of percentage sub-G1 populations among treatments described in A. C, immunoblot analyses examining the influence of PD0332991 pretreatment on the ability of RB-positive breast cancer cells to cleave PARP and modify levels of E2F target gene products. Lamin B served as a loading control. D, representative crystal violet stained outgrowth plates of cells treated as described in C. For populations treated with PD0332991. The drug was maintained in the media for all 7 days post paclitaxel wash out. E, representative crystal violet stained plates after the indicated times of cell outgrowth post 24 h exposure to the various drug treatment schedules described in C.
During the aforementioned flow cytometric analysis, a substantial sub-G1 population was observed in several treatments, indicative of cell death. These data were quantified and are represented in Fig. 2B. Importantly, PD0332991 treatment alone did not significantly alter cell death (as compared with untreated) in both RB-proficient and RB-deficient breast cancer cells. Among RB-positive populations, treatment with paclitaxel increased cell death ∼3-fold. While concurrent PD0332991 treatment reduced the overall percentage of sub-G1 cells, 24 h pretreatment with PD0332991 was able to fully rescue the cell death phenotype observed in paclitaxel-treated populations (Fig. 2B, left panel). The effects of paclitaxel were largely recapitulated in RB-deficient MDA-MB-468 cells, as a 6-fold increase in sub-G1 cells were observed upon treatment (Fig. 2B, right panel). However, neither concurrent nor PD0332991-pretreatment were able to prevent accumulation of these populations in an RB-deficient background.
Similar to results observed in Fig. 1, treatment of RB-proficient (MDA-MB-231 and Hs578t) cells with PD0332991 resulted in down-regulation of the E2F-regulated cyclin A, MCM7, and Rad51 proteins while failing to increase any appreciable PARP cleavage (Fig. 2C). Treatment with paclitaxel did not dramatically deregulate those same E2F-responsive genes, but did result in an increased amount of PARP cleavage within these populations. Interestingly, concurrent administration of PD0332991 and paclitaxel promoted a general RB-dependent down-regulation of E2F-mediated proteins, but was only able to partially prevent PARP cleavage. These results were exacerbated upon 24 h pretreatment with PD0332991, wherein the PARP cleavage observed upon treatment with paclitaxel was fully inhibited (Fig. 2C). Once again, CDK4/6 inhibition had no affect on RB-deficient MDA-MB-468 cells. Since taxanes are known to induce mitotic catastrophe, which contributes to their cytotoxicity MDA-MB-231 and MDA-MB-468 cells were treated with the regimens utilized in Fig. 2C and phospho-histone H3 (Ser-10) staining was performed to visualize mitotic figures. As depicted in supplemental Fig. S2, treatment with paclitaxel resulted in a higher proportion of cells demonstrating aberrant mitoses, regardless of RB status. This corresponds with a general decrease in cell number in both cell lines. Importantly, these phenomena are prevented only in RB-proficient MDA-MB-231 cells upon pre-exposure to PD0332991, suggesting that pre-arrest of these cells via CDK4/6 inhibition before exposure to paclitaxel can prevent mitotic catastrophe associated with such agents. Thus, while anthracyclines and taxanes operate via distinct mechanisms in different phases of the cell cycle, inhibition of cell cycle progression via CDK4/6 inhibition ultimately antagonizes the cytotoxic activity of both agents.
While pre-arresting RB-proficient cell populations was capable of modifying the acute response to paclitaxel, we next performed experiments to examine the ability of CDK4/6 inhibition to prevent cell death and modify cellular outgrowth in response to paclitaxel over a longer period of time. As depicted in Fig. 2D, exposure of MDA-MB-231 cells to 24 h of paclitaxel resulted in a decrease in cell number over the course of 7 days as compared with PD0332991 treatment alone. Among populations receiving continuous exposure to the CDK4/6 inhibitor, either before or concurrent with administration of the taxane, an increased number of cells was observed. These data suggest that maintenance of cell cycle arrest through continuous CDK4/6 inhibition is capable of preventing cell death in response to chemotherapeutic agents for an extended period of time.
Although the aforementioned protection-based studies were suggestive of an overall antagonistic effect between CDK4/6 inhibition and taxanes, we next examined the potential therapeutic benefit in using CDK4/6 inhibition to synchronize cell populations prior to release into paclitaxel. As demonstrated in Fig. 2E, while paclitaxel treatment alone results in fewer cells after 3 weeks of outgrowth (as compared with untreated and PD0332991-treated populations), pre-or concurrent administration of PD0332991 with 24 h of paclitaxel exposure results in little no to colony outgrowth. Thus, while continuous CDK4/6 inhibition prevents paclitaxel-induced cytotoxicity, acute synchronization with PD0332991 appears to result in enhanced cytotoxicity and ultimately decreased cell outgrowth.
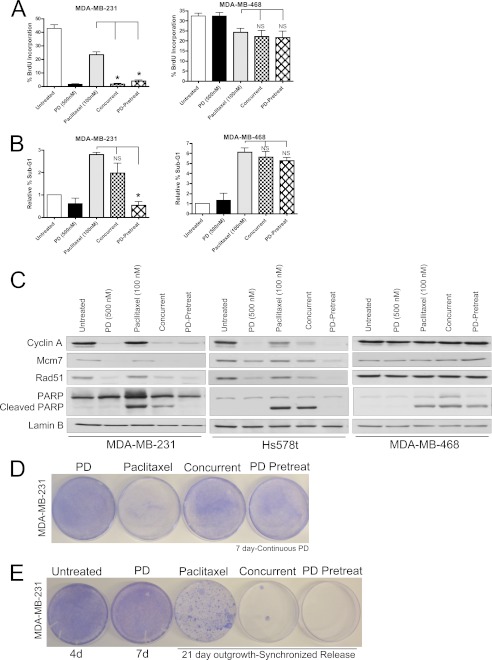
CDK4/6 Inhibition Prevents Cell Cycle Progression and Accumulation of Rad51 Foci in Vivo
To expand upon our in vitro studies, RB-proficient MDA-MB-231 xenografts were grown in nude mice and treated with vehicle, PD0332991, doxorubicin or a combination of PD0332991 and doxorubicin. Doxorubicin was administered as a single intraperitoneal injection, and PD0332991 was administered via oral gavage daily until sacrifice. Xenografts were harvested 7 days post-doxorubicin treatment. BrdU, Cyclin A, and MCM7 immunohistochemical staining, in addition to Rad51 immunofluorescence staining, was carried out on tumor sections, and representative images are shown in Fig. 3. In vivo, CDK4/6 inhibition was capable of decreasing the levels of BrdU incorporation, as well as expression levels of RB/E2F targets cyclin A and MCM7. In contrast, upon treatment with doxorubicin expression levels of BrdU, cyclin A, and MCM7 remained elevated. Additionally, Rad51 foci accumulation is observed, specifically with doxorubicin treatment, consistent with our in vitro studies. Importantly, pre-and continuous exposure to PD0332991 resulted in a clear reduction of BrdU incorporation, as well as cyclin A and MCM7 protein levels and prevented the induction of Rad51 foci associated with doxorubicin treatment. Overall, these data suggest that pretreatment with a CDK4/6 inhibitor prevents in vivo cell cycle progression in the presence of doxorubicin as measured by BrdU and E2F target gene activity. This cessation of cell cycle ultimately prevents doxorubicin-induced DNA damage response and repair mediated via Rad51, resulting in the absence of Rad51 foci in doxorubicin-treated tissues and reinforcing the notion of coordination between cell cycle progression and DNA damage repair.
FIGURE 3.
CDK4/6 inhibition modifies the therapeutic response of RB-positive breast cancer xenografts in vivo. A, representative α-BrdU immunohistochemical images of MDA-MB-231 xenografts treated with PD0332991, Doxorubicin or 72 h PD-pretreatment. Xenografts were harvested 7 days post treatment initiation. B, representative α-cyclin A and α-MCM7 immunohistochemical staining of tissues described in A. C, representative α-Rad51 immunofluorescence images in and quantification of MDA-MB-231 xenografts treated as described in A.
CDK4/6 Inhibition Does Not Alter the Rate of IR-induced DNA Damage Repair, but Does Reduce the Abundance of Proteins Necessary for Homologous Recombination in an RB-dependent Manner
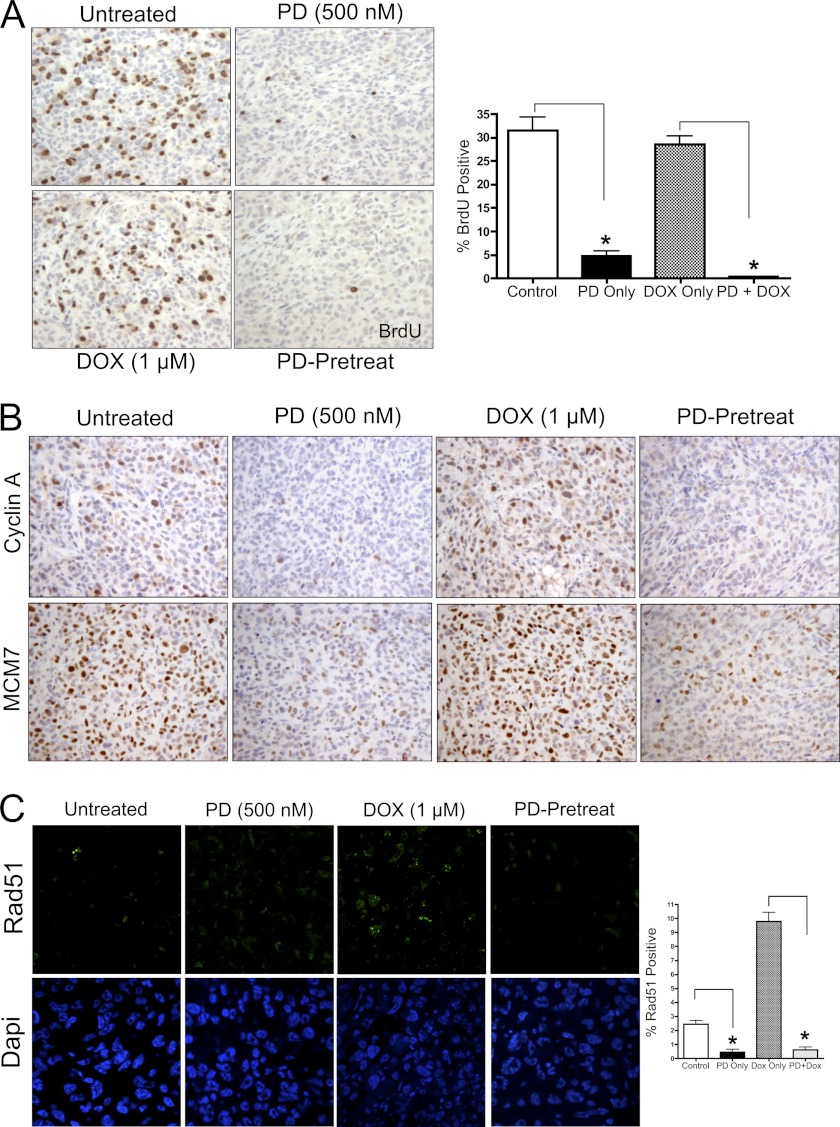
As much of the influence of CDK4/6 inhibition on chemotherapeutic response is ostensibly cell cycle dependent, we used ionizing radiation (IR) to induce DNA damage irrespective of cell cycle. Ionizing radiation is known to induce DNA damage regardless of cell cycle phase and is frequently employed in the clinical treatment of breast cancers. Initially, RB-proficient MDA-MB-231 breast cancer cells were exposed to 2Gy IR, and we observed the rate of damage repair by quantifying phospho-γ-H2AX (Ser-129) foci formation over 24 h postirradiation (Fig. 4A and supplemental Fig. S3). Previous studies have demonstrated phospho-γ-H2AX foci to be an accurate readout for IR-induced double strand DNA breaks (21, 22). Generally, the abundance of phospho-γ-H2AX foci peaked at ∼2 h post IR and returned to baseline levels near 24 h post IR in both the presence and absence of CDK4/6 inhibition. These results were surprising, as many DNA damage and repair proteins are observed in the RB/E2F gene signature (3), including those involved in double-strand break (DSB) repair (e.g. Rad51).
FIGURE 4.
CDK4/6 inhibition reduces Rad51 protein levels, but does not modify the rate of DNA damage repair in breast cancer cells treated with γ-irradiation. A, quantification of phospho-γ-H2AX foci over 24 h post IR in RB-proficient breast cancer cells treated with 2 Gy IR, 500 nm PD0332991 for 24 h followed by 2 Gy IR (PD-pretreatment) or 500 nm PD0332991 at the time of IR (Concurrent). Data are represented as the average number of phospho-γ-H2AX foci per cell as visualized by immunofluorescence. B, (left) representative immunofluorescence images of Rad51 protein among treatments described in A. RB-deficient MDA-MB-468 cells are included for comparison. Right, quantification of Rad51 foci and average Rad51 foci size 24 h post IR exposure. C, immunoblot analysis examining Rad51 protein levels and RB/E2F target gene product response in RB-proficient and RB-deficient breast cancer cell lines after treatment with 500 nm PD0332991, 2 Gy IR or 24 h 500 nm PD0332991 treatment followed by 2 Gy IR (PD-pretreat). Lamin B served as a loading control. D, immunoblot analysis of Rad51 protein levels in matched RB-proficient and RB-knockdown breast cancer cells after 500 nm PD0332991 treatment, 2 Gy IR or exposure to 500 nm PD0332991 24 h prior to IR (PD-pretreatment).
While the overall rate of DNA damage repair remained largely unaffected by CDK4/6 inhibition, we surmised that the specific mechanism through which DNA damage repair was occurring may be altered due to the tight correlation of essential repair proteins to E2F-mediated transcription. To this end, we examined the accumulation of Rad51 foci in response to combined CDK4/6 inhibition and IR exposure. In response to ionizing radiation, Rad51 form sub-nuclear foci that are easily visualized by immunofluorescence microscopy (23). As shown in Fig. 4B (top row), 2 Gy IR exposure promotes formation of Rad51 foci as measured by immunofluorescence. The abundance of Rad51 foci was not appreciably altered through concurrent administration of PD0332991 (middle row); however, PD0332991-pretreatment resulted in complete inhibition of Rad51 foci formation throughout the 24 h repair window (Fig. 4B, bottom row). Importantly, RB-deficient MDA-MB-468 cells demonstrated no appreciable down-regulation of Rad51 foci in response to CDK4/6 inhibition pre IR exposure (Fig. 4B, right column). Quantification of average foci per cell and average foci size per positive cell were calculated, and demonstrated that only pretreatment with PD0332991 in an RB proficient setting resulted in significant inhibition of Rad51 foci accumulation as compared with the IR-only treatments (Fig. 4B, right panels). To confirm that the regulation of Rad51 protein abundance was not cell line specific, immunoblot analysis was employed to examine Rad51 protein abundance in PD0332991-only, 2 Gy IR and PD0332991-pretreatment conditions across additional cell lines. As shown in Fig. 4C, RB-proficient breast cancer cell lines (MDA-MB-231, MCF7, and Hs578t) demonstrate a dramatic down-regulation of Rad51 specifically in response to PD0332991 treatment. This corresponds to inhibition of cell cycle progression as demonstrated by reduction of cyclin A, and reinforces the concept that these proteins are regulated in an RB-dependent manner. Importantly, among RB-deficient populations (MDA-MB-468) the intracellular levels of Rad51 and cyclin A were not appreciably altered through CDK4/6 inhibition.
Given the previous inclusion of Rad51 in an RB/E2F gene signature and co-regulation with cyclin A, knockdown of RB transcript and protein using shRNA was employed to generate isogenic breast cancer cells that could be used to confirm specificity of RB status (Fig. 4D). miNS (RB-proficient) populations display a pattern of Rad51 protein abundance as previously described in Fig. 4C, wherein the presence of PD prevents accumulation of Rad51 regardless of IR exposure. Correspondingly, miRB (RB knockdown) populations demonstrate rescue of Rad51 protein in the presence of CDK4/6 inhibition. While Rad51 abundance is not completely restored to that of untreated IR-only levels, this is likely due to incomplete knockdown of RB transcript and a partial dependence on CDK4/6 activity. Therefore, the response of Rad51 protein to CDK4/6 inhibition is occurring in an RB-dependent manner.
CDK4/6 Inhibition Alters the Mechanism through Which IR-induced DSBs Are Repaired
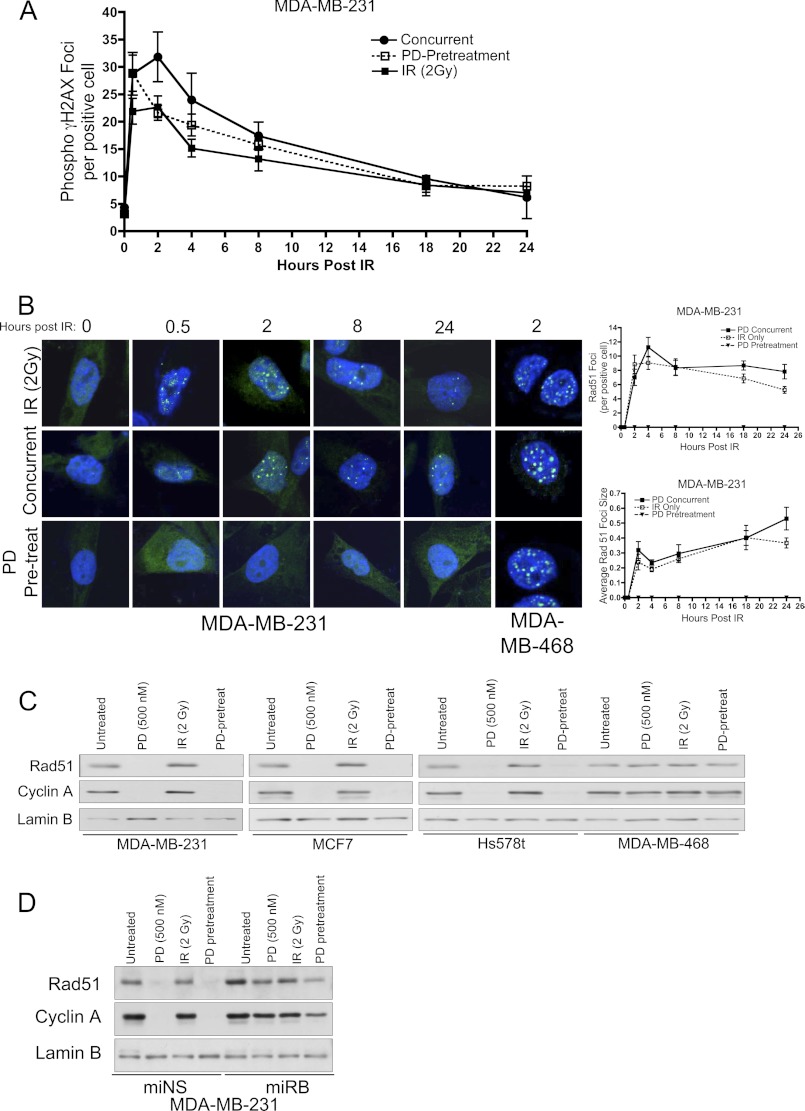
The observed decrease in Rad51 protein combined with no significant difference in overall DSB repair as measured via H2AX foci (Fig. 4), suggests that repair is occurring in an HR-independent manner. The other primary mechanism for cellular DSB repair is the NHEJ pathway. To test the hypothesis that CDK4/6 inhibition could specifically modify the mechanisms through which IR-induced DNA damage is repaired (24), we first performed a biochemical analysis of proteins central to HR versus NHEJ in cells irradiated in the presence or absence of CDK4/6 inhibition (Fig. 5A). Initially, Rad51 protein levels were confirmed to be undetectable among PD0332991 pretreated RB-proficient cell populations, which were again correlated with decreased levels of cyclin A, indicative of a cell cycle arrest in G1-phase. Interestingly, the abundance of CHK1 protein was also decreased in the PD0332991-pretreated condition. As CHK1 is known to inactivate CDC25 phosphatases in response to stalled replication forks and prevent entry into mitosis, a lack of CHK1 protein ultimately suggests that in the presence of CDK4/6 inhibition, cell cycle does not appreciably pass beyond the restriction point of the cell cycle, thus negating the need for negative regulation of entry into mitosis (25). Importantly, phosphorylation of CHK2 on the Thr68 residue was observed to be elevated for several hours after irradiation regardless of RB status and/or CDK4/6 inhibition. Phosphorylation of CHK2 has been previously demonstrated to be directly responsive to IR exposure (26–28). Our data suggest that the sensing of DNA damage by ATM/ATR was intact and largely independent of RB in these cells.
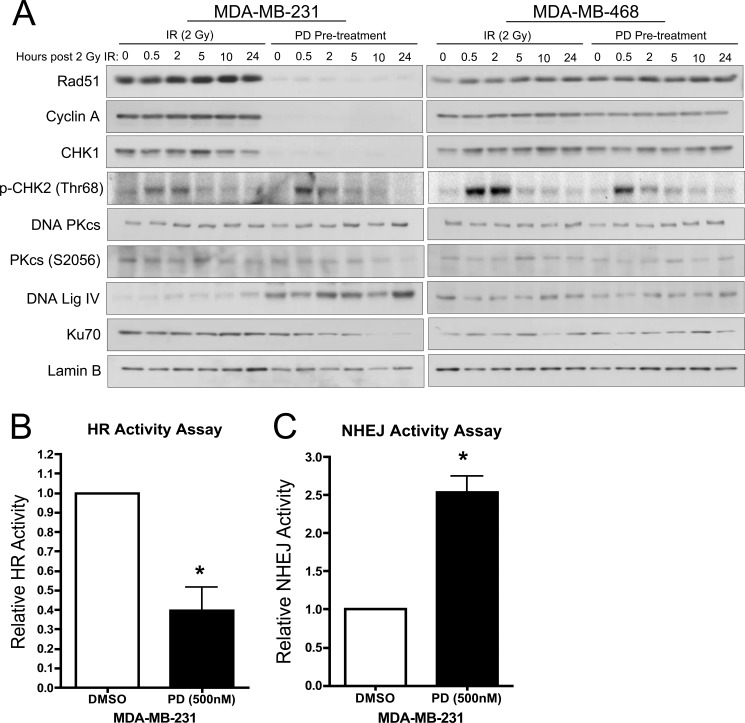
FIGURE 5.
CDK4/6 inhibition alters the mechanism through which IR-induced DSBs are repaired. A, immunoblot time course analysis examining the influence of PD0332991 pretreatment on proteins involved in homologous recombination and non-homologous end joining DNA damage repair pathways up to 24 h post IR in RB-proficient (MDA-MB-231) and RB-deficient (MDA-MB468) cells. Cyclin A is included as an overall readout of E2F activity. Lamin B served as a loading control. B, relative GFP reporter activity for homologous recombination-mediated activity assay in DMSO and 24 h PD0332991 pretreated MDA-MB-231 cells. C, relative GFP reporter activity for non-homologous end joining repair assay in DMSO and 24 h PD0332991 pretreated MDA-MB-231 cells.
In the context of factors involved in NHEJ, immunoblot analysis demonstrates no appreciable change in total DNA PKcs or its phosphorylation on Ser-2056 upon CDK4/6 inhibition in RB-proficient populations (Fig. 5A). DNA PKcs is a cellular kinase integral to NHEJ biology, and its phosphorylation has been previously demonstrated to be essential for overall NHEJ activity (29). Interestingly, protein levels of the downstream NHEJ DNA ligase (DNA Lig IV) were observed to be uniformly increased upon CDK4/6 inhibition, as compared with IR alone, and in an RB-dependent manner. This could indicate increased ligase activity from the protein that constitutes the final step in NHEJ. Lastly, abundance of Ku70 protein was observed to be largely unchanged in response to IR alone. However, upon exposure to PD0332991, the abundance of Ku70 was steadily diminished to near undetectable levels by 10 h post IR (Fig. 5A). Having been recently described as central to the NHEJ process, the gradual reduction across the window of repair (24 h) may indicate a utilization of all available Ku70 protein and a failure to replenish cellular pools under conditions of CDK4/6 inhibition (30). Importantly, among RB-deficient populations (Fig. 5A, right panel) protein levels of all examined components of HR and NHEJ are unaffected by CDK4/6 inhibition, reinforcing the concept that these pathways are responsive to RB phosphorylation via CDKs 4/6.
To functionally interrogate the DNA repair mechanisms utilized, reporter plasmids for HR and NHEJ activity were employed. To directly measure homologous recombination activity, MDA-MB-231 cells pretreated with PD0332991 or DMSO for 24 h were irradiated and transfected with a linear (ISCE-1 digested) DR-GFP reporter plasmid alongside a DsRed expression vector to control for transfection efficiency. GFP fluorescence was evaluated 72 h post-transfection via flow cytometry and is reported as relative to the untreated control (Fig. 5B). As depicted, CDK4/6 inhibition resulted in an ∼60% decrease in HR-mediated DNA damage repair. Thus, the decrease in Rad51 protein observed upon exposure to PD0332991 correlates with a significant decrease in HR-mediated DSB repair. Conversely, pEGFP-PEM1-Ad2 reporter activity demonstrated NHEJ activity was increased ∼2.5-fold under identical conditions (Fig. 5C). Taken together, these data indicate that CDK4/6 inhibition can alter the mechanisms by which cells signal the presence of DNA damage and ultimately the pathways utilized to repair therapeutically induced DSBs.
DISCUSSION
While the treatment of human malignancy has greatly benefited in recent years through the development of targeted therapies, the vast majority of advanced cancers are still treated with classical chemotherapeutic agents. In the case of breast cancer, subtypes that initially respond to targeted interventions (e.g. ER and Her2 antagonists) frequently evolve to acquire resistance to these agents. Additionally, triple negative breast cancers (TNBCs), which lack established markers to direct targeted intervention, are virtually always treated with cytotoxic chemotherapy regimens. Given the long-term and ubiquitous application of these agents, there is a wealth of information regarding their mechanisms of action, as well as their far-reaching clinical side effects. Thus, there is a clear need for novel therapeutic agents in the treatment of advanced cancers that lack the targets for, or are unresponsive to, current targeted therapies. In this context, CDK4/6 inhibitors (e.g. PD0332991) are a novel class of targeted agents that elicit a potent cytostatic response in cells that harbor a functional RB pathway. As RB is downstream of most mitogenic signaling pathways (e.g. ER, HER2), therapeutic CDK4/6 inhibition represents an effective means to target those cancers that are unresponsive to established targeted therapies. Furthermore, results from Phase I clinical trials utilizing PD0332991 have demonstrated a low toxicity profile, which suggests the potential for combining CDK4/6 inhibitors with current utilized chemotherapy regimens. In fact, PD0332991 is currently undergoing a phase I/II study of patients with advanced breast cancers in combination with paclitaxel.
As the RB/E2F axis is considered the point at which mitogenic signaling interacts with cell cycle machinery, input from these signaling cascades directly influences a wide range of transcriptional networks. Recently, the RB pathway has been profiled in greater detail, and has been shown to regulate the transcription of numerous genes involved in cell cycle progression, DNA repair, and apoptosis (3). Standard chemotherapeutic agents exploit the deregulation of these aforementioned processes in tumor cells to elicit their cytotoxic effects. Considering the well-documented mechanism of action of CDK4/6 inhibitors through the RB pathway, it is unclear whether a cytostatic agent such as PD0332991 would ultimately function cooperatively or antagonistically with cytotoxic chemotherapies. Recently, studies have demonstrated an importance for CDK4/6 activity in cellular senescence mediated by FOXM1 (31). While this systematic screen identified the first potentially relevant targets for CDK4/6 inhibition outside of the RB protein, FOXM1 protein levels were found to be regulated in an RB-dependent manner within our model systems (supplemental Fig. S4). Ultimately, these data reinforce the notion that CDK4/6 inhibition was driving cell cycle arrest through RB, and that this influence could modify the response to therapeutic agents requiring active cell cycle. To interrogate this question in a preclinical setting, we combined PD0332991 with frequently utilized genotoxic chemotherapies in models of TNBC.
In the context of anthracyclines and taxanes, RB-proficient cells that were pretreated with the CDK4/6 inhibitor demonstrated a prominent G1 cell cycle arrest, and a corresponding cell cycle distribution that was indistinguishable from cells treated with the CDK4/6 inhibitor alone. These results indicate that the inhibition of cell cycle progression via PD0332991 blocked cells from cycling through the chemotherapy-induced lesions and ultimately prevented cell death. Indeed, protein expression patterns of E2F-regulated genes among in vitro and in vivo populations exposed to the CDK4/6 inhibitor prior to chemotherapy administration reinforced the conclusion that RB-mediated transcriptional repression was arresting cell cycle progression prior to the induction of DNA damage. Moreover, RB-proficient populations pretreated with the CDK4/6 inhibitor did not effectively signal for PARP cleavage in response to genotoxic insult, suggesting that a cell cycle arrest by PD0332991 was ultimately able to prevent a cytotoxic apoptotic response. This notion was reinforced through long term cell growth studies wherein continuous PD0332991 treatment promoted increased survival after exposure to paclitaxel over a course of 7 days. Interestingly, a very different result was observed during cell outgrowth experiments wherein PD0332991 treatment was used to synchronize cells for 24 h prior to paclitaxel treatment. In this setting, the acute exposure to the CDK4/6 inhibitor resulted in increased cytotoxicity. These data suggest that the cycling of PD0332991, or any other CDK4/6 inhibitor, could be particularly effective in a metronomic setting with cytotoxic chemotherapies. Clearly, more in depth studies are required with specific agents to determine the clinical utility of a synchronized release strategy.
Mechanistically, while anthracyclines and taxanes are known to act through distinct mechanisms that are dependent on cell proliferation, one commonality of virtually all genotoxic agents, including γ irradiation (IR), is that they promote cell death through the induction of DNA damage. This damage is frequently in the form of double strand DNA breaks (DSBs), which can be repaired by the cell through one of two pathways, Homologous Recombination (HR) or Non-Homologous End Joining (NHEJ) (32). Importantly, HR and NHEJ pathways are comprised of unique protein components and complexes, many of which have been recently reported in an RB/E2F gene signature (3). To determine whether CDK4/6 inhibition can directly modulate the induction and response to DNA damage independent of the obvious effects on cell cycle progression, we evaluated the impact of CDK4/6 inhibition in combination with IR exposure. Upon initially surveying cellular components specifically responsible for sensing IR-induced DNA damage, it was observed that Chk2 phosphorylation at Thr68 was universal in response to IR, and was not altered through the presence of CDK4/6 inhibition or RB status. Interestingly, while the amount of DNA damage generated and repaired was virtually indistinguishable under all treatment conditions, we found that the repair was occurring in the absence of Rad51, a critical component of the HR-pathway (33). Moreover, a significant decrease in HR activity was observed in the presence of CDK4/6 inhibition. While cell cycle independent, this loss of Rad51 expression was demonstrated to be RB-dependent. Therefore, while CDK4/6 inhibition did not prevent the generation of DNA damage induced by exposure to IR, the mechanism by which the damage is repaired was clearly altered in an RB-dependent but cell cycle-independent manner.
Upon examination of components central to NHEJ, it was clear that the burden of DNA repair in cells treated with PD0332991 was shifted from the HR to NHEJ pathway. Components of NHEJ are not known to be regulated by RB and were not affected by CDK4/6 inhibition alone. However, upon exposure to PD0332991 and IR, levels of certain NHEJ components, as well as NHEJ activity, were elevated. Thus, repression of critical factors of HR (i.e. Rad51) via CDK4/6 inhibition results in a compensatory mechanism of DNA repair through NHEJ. This shift in DNA repair mechanisms could have significant impact in cells treated with combination regimens of CDK4/6 inhibitors and genotoxic agents such as IR, as NHEJ has been previously demonstrated to promote increased rates of genomic rearrangements, as compared with HR (34, 35). In fact, overexpression of Rad51 protein has been previously demonstrated to be sufficient to reverse recombination defects that occur in an HR-deficient setting, suggesting a critical role for Rad51 in preventing genomic instability (36). This is particularly relevant in the context of human cancers, as genomic instability is known to be one of the canonical hallmarks of cancer. While much effort is being concentrated on elucidating the specific contributions that genomic instability plays on the genesis and progression of human cancers, it is not hard to imagine that forcing populations of cancer cells to repair clinically-induced DNA damage through the error-prone NHEJ pathway could promote increased rates of genomic rearrangement and ultimately provide mutations, which influence both response to treatment and disease recurrence.
As novel targeted agents such as CDK4/6 inhibitors progress to clinical utility, particularly when considering combination regimens with currently used chemotherapies, it will be critical to consider the unique mechanistic requirements of each therapy (i.e. cell cycle), as well as alterations to the transcriptional landscape of cells that may ultimately modify therapeutic response. Clearly, the RB/E2F signaling axis is responsible for the transcription of a diverse array of gene products, and given its central role in cell cycle progression, it would be prudent to consider availability of therapeutic targets and components of required cellular processes prior to utilizing CDK4/6 inhibitors in combination with current and future chemotherapeutic agents.
Supplementary Material

This article contains supplemental Figs. S1–S4.
- RB
- retinoblastoma tumor suppressor
- CDK
- cyclin-dependent kinase
- HR
- homologous recombination
- NHEJ
- non-homologous end joining
- IR
- β-irradiation
- PARP
- poly-ADP ribose polymerase
- TNBC
- triple negative breast cancer
- DSB
- double strand DNA break
- MCM7
- mini-chromosome maintenance 7 protein
- BrdU
- bromodeoxyuridine (5-bromo-2′-deoxyuridine).
REFERENCES
- 1. Knudsen E. S., Knudsen K. E. (2008) Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer 8, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Viatour P., Sage J. (2011) Newly identified aspects of tumor suppression by RB. Dis. Model Mech. 4, 581–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ertel A., Dean J. L., Rui H., Liu C., Witkiewicz A. K., Knudsen K. E., Knudsen E. S. (2010) RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle 9, 4153–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dean J. L., Thangavel C., McClendon A. K., Reed C. A., Knudsen E. S. (2010) Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene 29, 4018–4032 [DOI] [PubMed] [Google Scholar]
- 5. Finn R. S., Dering J., Conklin D., Kalous O., Cohen D. J., Desai A. J., Ginther C., Atefi M., Chen I., Fowst C., Los G., Slamon D. J. (2009) PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 11, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fry D. W., Bedford D. C., Harvey P. H., Fritsch A., Keller P. R., Wu Z., Dobrusin E., Leopold W. R., Fattaey A., Garrett M. D. (2001) Cell cycle and biochemical effects of PD 0183812. A potent inhibitor of the cyclin D-dependent kinases CDK4 and CDK6. J. Biol. Chem. 276, 16617–16623 [DOI] [PubMed] [Google Scholar]
- 7. Fry D. W., Harvey P. J., Keller P. R., Elliott W. L., Meade M., Trachet E., Albassam M., Zheng X., Leopold W. R., Pryer N. K., Toogood P. L. (2004) Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3, 1427–1438 [PubMed] [Google Scholar]
- 8. Michaud K., Solomon D. A., Oermann E., Kim J. S., Zhong W. Z., Prados M. D., Ozawa T., James C. D., Waldman T. (2010) Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 70, 3228–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Konecny G. E., Winterhoff B., Kolarova T., Qi J., Manivong K., Dering J., Yang G., Chalukya M., Wang H. J., Anderson L., Kalli K. R., Finn R. S., Ginther C., Jones S., Velculescu V. E., Riehle D., Cliby W. A., Randolph S., Koehler M., Hartmann L. C., Slamon D. J. (2011) Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin. Cancer Res. 17, 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thangavel C., Dean J. L., Ertel A., Knudsen K. E., Aldaz C. M., Witkiewicz A. K., Clarke R., Knudsen E. S. (2011) Therapeutically activating RB: reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr. Relat. Cancer 18, 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiedemeyer W. R., Dunn I. F., Quayle S. N., Zhang J., Chheda M. G., Dunn G. P., Zhuang L., Rosenbluh J., Chen S., Xiao Y., Shapiro G. I., Hahn W. C., Chin L. (2010) Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc. Natl. Acad. Sci. U.S.A. 107, 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Musgrove E. A., Caldon C. E., Barraclough J., Stone A., Sutherland R. L. (2011) Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572 [DOI] [PubMed] [Google Scholar]
- 13. Schwartz G. K., LoRusso P. M., Dickson M. A., Randolph S. S., Shaik M. N., Wilner K. D., Courtney R., O'Dwyer P. J. (2011) Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1). Br. J. Cancer 104, 1862–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Treré D., Brighenti E., Donati G., Ceccarelli C., Santini D., Taffurelli M., Montanaro L., Derenzini M. (2009) High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Ann. Oncol. 20, 1818–1823 [DOI] [PubMed] [Google Scholar]
- 15. Zagorski W. A., Knudsen E. S., Reed M. F. (2007) Retinoblastoma deficiency increases chemosensitivity in lung cancer. Cancer Res. 67, 8264–8273 [DOI] [PubMed] [Google Scholar]
- 16. Saeki H., Siaud N., Christ N., Wiegant W. W., van Buul P. P., Han M., Zdzienicka M. Z., Stark J. M., Jasin M. (2006) Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. U.S.A. 103, 8768–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seluanov A., Mittelman D., Pereira-Smith O. M., Wilson J. H., Gorbunova V. (2004) DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 101, 7624–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang M., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G. (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McClendon A. K., Dean J. L., Ertel A., Fu Z., Rivadeneira D. B., Reed C. A., Bourgo R. J., Witkiewicz A., Addya S., Mayhew C. N., Grimes H. L., Fortina P., Knudsen E. S. (2011) RB and p53 cooperate to prevent liver tumorigenesis in response to tissue damage. Gastroenterology 141, 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. (2004) Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 56, 185–229 [DOI] [PubMed] [Google Scholar]
- 21. Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. (2004) H2AX: the histone guardian of the genome. DNA Repair 3, 959–967 [DOI] [PubMed] [Google Scholar]
- 22. Löbrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A. A., Barton O., Jeggo P. A. (2010) γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 9, 662–669 [DOI] [PubMed] [Google Scholar]
- 23. McCabe N., Turner N. C., Lord C. J., Kluzek K., Bialkowska A., Swift S., Giavara S., O'Connor M. J., Tutt A. N., Zdzienicka M. Z., Smith G. C., Ashworth A. (2006) Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115 [DOI] [PubMed] [Google Scholar]
- 24. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 25. Dai Y., Grant S. (2010) New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. 16, 376–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahn J. Y., Schwarz J. K., Piwnica-Worms H., Canman C. E. (2000) Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60, 5934–5936 [PubMed] [Google Scholar]
- 27. Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. (2000) Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 97, 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melchionna R., Chen X. B., Blasina A., McGowan C. H. (2000) Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2, 762–765 [DOI] [PubMed] [Google Scholar]
- 29. Smith G. C., Jackson S. P. (1999) The DNA-dependent protein kinase. Genes Dev. 13, 916–934 [DOI] [PubMed] [Google Scholar]
- 30. Fattah F., Lee E. H., Weisensel N., Wang Y., Lichter N., Hendrickson E. A. (2010) Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 6, e1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anders L., Ke N., Hydbring P., Choi Y. J., Widlund H. R., Chick J. M., Zhai H., Vidal M., Gygi S. P., Braun P., Sicinski P. (2011) A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20, 620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pardo B., Gómez-González B., Aguilera A. (2009) DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol. Life. Sci. 66, 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sonoda E., Sasaki M. S., Buerstedde J. M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Campbell P. J., Stephens P. J., Pleasance E. D., O'Meara S., Li H., Santarius T., Stebbings L. A., Leroy C., Edkins S., Hardy C., Teague J. W., Menzies A., Goodhead I., Turner D. J., Clee C. M., Quail M. A., Cox A., Brown C., Durbin R., Hurles M. E., Edwards P. A., Bignell G. R., Stratton M. R., Futreal P. A. (2008) Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lieber M. R., Yu K., Raghavan S. C. (2006) Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair 5, 1234–1245 [DOI] [PubMed] [Google Scholar]
- 36. Schild D., Wiese C. (2010) Overexpression of RAD51 suppresses recombination defects: a possible mechanism to reverse genomic instability. Nucleic Acids Res. 38, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.