Background: The enzyme β4Gal-T1 synthesizes the LacNAc moiety of glycans.
Results: The extended oligosaccharide moiety of β1–6-branched GlcNAc acceptors binds to a different region on the enzyme.
Conclusion: β4Gal-T1 has two different oligosaccharide binding regions for extended oligosaccharide moieties of different acceptor substrates.
Significance: Multiple carbohydrate acceptor binding regions are observed on a glycosyltransferase.
Keywords: Enzyme Kinetics, Enzyme Structure, Glycosyltransferases, Oligosaccharide, Structural Biology, Multiple Carbohydrate Binding Site, Beta-4Gal-T1, i/I-Antigen Synthesis
Abstract
N-Acetyllactosamine is the most prevalent disaccharide moiety in the glycans on the surface of mammalian cells and often found as repeat units in the linear and branched polylactosamines, known as i- and I-antigen, respectively. The β1–4-galactosyltransferase-I (β4Gal-T1) enzyme is responsible for the synthesis of the N-acetyllactosamine moiety. To understand its oligosaccharide acceptor specificity, we have previously investigated the binding of tri- and pentasaccharides of N-glycan with a GlcNAc at their nonreducing end and found that the extended sugar moiety in these acceptor substrates binds to the crevice present at the acceptor substrate binding site of the β4Gal-T1 molecule. Here we report seven crystal structures of β4Gal-T1 in complex with an oligosaccharide acceptor with a nonreducing end GlcNAc that has a β1–6-glycosidic link and that are analogous to either N-glycan or i/I-antigen. In the crystal structure of the complex of β4Gal-T1 with I-antigen analog pentasaccharide, the β1–6-branched GlcNAc moiety is bound to the sugar acceptor binding site of the β4Gal-T1 molecule in a way similar to the crystal structures described previously; however, the extended linear tetrasaccharide moiety does not interact with the previously found extended sugar binding site on the β4Gal-T1 molecule. Instead, it interacts with the different hydrophobic surface of the protein molecule formed by the residues Tyr-276, Trp-310, and Phe-356. Results from the present and previous studies suggest that β4Gal-T1 molecule has two different oligosaccharide binding regions for the binding of the extended oligosaccharide moiety of the acceptor substrate.
Introduction
The cell surface glycans play an important role in several cellular functions (1). The N-acetyllactosamine (LacNAc)2 moiety (Galβ1–4GlcNAcβ) is the most prevalent glycan moiety found on the mammalian cell surface. In N-glycans, it is linked to an α-mannose (α-Man) in a β1–2-, β1–4-, or β1–6-glycosidic linkage, whereas in the O-glycans in core 2 and core 4 structures, it is linked to an α-N-acetylgalactosamine (α-GalNAc) in β1–3- and β1–6-glycosidic linkages. Interestingly, the N-acetyllactosamine is also found repeated in linear and branched polylactosamines, characterized as i and I antigens, respectively (2, 3) (supplemental Fig. S-F1). In linear repeats (i-antigen), LacNAc is linked to β-Gal in a β1–3-glycosidic linkage forming a linear structure such as (Galβ1-4GlcNAcβ)n1-3Galβ1-4GlcNAcβ)-R, and in branched repeats (I-antigen), it is linked to a β-Gal residue in the linear poly-LacNAc in a β1-6-glycosidic linkage, such as the (Galβ1-4GlcNAcβ)n1-3 (Galβ1-4GlcNAcβ1-6))Galβ1-4GlcNAcβ-R structure (underline indicates nonreducing end GlcNAc residue). These linear repeats of the LacNAc moiety are found to have Lewis-type carbohydrate moieties at their nonreducing end, and they are known to play an important role in several cellular recognition processes by binding to Sialic acid-recognizing immunoglobulin superfamily lectin (4). Interestingly, the LacNAc moiety is linked to a β-sugar in i/I antigens, whereas in others, such as in N-/O-glycans, it is linked to α-sugars.
Glycosyltransferases are Golgi-resident, type II membrane proteins, and they are responsible for the synthesis of most glycans on the cell surface (5). Among these transferases, β1–4-galactosyltransferase I (β4Gal-T1) is involved in the synthesis of the LacNAc moiety. In the presence of manganese, it transfers galactose (Gal) from UDP-Gal to a nonreducing end β-N-acetylglucosamine (GlcNAcβ) acceptor sugar moiety of glycoconjugates, thus synthesizing the LacNAc (Galβ1–4GlcNAcβ) moiety (6, 7). Although this enzyme transfers Gal to a monosaccharide β-GlcNAc acceptor, a β-GlcNAc at the nonreducing end of an oligosaccharide is a preferred acceptor substrate, as judged from their kinetic constants (Km) (8, 9). The structure and function of this enzyme have been well characterized by both the single crystal structure determination and the enzyme kinetics studies (10, 11). It has been found from the crystal structure studies that in addition to the acceptor monosaccharide binding site, an extended sugar binding site was first predicted and in later studies found to entertain the binding of the extended sugars of the acceptor sugar GlcNAc (12, 13). Furthermore, the oligosaccharide preference of this extended sugar binding site has also been elucidated by these studies. However, these studies have been carried out mostly with the oligosaccharide acceptor substrates that are part of the N-glycan structure, particularly GlcNAc residues linked to α-mannose in β1–2-, β1–3-, and β1–4-glycosidic linkages. The binding of the nonreducing end GlcNAc with a β1–6-glycosidic linkage to a linear saccharide or to a branch from a poly-LacNAc glycan was not known and has been investigated here. The β1–6-glycosidic linkage differs from β1–2-, β1–3-, and β1–4-glycosidic linkages by having a CH2 group between the glycosidic bond and the sugar ring. The CH2-group present in the β1–6-glycosidic-linked disaccharide gives more conformational flexibility for the disaccharide when compared with the others. A GlcNAc residue β1–6-linked to α-Man sugar moiety of an N-glycan is abundantly found in cancer cells (14). Here we present the crystal structure studies on the binding of oligosaccharide acceptors with β1–6-linked GlcNAc at the nonreducing end that is analogous to i/I antigens and an N-glycan structure to the human β4Gal-T1 molecule (see Fig. 1 and supplemental Fig. S-F2). We find that the extended sugar moiety attached to the acceptor β1–6-branched GlcNAc residue in the I-antigen analog binds to a new region of the acceptor binding site of human β4Gal-T1 enzyme that has not been previously observed or predicted.
FIGURE 1.
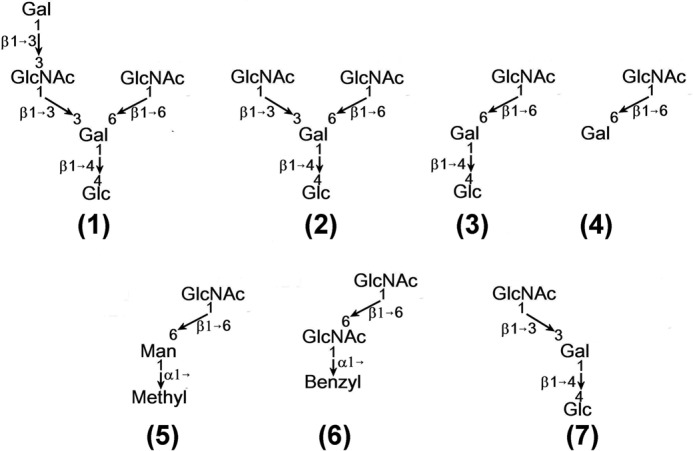
The seven oligosaccharide acceptors of β4Gal-T1 enzyme used in the present crystal structure determination studies are shown. Acceptors 1, 2, and 7 are generated from the neo-glycosides, lacto-neohexaose, lacto-N-neohexaose, and lacto-neotetraose, respectively.
EXPERIMENTAL PROCEDURES
The acceptor carbohydrates were purchased from Sigma and ProZyme. The β1–4-galactosidase enzyme was purchased from Calbiochem. In a typical degalactosylating reaction, a 10 mm oligosaccharide solution from 1 mg was treated overnight with 2 milliunits of β1–4-galactosidase from Streptococcus pneumoniae (Calbiochem) at room temperature. The Acceptor 1 substrate was made by selective degalactosylation of lacto-neohexaose by treating it with β1–4-galactosidase, which degalactosylated the LacNAc moiety at the β1–6 branch (supplemental Fig. S-F3). Similarly, the Acceptor 2 and Acceptor 3 acceptor substrates were made by treating the lacto-N-neohexaose and lacto-neotetraose with β1–4-galactosidase, respectively (supplemental Fig. S-F3). These degalactosylated oligosaccharides were used in our crystallization without further purifications.
The expression and purification of the human C338T-M340H-β4Gal-T1 protein are similar to that previously described (13). The protein complex with the acceptor substrates was grown at 4 °C in the presence of 5 mm Mn2+ and 5 mm UDP-hexanolamine. The three-dimensional diffraction data from the single crystals were collected in-house at a data collection facility equipped with a Mar345 area detector and processed using HKL2000. The data collection statistics are given in Table 1. The crystals are isomorphous to the previous crystals of the same protein complex with the different oligosaccharide acceptors; therefore, the crystal structures were solved by molecular placement methods using only protein atoms without any substrates. The substrates Mn2+, UDP-hexanolamine, and the acceptor were located from the difference Fourier electron density maps. The structures were refined using REFMAC5, and WinCoot was used for model correction and solvent locations (15, 16). The final refinement statistics are given in Table 1. The final coordinates and the diffraction data have been deposited in the Protein Data Bank (PDB).
TABLE 1.
X-ray data collection and refinement statistics for human C338T-M340H-β4Gal-T1 protein complex with UDP·Mn2+ and the oligosaccharide
Values shown in parenthesis correspond to high resolution shell.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Crystal data | |||||||
| a = (Å) | 107.6 | 107.3 | 107.3 | 107.4 | 107.4 | 107.8 | 107.3 |
| b = (Å) | 195.2 | 196.0 | 196.2 | 195.5 | 196.4 | 198.2 | 195.3 |
| c = (Å) | 143.7 | 143.8 | 143.6 | 143.7 | 143.8 | 143.7 | 143.9 |
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 |
| X-ray data collection data | |||||||
| Resolution (Å) | 2.3 | 1.95 | 2.0 | 2.2 | 2.2 | 2.3 | 2.2 |
| Unique reflections | 60,823 | 97,326 | 96,684 | 70,184 | 72,510 | 64,732 | 70,949 |
| Data redundancy | 6.4 (5.3) | 3.5 (3.1) | 6.0 (4.9) | 4.1 (3.4) | 5.6 (4.5) | 3.4 (3.2) | 7.4 (6.6) |
| Completeness | 91 (67) | 94 (64) | 100 (99) | 96 (78) | 99 (96) | 95 (97) | 98 (96) |
| Rsym (%) | 9.2 (37) | 4.0 (27) | 4.9 (47) | 8.2 (40) | 7.4 (35) | 5.8 (35) | 7.0 (54) |
| Refinement statistics | |||||||
| Refinement parameters | |||||||
| Rfinal (%) | 18.7 (26) | 19.5 (28) | 20.0 (33) | 19.9 (29) | 19.2 (27) | 20.3 (29) | 20.3 (28) |
| Rfree (%) | 24 (34) | 23 (33) | 24 (36) | 24 (33) | 24 (32) | 25 (39) | 25 (32) |
| r.m.s.a deviation | |||||||
| Bond length (Å) | 0.019 | 0.022 | 0.018 | 0.017 | 0.016 | 0.019 | 0.015 |
| Bond angle (°) | 1.948 | 2.004 | 1.822 | 1.834 | 1.759 | 1.755 | 1.732 |
| Ramachandran map (using Procheck) | |||||||
| Preferred/Allowed region | 87/13 | 89.2/10.8 | 88.8/11.2 | 87.4/12.6 | 88.5/11.5 | 88.5/11.5 | 88.7/11.3 |
| PDB entry number | 4EE3 | 4EE4 | 4EEA | 4EEG | 4EEM | 4EEO | 4EE5 |
a r.m.s., root mean square.
The catalytic activity using Acceptors 1 and 2 were carried out using different acceptor substrate concentrations with 0.5 mm UDP-Gal was performed as described earlier (13). The apparent Km values for these acceptor substrates were determined based on their enzyme activity curve fitted with SigmaPlot using an equation defining ligand binding to one site saturation without inhibition (supplemental Fig. S-F5). The MALDI-TOF analyses at different time points were also performed as described earlier (supplemental Fig. S-F6) (17).
RESULTS AND DISCUSSION
Earlier we reported the crystal structure of the catalytic domain of human C338T-M340H-β4Gal-T1 mutant protein in the presence of Mn2+ and UDP-hexanolamine with several oligosaccharide acceptor substrates with GlcNAc at the nonreducing end (13). The mutation of Cys-338 to Thr-338 in human β4Gal-T1 (Cys-342 in bovine β4Gal-T1) gives stability to the enzyme (18), whereas the mutation of Met-340 (Met-344 in bovine β4Gal-T1) to His-340 helps the mutant enzyme crystallize readily in the closed conformation in the presence of Mn2+ and UDP-hexanolamine (19). This allows the binding of the oligosaccharide acceptor substrates and the crystallization of the complexes (13). Here we report the crystal structures of the mutant protein C338T-M340H-β4Gal-T1 in complex with closely related seven oligosaccharide acceptor substrates (Fig. 1 and supplemental Fig. S-F2).
In all the present crystal structures, three C338T-M340H-β4Gal-T1 protein molecules are found in the asymmetric unit, and they all adopt the closed conformation. As expected, each molecule has a bound Mn2+ ion and a UDP-hexanolamine molecule. Although the uridine nucleoside moiety is clearly seen from the electron density maps in these crystal structures, the electron densities for the phosphate and hexanolamine groups are not clearly observed, suggesting that these groups are disordered. The Mn2+ ion forms coordination bonds with the residues Asp-250, His-340, and His-343. In the closed conformation, each phosphate group oxygen atom of the diphosphate moiety forms a coordination bond with the Mn2+ ion. However, in the present crystal structures, due to the disordered nature of the diphosphate groups, their coordination with the Mn2+ ion could not be clearly determined. This is similar to the crystal structure of the bovine W314A-β4Gal-T1 complex with α-lactalbumin protein, where the β4Gal-T1 molecule was found in the closed conformation, with disordered phosphate groups and Mn2+ coordination (20). In all the crystal structures, the respectively bound acceptor substrates could be clearly seen in all three molecules in the asymmetric unit from the electron density maps (supplemental Fig. S-F4). The molecular interactions between the acceptor GlcNAc moiety and the protein molecule are very similar to those found in the previous crystal structures (10, 13). However, the extended sugar interactions with the protein molecules vary; these interactions are discussed below.
Interactions of the Pentasaccharide Galβ1–3GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc (Acceptor 1) with the β4Gal-T1
The true minimum size I-antigen acceptor substrate for the β4Gal-T1 enzyme is a pentasaccharide containing a linear di-LacNAc tetrasaccharide with a nonreducing end GlcNAc residue (underlined) in a β1–6-glycosidic linkage with the core Gal, such as Galβ1–4GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1-4GlcNAc. This pentasaccharide is not commercially available. The Acceptor 1 pentasaccharide, Galβ1–3GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc, is similar to the true I-antigen acceptor pentasaccharide, except that it has a Glc residue instead of a GlcNAc residue at the reducing end and therefore was used for crystallization with the human C338T-M340H-β4Gal-T1 protein in the presence of Mn2+ and UDP-hexanolamine.
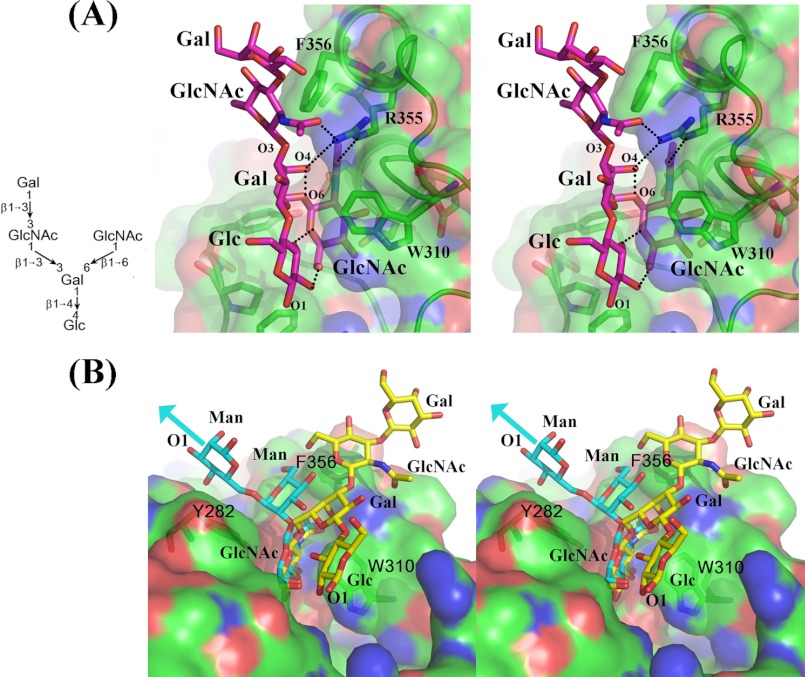
In the crystal structure of the pentasaccharide Acceptor 1 complex with the β4Gal-T1 molecule, the nonreducing end β1–6 branch GlcNAc moiety is bound in the acceptor binding site of the β4Gal-T1 molecule (Fig. 2A). The molecular interactions between this GlcNAc moiety and the protein molecule are very similar to those found in the earlier crystal structures (10, 13). However, the binding of the extended tetrasaccharide moiety to the protein molecule is quite different. The extended tetrasaccharide forms a linear sheet structure and forms a number of intramolecular hydrogen-bonding interactions with the β1–6-branched GlcNAc moiety (Fig. 2A). In addition, the extended tetrasaccharide also has hydrophobic interactions with the β4Gal-T1 molecule. The reducing end Glc, the core Gal, and the β1–3-linked GlcNAc residues form hydrophobic interactions with the aromatic side chains of the Phe-276, Trp-310, and Phe-356 residues, respectively. Although in the electron density maps the electron density for the glycosidic bond of the nonreducing end β1–3-linked Gal moiety is clearly observed, it extends only to the cyclic atoms of the Gal moiety. This may be due to the fact that this reducing end Gal moiety does not have any molecular interactions with the protein molecule. The side-chain guanidine nitrogen atoms of the residue Arg-355 form a hydrogen bond with the carbonyl oxygen atom of the N-acetyl moiety of the GlcNAc acceptor and the β1–3-linked GlcNAc residues; in addition, they form a hydrogen bond with the O4 oxygen atom of the core Gal residue. This hydrogen-bonding network specifically recognizes the trisaccharide moiety, GlcNAcβ1–6Galβ1–3GlcNAc. In the previously determined β4Gal-T1 complex with the oligosaccharide complex crystal structures, the extended sugar moieties interacted with the hydrophobic region created by the side chains of the residues Tyr-282, Phe-356, and Ile-358 (Fig. 2B). However, in the present crystal structure, the extended sugars do not make any molecular interactions with the side chains of the Tyr-282 and Ile-358 residues. Thus, the binding of the extended tetrasaccharide in the present crystal structure is distinctly different from the binding of the oligosaccharide from the N-glycan determined previously (Fig. 2B). The apparent Km value for the Acceptor 1 is 0.19 ± 0.04 mm, and it is comparable with the binding of different N-glycan trisaccharides to the enzyme (13).
FIGURE 2.
A, stereo diagram showing the binding of the pentasaccharide acceptor (Acceptor 1) bound to the β4Gal-T1 molecule. The β4Gal-T1 molecule is shown in the surface diagram with an embedded graphic diagram, and the key protein residues are shown in the stick diagram. The pentasaccharide is shown in the cyan-colored stick diagram, and the hydrogen bonds are shown in dotted black lines. The O2 and O3 hydroxyl group of the Glc residue forms a hydrogen bond with the O6 and O5 oxygen atoms of the nonreducing end GlcNAc molecule. Also, the side-chain amino groups of the Arg-355 form hydrogen bonds with both the GlcNAc and the core Gal residues, specifically recognizing the GlcNAcβ1–6Galβ1–3GlcNAc motif. B, stereo diagram showing the comparison of the binding of the I-antigen analog pentasaccharide (Acceptor 1, shown in yellow sticks) with the previously studied N-glycan trisaccharide, GlcNAcβ1–2Manα1–6Manβ- (shown in cyan-colored sticks; PDB 2AEC) to the β4Gal-T1 molecule (shown in green-colored surface diagram). The extended saccharide in the N-glycan (cyan) is found to interact with the Tyr-282 residue, whereas the extended tetrasaccharide in the I-antigen analog (yellow stick) is extended away from the N-glycan toward the residue Trp-310.
Interactions of the Tetrasaccharide GlcNAcβ1–3 (GlcNAcβ1-6)Galβ1–4Glc (Acceptor 2) with the β4Gal-T1
Althoughthe nonreducing end Gal moiety does not have any direct molecular interactions with the protein molecule, this pentasaccharide differs from the true I-antigen pentasaccharide acceptor by the glycosidic linkage of this Gal residue and therefore raises the possibility that the observed different binding mode for the extended tetrasaccharide may be due to this difference. Thus, the binding of the tetrasaccharide Acceptor 2, GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc (Fig. 1), without the nonreducing end β1–3-linked Gal residue that is present in the pentasaccharide Acceptor 1 to β4Gal-T1, was investigated. This tetrasaccharide acceptor has two GlcNAc residues at its nonreducing end, one that is linked by a β1–3- and the other that is linked by a β1–6-glycosidic linkage. Although there are two GlcNAc moieties available in the tetrasaccharide that can bind to the acceptor site in the β4Gal-T1 molecule, it is the β1–6-branched GlcNAc moiety that is bound to the acceptor binding site in the crystal structure of the C338T-M340H-β4Gal-T1 complex, whereas the β1–3-glycosidic-linked GlcNAc moiety forms hydrophobic interactions with the aromatic side chain of the residue Phe-356 (Fig. 3A). The molecular interactions between the extended trisaccharide to the side-chain amino groups of the Arg-355 are quite similar to those found in the pentasaccharide complex (Fig. 3A). In addition, the apparent Km value for the Acceptor 2 (0.29 ± 0.07 mm) is comparable with the Km value for the Acceptor 1 (supplemental Fig. S-F5) Thus, the present structure suggests that the presence of the nonreducing end β1–3-linked Gal in the pentasaccharide Acceptor 1 was not responsible for the observed different binding mode of the extended sugar moiety of the pentasaccharide.
FIGURE 3.
A–C, the binding of the tetrasaccharide Acceptor 2 (A), trisaccharide Acceptor 3 (B), and the disaccharide Acceptor 4 to the β4Gal-T1 molecule (C). The β4Gal-T1 molecule is shown in the surface diagram with the important residues shown in sticks, whereas the bound acceptor substrates are shown in cyan-colored sticks. The binding of the acceptor substrates 2 and 3 is similar to its precursor, Acceptor 1, whereas the Acceptor 4 binds to the β4Gal-T1 quite differently (C). The extended sugar moiety Gal (shown in cyan) in Acceptor 4 interacts with the residue Tyr-282 away from Trp-310, whereas the same residue in Acceptor 3 interacts with residue Trp-310 away from residue Tyr-282. The binding of Acceptor 4 is similar to the N-glycan oligosaccharides, whose structures have been previously reported (13). Thus, these structures suggest that the different binding mode of the extended saccharide is due to the presence of the reducing end Glc residue.
To see whether the β1–3-linked nonreducing end GlcNAc moiety was bound in the acceptor binding site of the human C338T-M340H-β4Gal-T1 molecule in any other crystals of the same complex, we collected x-ray diffraction data from more crystals of the same tetrasaccharide Acceptor 2 complex. We did not find any crystals that had the β1–3-glycosidic-linked nonreducing end GlcNAc moiety bound to the β4Gal-T1 molecule. This suggests that this different binding mode could be due to the strong hydrophobic interactions between the β1–3-linked nonreducing ends GlcNAc moiety with the aromatic side chain of the residue Phe-356 and its hydrogen-bonding interactions with the residue Arg-355. Therefore, a trisaccharide Acceptor 3, GlcNAcβ1–6Galβ1–4Glc (Fig. 1), which is without the β1–3-linked GlcNAc at the reducing end complex, was crystallized with β4Gal-T1, and the crystal structure was analyzed.
Interactions of the Trisaccharide GlcNAcβ1–6Galβ1–4Glc (Acceptor 3) with the β4Gal-T1
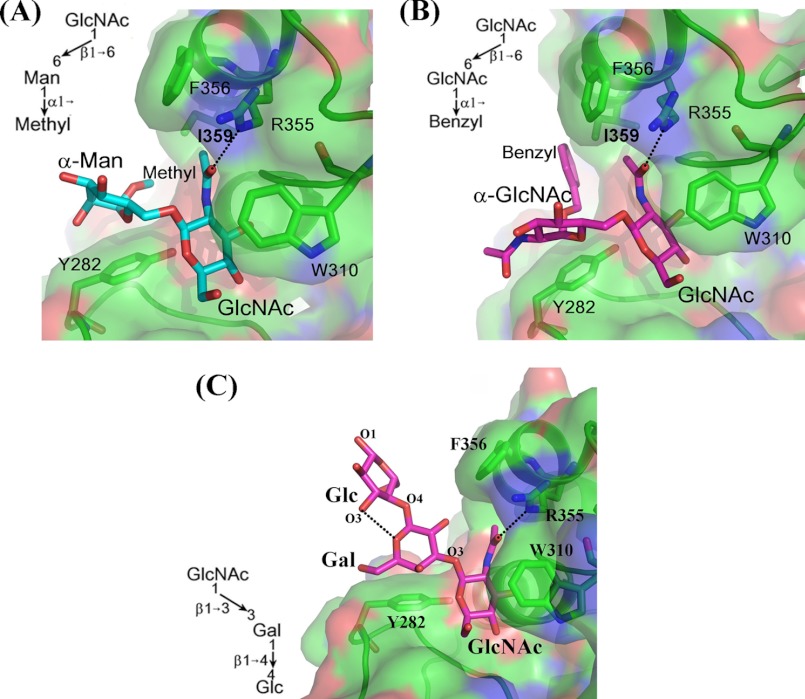
The trisaccharide Acceptor 3, GlcNAcβ1–6Galβ1–4Glc, was commercially available, and we have determined the crystal structure of its complex with the human C338T-M340H-β4Gal-T1 protein (Fig. 3B). In the crystal structure, we observed that the trisaccharide acceptor is bound to the β4Gal-T1 molecule in a way that is very similar to its precursor tetra- and pentasaccharide acceptor substrates. The intramolecular hydrogen bond between the Glc and the β1–6-branched GlcNAc sugars observed in the crystal structures of penta- and tetrasaccharide complexes is still present in the trisaccharide complex. In addition, the hydrogen bond between the O4 of the Gal to the side-chain nitrogen atom of the Arg-355 is also observed (Fig. 3B). This suggests that the different binding mode might be either inherently due to the β1–6 branch or due to the reducing end Glc moiety and its hydrogen-bonding interactions with the GlcNAc residue. Therefore, it is important to determine the crystal structure of the disaccharide GlcNAcβ1–6Gal (Fig. 1, Acceptor 4), bound to β4Gal-T1.
Interactions of the Disaccharide GlcNAcβ1–6Gal (Acceptor 4) with the β4Gal-T1
In the crystal structure of the disaccharide GlcNAcβ1–6Gal (Acceptor 4) in complex with the human C338T-M340H-β4Gal-T1 molecule (Fig. 3C), three protein molecules are present in the asymmetric unit. Although the electron density for the reducing end GlcNAc moiety is well observed in all three molecules, only in two molecules (molecules A and B) is the electron density for the Gal moiety clearly observed. The exact configuration (α- or β-) of the reducing end Gal moiety could not be clearly determined from the electron density maps, but it fits better as β-Gal than as α-Gal into the electron density maps. In all three molecules, the GlcNAc moiety is bound to the acceptor binding site of the β4Gal-T1 molecule, whereas the Gal moiety makes a hydrophobic interaction with the aromatic side chain of the residue Tyr-282, which is well away from the residue Trp-310. The interactions between the Gal moiety and the residue Tyr-282 in molecules A and B are slightly different. Although the binding of the reducing end GlcNAc moiety of the acceptor substrates 3 (GlcNAcβ1–6Galβ1–4Glc) and 4 (GlcNAcβ1–6Gal) to the β4Gal-T1 molecule is quite similar, the binding of their Gal moiety is quite different, and it is due to the different torsion angles about the C5-C6 (χ) and C6-O6 (Ψ) bonds of the Gal moiety. The torsion angles (χ,Ψ) of the Gal moiety in the acceptor substrates 3 and 4 are (-65°, 148°) and (138°, -114°), respectively. Simple modeling of the acceptor substrate 3 with the (χ,Ψ) torsion angle of the Acceptor 4 shows a severe steric hindrance between the reducing end Glc moiety and the residue Tyr-282, suggesting that the Acceptor 3 cannot bind to the β4Gal-T1 molecule similar to the Acceptor 4, and this is due to the presence of the β-linked reducing sugar moiety Glc. Furthermore, it is not clear whether the intramolecular H-bonding interactions observed between the Glc and the GlcNAc moiety in the tri, tetra-, and pentasaccharide complex structures also exist in a free state and therefore influence the moiety binding to the protein molecule. To confirm this observation, the structure of a trisaccharide GlcNAcβ1–6Galα1–4Glc in complex with the β4Gal-T1 molecule is essential. However, no such trisaccharide is commercially available. Instead, the disaccharide acceptor substrates, GlcNAcβ1–6Manα-methyl (Acceptor 5) and GlcNAcβ1–6GlcNAcα1-benzyl (Acceptor 6), were commercially available, which are similar to GlcNAcβ1–6 linked to a sugar moiety that is α1-linked to a methyl or benzyl moiety. Therefore, we have determined the crystal structure of these disaccharides complexed with the human C338T-M340H-β4Gal-T1 molecule.
Interactions of the Disaccharide GlcNAcβ1–6Manα-Methyl (Acceptor 5) and GlcNAcβ1–6GlcNAcα-Benzyl (Acceptor 6) with the β4Gal-T1
In the crystal structure of the disaccharide Acceptor 5, GlcNAcβ1–6Manα-methyl, in complex with the β4Gal-T1 molecule, the GlcNAc moiety is bound to the acceptor binding site of the β4Gal-T1 molecule, whereas the Man residue exhibits extensive hydrophobic interactions with the aromatic side chain of the Tyr-282 residue and its α-methyl group, which is found to interact with the hydrophobic side chain of the residue Ile-359 (Fig. 4A). In the crystal structure of the disaccharide Acceptor 6, GlcNAcβ1–6GlcNAcα-benzyl, in complex with the β4Gal-T1 molecule, the binding of the disaccharide moiety is quite similar to that of the binding of GlcNAcβ1–6Manα-methyl disaccharide Acceptor 5 (Fig. 4B). In this crystal structure, the benzyl group can be clearly located in the electron density maps, and it is found to interact with the hydrophobic side chain of the residue Ile-359. It is interesting to note that the disaccharide GlcNAcβ1–6Manα-methyl is part of the N-glycan branch, similar to GlcNAcβ1-6Manα1–6Manβ-GlcNAcβ1–4GlcNAc-Asn. The synthesis of this 1–6 branch in N-glycan is initiated by the N-acetyl-glucosaminyl transferase V that transfers a GlcNAc residue in a β1–6 linkage to the α-mannose in the 1–6 arm of the N-glycan and that these 1–6-branched glycans are found in abundance in cancer cells (15). Thus, it seems that the configuration of the core sugar (α-sugar/β-sugar) to which the branched β1–6-linked acceptor GlcNAc is attached determines the binding mode of the extended oligosaccharide moiety of the acceptor substrates. Furthermore, the present study clearly shows that the I-antigen acceptor binding to the β4Gal-T1 molecule is similar to the binding of the pentasaccharide Acceptor 1 and that this binding is quite different from the N-glycan binding to the β4Gal-T1.
FIGURE 4.
A–C show the binding of Acceptors 5, 6, and 7 (shown in cyan sticks) to the β4Gal-T1 molecule (shown in the surface diagram with important residues in sticks), respectively. The binding of the two acceptor substrates 5 and 6 are very similar, and their reducing end hydrophobic groups interact with the hydrophobic side chain of Ile-359. Also, the binding of their extended moieties is similar to the binding of the extended sugar moiety of the N-glycan acceptor sugars. C, in the crystal structure, the acceptor substrate 7 is bound in a way similar to the binding of chitotriose, where the acceptor GlcNAc residue is bound in the acceptor binding site, whereas the core sugar makes hydrophobic interactions with the aromatic side chain of the Tyr-282, and the reducing end sugar is exposed to the solvent without any molecular interactions with the protein molecule. The intramolecular hydrogen bond between the Gal and the Glc moieties is shown in a black dotted line. Acceptor 7 differs from a true i-antigen by this reducing end Glc residue, where it is replaced by a GlcNAc residue. Because this reducing end does not interact with a protein molecule, the binding of Acceptor 7 represents the true i-antigen acceptor binding to the β4Gal-T1 molecule.
In the complex of tetrasaccharide GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc (Acceptor 2) with the β4Gal-T1 molecule, only the GlcNAc moiety β1–6-linked to Gal is bound to the acceptor binding site in the β4Gal-T1 molecule, suggesting that this moiety may have a higher affinity to the β4Gal-T1 molecule than the GlcNAc moiety β1–3-linked to Gal. Our attempts to get crystals of the β4Gal-T1 complex with the tetrasaccharide Acceptor 2, where GlcNAc moiety is β1–3-linked to Gal, which is bound in the acceptor binding site of β4Gal-T1, have not been successful. The catalytic activity at different time points using the acceptor substrate 2 together with the mass spectroscopy (MALDI-TOF) analysis shows that the tetrasaccharide acceptor is rapidly converted into a pentasaccharide (supplemental Fig. S-F6). The conversion of the pentasaccharide to a hexasaccharide is very slow. This suggests that although there are two acceptor GlcNAc moieties present in the tetrasaccharide acceptor substrate, only one GlcNAc moiety is readily available for the catalytic activity, thus rapidly converting the tetrasaccharide to a pentasaccharide. Therefore, it seems that the GlcNAc residue β1–6-linked to Gal in the linear tetrasaccharide GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1-4Glc (Acceptor 2) may hinder the binding of the GlcNAc moiety β1–3-linked to Gal and hence to the β4Gal-T1 molecule. To test this hypothesis, the binding of the trisaccharide Acceptor 7, GlcNAcβ1–3 Galβ1–4Glc (Fig. 1), without the β1–6-branched GlcNAc moiety, to β4Gal-T1 was investigated. The trisaccharide GlcNAcβ1–3 Galβ1–4Glc (Acceptor 7) is analogous to i-antigen and differs from it by the reducing end Glc moiety. Crystal of this trisaccharide Acceptor 7 complex with the human C338T-M340H-β4Gal-T1 protein could be readily grown. In the crystal structure, the binding of the trisaccharide to the β4Gal-T1 molecule, the nonreducing end GlcNAc moiety of the trisaccharide 7, was bound to the acceptor binding site of the β4Gal-T1 molecule (Fig. 4C). This binding is similar to the previously described chitotriose binding to β4Gal-T1 protein (13). In the extended sugar moiety, the Gal residue forms hydrophobic interactions with the aromatic side chain of Tyr-282, whereas the reducing end Glc moiety does not make any molecular interactions with the protein molecule. The O6 hydroxyl group of the Gal moiety is found to have two disordered positions, and it also exhibits van der Waals interactions with the residue Tyr-282. Therefore, it seems that any β1–6 branch at the Gal residue may cause steric hindrance, thus offering an explanation for not observing the binding of GlcNAc β1–3-linked to Gal of the branched tetrasaccharide GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc (Acceptor 2) in the acceptor binding site of the β4Gal-T1 molecule.
Previously, we determined the crystal structure of the complex of acceptor substrate inhibitor GlcNAcβ1–3Galβ-naphthalenemethanol with the human C338T-M340H-β4Gal-T1 protein molecule (21). In the crystal structure of the β4Gal-T1 complex with the Galβ-naphthalenemethanol molecule, the naphthalene moiety forms hydrophobic interactions with the N-glycan binding site on the β4Gal-T1 molecule, whereas in the crystal structure of the linear trisaccharide Acceptor 7 complex with β4Gal-T1, the Glc moiety is exposed to the solvent region with no interactions with the β4Gal-T1 molecule.
The pentasaccharide Acceptor 1, Galβ1–3GlcNAcβ1–3 (GlcNAcβ1–6)Galβ1–4Glc, differs from the true I antigen acceptor substrates largely by the linkage of its nonreducing Gal and the reducing end Glc. The nonreducing Gal moiety may not influence the oligosaccharide binding because it is not involved in any molecular interactions with the protein molecule. However, replacing the O2 of the reducing end Glc with an N-acetyl could reasonably affect the binding of the true I-antigen acceptor binding to the enzyme. Similarly, the trisaccharide Acceptor 7, GlcNAcβ1–3 Galβ1–4Glc, differs from a true i-antigen by this reducing end Glc residue, where it is replaced by a GlcNAc residue (supplemental Fig. S-F1). Because this reducing end does not interact with the protein molecule, the binding of Acceptor 7 therefore most likely represents the binding mode of the true i-antigen acceptor binding to the β4Gal-T1 molecule. Once the I-antigen acceptor (Acceptor 1) is galactosylated by the β4Gal-T1 enzyme, the product β1–6LacNAc is extended with a β1–3-linked GlcNAc moiety by the β3GlcNAc-T enzyme, and then it becomes an i-antigen acceptor (Acceptor 7) for the β4Gal-T1 enzyme, thus synthesizing the LacNAc moiety of I-/i-antigen.
In nature, LacNAc moiety is found in a variety of glycans. The β4Gal-T1 enzyme, one of the seven members of the β4Gal-T family, is considered to be responsible for the synthesis of these LacNAc moieties (22). The β4Gal-T1 enzyme does not have high affinity for the monosaccharide GlcNAc (Km = 8–10 mm), in contrast to many glycosyltransferases that exhibit relatively high affinity (Km <1 mm) toward their nonreducing end acceptor sugar residues without any extended oligosaccharide moiety (23, 24). However, β4Gal-T1 shows high affinity toward the acceptor substrates with GlcNAc at nonreducing end of an oligosaccharide (Km < 1 mm). Therefore, it is expected to facilitate the binding of the extended oligosaccharide moieties of these glycans during the galactosylation of their nonreducing end GlcNAc residue. Similar to β4Gal-T1, β2GlcNAc-T1 enzyme is also known to exhibit high affinity for the oligosaccharide acceptor substrate (25). Oligosaccharides exhibit a wide range of conformational flexibility depending on the nature of the sugars and their glycosidic linkages (26). Thus, to accommodate their inherent conformational flexibility, it is not surprising to find different binding regions for different extended oligosaccharides of the acceptor substrates. This is the first time a second carbohydrate binding region on a glycosyltransferase has been observed, although a secondary carbohydrate binding site has been earlier observed in many carbohydrate-degrading enzymes (27).
In mammals, during lactation in the mammary gland, α-lactalbumin protein is produced in large quantities (28). It modulates the acceptor sugar specificity of β4Gal-T1 enzyme from an oligosaccharide with a GlcNAc at the nonreducing end to a monosaccharide, Glc molecule (29, 30). The crystal structures of β4Gal-T1 in complex with α-lactalbumin and with different substrates have clearly explained the modulation mechanism of α-lactalbumin (10, 11). In the crystal structure of the complex, the α-lactalbumin molecule is bound to the bovine β4Gal-T1 molecule in the closed conformation through many hydrogen-bonding and hydrophobic interactions, particularly with the residues Tyr-286, Phe-360, and Ile-363 (10). The corresponding residues in human β4Gal-T1 are Tyr-282, Phe-356, and Ile-359. The α-lactalbumin binding to the hydrophobic region on the β4Gal-T1 molecule is utilized by the extended sugar moieties linked to the acceptor GlcNAc residue for maximizing their interactions with the β4Gal-T1 molecule. Because most of the extended oligosaccharide binding residues have been conserved in β4Gal-T1 molecules of non-mammal vertebrates, many of which are also known to make i/I-antigen (31), the observed binding of the extended oligosaccharides to the α-lactalbumin binding region of the β4Gal-T1 molecule is not a coincidence. In fact, the presence of the α-lactalbumin binding site in non-mammal β4Gal-T1 molecules such as from chicken and in its ortholog invertebrate β4GalNAc-T enzyme is a coincidence because α-lactalbumin is not found in these species (31–34). Therefore, the observation of this second carbohydrate binding region for the extended oligosaccharide of the acceptor substrate must be physiologically significant. The β4Gal-T1 enzyme is also involved in galactosylating the GlcNAc residue of short glycoconjugates that are O-linked to Ser/Thr residues of proteins, such as GlcNAcβ1-3Fucα-O-Ser- or GlcNAcβ1–2Manα-O-Ser- (35). It will be of interest to see whether β4Gal-T1 enzyme utilizes its α-lactalbumin binding site for the binding of these proteins to enhance its affinity toward these glycans.
Conclusion
In the present study, we find that if the extended sugar, such as the LacNAc moiety, is linked to the core sugar via α-configuration, the LacNAc moiety tends to bind to the previously observed N-glycan binding crevice on the β4Gal-T1 enzyme, whereas the poly-LacNAc moiety of the branched β1–6GlcNAc linked via β-configuration binds to the new binding region on the β4Gal-T1 molecule. Because LacNAc moiety is found in a variety of glycans, the β4Gal-T1 molecule is expected to facilitate the binding of these oligosaccharide moieties that are part of the extended oligosaccharides of the acceptor GlcNAc moiety. It is, therefore, not surprising to find a second carbohydrate binding region in β4Gal-T1 molecule. Attempts are being made in many laboratories to design better acceptor-based inhibitors for the β4Gal-T1 molecule by taking into account the hydrophobic N-glycan binding site on the β4Gal-T1 molecule. The present observation of the new binding region for the extended sugar not only enhances our understanding of the oligosaccharide specificity of the β4Gal-T1 enzyme but also aids one to design a better acceptor-based inhibitor for the enzyme.
Supplementary Material
Acknowledgments
We thank Dr. Alexander Wlodawer and Mi Li of Macromolecular Crystallography Laboratory, Frederick National Laboratory for Cancer Research for the x-ray data collection.
This work was supported, in whole or in part, by National Institutes of Health Contract HHSN261200800001E through the NCI. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory for Cancer Research.

This article contains supplemental Figs. S-F1–S-F6.
- LacNAc
- N-acetyllactosamine
- β4Gal-T1
- β1–4-galactosyltransferase-I.
REFERENCES
- 1. Varki A., Lowe J. B. (2009) in Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 75–88, NY [Google Scholar]
- 2. Renkonen O. (2000) Enzymatic in vitro synthesis of I-branches of mammalian polylactosamines: generation of scaffolds for multiple selectin binding saccharide determinants. Cell Mol. Life Sci. 57, 1423–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hakomori S. (1999) Antigen structure and genetic basis of histo-blood groups A, B, and O: their changes associated with human cancer. Biochim. Biophys. Acta. 1473, 247–266 [DOI] [PubMed] [Google Scholar]
- 4. Hakomori S. (2000) Traveling for the glycosphingolipid path. Glycoconj. J. 17, 627–647 [DOI] [PubMed] [Google Scholar]
- 5. Stanley P. (2011) Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 3, a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brew K., Vanaman T. C., Hill R. L. (1968) The role of α-lactalbumin and the A protein in lactose synthetase: a unique mechanism for the control of a biological reaction. Proc. Natl. Acad. Sci. U.S.A. 59, 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morrison J. F., Ebner K. E. (1971) Studies on galactosyltransferase: kinetic investigations with N-acetylglucosamine as the galactosyl group acceptor. J. Biol. Chem. 246, 3977–3984 [PubMed] [Google Scholar]
- 8. Prieels J. P., Dolmans M., Schindler M., Sharon N. (1976) The binding of glycoconjugates to human milk d-galactosyltransferase. Eur. J. Biochem. 66, 579–582 [DOI] [PubMed] [Google Scholar]
- 9. Powell J. T., Brew K. (1976) A comparison of the interactions of galactosyltransferase with a glycoprotein substrate (ovalbumin) and with α-lactalbumin. J. Biol. Chem. 251, 3653–3663 [PubMed] [Google Scholar]
- 10. Ramakrishnan B., Qasba P. K. (2001) Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the β1–4-galactosyltransferase-I. J. Mol. Biol. 310, 205–218 [DOI] [PubMed] [Google Scholar]
- 11. Qasba P. K., Ramakrishnan B., Boeggeman E. (2008) Structure and function of β1–4-galactosyltransferase. Curr. Drug Targets 9, 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramakrishnan B., Balaji P. V., Qasba P. K. (2002) Crystal structure of β1–4-galactosyltransferase complex with UDP-Gal reveals an oligosaccharide acceptor binding site. J. Mol. Biol. 318, 491–502 [DOI] [PubMed] [Google Scholar]
- 13. Ramasamy V., Ramakrishnan B., Boeggeman E., Ratner D. M., Seeberger P. H., Qasba P. K. (2005) Oligosaccharide preferences of β1–4-galactosyltransferase-I: crystal structures of M340H mutant of human β1–4-galactosyltransferase-I with a pentasaccharide and trisaccharides of the N-glycan moiety. J. Mol. Biol. 353, 53–67 [DOI] [PubMed] [Google Scholar]
- 14. Chakraborty A. K., Pawelek J. M. (2003) GnT-V, macrophage, and cancer metastasis: a common link. Clin. Exp. Metastasis 20, 365–373 [DOI] [PubMed] [Google Scholar]
- 15. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boeggeman E., Ramakrishnan B., Pasek M., Manzoni M., Puri A., Loomis K. H., Waybright T. J., Qasba P. K. (2009) Site-specific conjugation of fluoroprobes to the remodeled Fc N-glycans of monoclonal antibodies using mutant glycosyltransferases: application for cell surface antigen detection. Bioconjug. Chem. 20, 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramakrishnan B., Shah P. S., Qasba P. K. (2001) α-Lactalbumin (LA) stimulates milk β1–4-galactosyltransferase I (β4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine: crystal structure of β4Gal-T1·LA complex with UDP-Glc. J. Biol. Chem. 276, 37665–37671 [DOI] [PubMed] [Google Scholar]
- 19. Ramakrishnan B., Boeggeman E., Qasba P. K. (2004) Effect of the M344s mutation on the conformational dynamics of bovine β1–4-galactosyltransferase: crystal structure of the M344H mutant in complex with chitobiose. Biochemistry 43, 12513–12522 [DOI] [PubMed] [Google Scholar]
- 20. Ramasamy V., Ramakrishnan B., Boeggeman E., Qasba P. K. (2003) The role of tryptophan 314 in the conformational changes of β1–4-galactosyltransferase-I. J. Mol. Biol. 331, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 21. Brown J. R., Yang F., Sinha A., Ramakrishnan B., Tor Y., Qasba P. K., Esko J. D. (2009) Deoxygenated disaccharide analogs as specific inhibitors of β1–4-galactosyltransferase 1 and selectin-mediated tumor metastasis. J. Biol. Chem. 284, 4952–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amado M., Almeida R., Schwientek T., Clausen H. (1999) Identification and characterization of large galactosyltransferase gene families: galactosyltransferases for all functions. Biochim. Biophys. Acta 1473, 35–53 [DOI] [PubMed] [Google Scholar]
- 23. Marcus S. L., Polakowski R., Seto N. O., Leinala E., Borisova S., Blancher A., Roubinet F., Evans S. V., Palcic M. M. (2003) A single point mutation reverses the donor specificity of human blood group B-synthesizing galactosyltransferase. J. Biol. Chem. 278, 12403–12405 [DOI] [PubMed] [Google Scholar]
- 24. Ouzzine M., Gulberti S., Levoin N., Netter P., Magdalou J., Fournel-Gigleux S. (2002) The donor substrate specificity of the human β1–3-glucuronosyltransferase I toward UDP-glucuronic acid is determined by two crucial histidine and arginine residues. J. Biol. Chem. 277, 25439–25445 [DOI] [PubMed] [Google Scholar]
- 25. Nishikawa Y., Pegg W., Paulsen H., Schachter H. (1988) Control of glycoprotein synthesis: purification and characterization of rabbit liver UDP-N-acetylglucosamine:α-3-d-mannoside β1–2-N-acetylglucosaminyltransferase I. J. Biol. Chem. 263, 8270–8281 [PubMed] [Google Scholar]
- 26. Rao V. S., Qasba P. K., Balaji P. V., Chandrasekaran R. (1998) Conformations of Carbohydrates, Harwood Academic Publishers, pp. 131–189, The Netherlands [Google Scholar]
- 27. Nielsen M. M., Bozonnet S., Seo E. S., Mótyán J. A., Andersen J. M., Dilokpimol A., Abou Hachem M., Gyémánt G., Naested H., Kandra L., Sigurskjold B. W., Svensson B. (2009) Two secondary carbohydrate binding sites on the surface of barley α-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry 48, 7686–7697 [DOI] [PubMed] [Google Scholar]
- 28. Qasba P. K., Kumar S. (1997) Molecular divergence of lysozymes and α-lactalbumin. Crit. Rev. Biochem. Mol. Biol. 32, 255–306 [DOI] [PubMed] [Google Scholar]
- 29. Brodbeck U., Denton W. L., Tanahashi N., Ebner K. E. (1967) The isolation and identification of the B protein of lactose synthetase as α-lactalbumin. J. Biol. Chem. 242, 1391–1397 [PubMed] [Google Scholar]
- 30. Klee W. A., Klee C. B. (1970) The role of α-lactalbumin in lactose synthetase. Biochem. Biophys. Res. Commun. 39, 833–841 [DOI] [PubMed] [Google Scholar]
- 31. Shaper N. L., Meurer J. A., Joziasse D. H., Chou T. D., Smith E. J., Schnaar R. L., Shaper J. H. (1997) The chicken genome contains two functional nonallelic β1–4-galactosyltransferase genes: chromosomal assignment to syntenic regions tracks fate of the two gene lineages in the human genome. J. Biol. Chem. 272, 31389–31399 [DOI] [PubMed] [Google Scholar]
- 32. Kawar Z. S., Van Die I., Cummings R. D. (2002) Molecular cloning and enzymatic characterization of a UDP-GalNAc:GlcNAc(β)-R 1–4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J. Biol. Chem. 277, 34924–34932 [DOI] [PubMed] [Google Scholar]
- 33. Ramakrishnan B., Qasba P. K. (2007) Role of a single amino acid in the evolution of glycans of invertebrates and vertebrates. J. Mol. Biol. 365, 570–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramakrishnan B., Qasba P. K. (2010) Structure-based evolutionary relationship of glycosyltransferases: a case study of vertebrate β1–4-galactosyltransferase, invertebrate β1–4-N-acetylgalactosaminyltransferase, and α-polypeptidyl-N-acetylgalactosaminyltransferase. Curr. Opin. Struct. Biol. 20, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haltiwanger R. S., Lowe J. B. (2004) Role of glycosylation in development. Annu. Rev. Biochem. 73, 491–537 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.