Background: DNA strand exchange proteins Dmc1 and Rad51 and translocases Tid1 and Rad54 function in DNA break repair during meiosis.
Results: We biochemically demonstrate that Dmc1 and Rad51 are specifically stimulated by Tid1 and Rad54, respectively.
Conclusion: Dmc1-Tid1 and Rad51-Rad54 represent functional pairs for DNA pairing and joint molecule formation.
Significance: The separate and independent functioning of these proteins offers insight into DNA pairing in meiosis.
Keywords: Atomic Force Microscopy, ATPases, DNA Enzymes, DNA Recombination, DNA-Protein Interaction, Electron Microscopy (EM), DNA Translocases
Abstract
The Saccharomyces cerevisiae Dmc1 and Tid1 proteins are required for the pairing of homologous chromosomes during meiotic recombination. This pairing is the precursor to the formation of crossovers between homologs, an event that is necessary for the accurate segregation of chromosomes. Failure to form crossovers can have serious consequences and may lead to chromosomal imbalance. Dmc1, a meiosis-specific paralog of Rad51, mediates the pairing of homologous chromosomes. Tid1, a Rad54 paralog, although not meiosis-specific, interacts with Dmc1 and promotes crossover formation between homologs. In this study, we show that purified Dmc1 and Tid1 interact physically and functionally. Dmc1 forms stable nucleoprotein filaments that can mediate DNA strand invasion. Tid1 stimulates Dmc1-mediated formation of joint molecules. Under conditions optimal for Dmc1 reactions, Rad51 is specifically stimulated by Rad54, establishing that Dmc1-Tid1 and Rad51-Rad54 function as specific pairs. Physical interaction studies show that specificity in function is not dictated by direct interactions between the proteins. Our data are consistent with the hypothesis that Rad51-Rad54 function together to promote intersister DNA strand exchange, whereas Dmc1-Tid1 tilt the bias toward interhomolog DNA strand exchange.
Introduction
Eukaryotes undergo a specialized type of cell division termed meiosis (Greek for “to reduce”). Meiosis results in the generation of haploid gametes. For this reduction of the genetic complement to take place, the cell undergoes one round of replication followed by two successive cell divisions (meiosis I and meiosis II). During meiosis I, the homologous chromosomes form a tripartite proteinaceous structure termed the synaptonemal complex and undergo crossover recombination at frequencies 100–1000-fold higher than in vegetative cells (1). These recombination events lead to a physical connection between homologous chromosomes, a joining that is needed for accurate segregation of chromosomes (2). Meiosis I is followed by meiosis II, which is analogous to mitosis and leads to segregation of sister chromatids. Diploidy is subsequently restored upon fertilization. In the absence of meiosis, sexual reproduction would lead to an exponential increase of the chromosome content that would cause severe genome instability (3).
In Saccharomyces cerevisiae, homologous recombination during the first meiotic division plays a crucial role in ensuring accurate segregation of chromosomes. Whereas the primary function of homologous recombination in vegetative cells is the repair of DNA breaks, in germ line cells, homologous recombination plays a defining role in bridging the homologous chromosomes (2, 4, 5). The steps involved during meiotic homologous recombination in S. cerevisiae are well defined (6). Recombination is initiated by programmed DNA breaks catalyzed by the meiosis-specific endonuclease, Spo11 (7). The broken ends are processed by the resection machinery (8) to produce 3′-ended single-strand DNA (ssDNA)3 tails that are ≥500 nucleotides long (9, 10). The resultant ssDNA is first coated with replication protein A. A number of proteins that are classified as “mediators” act as catalysts to replace replication protein A with the DNA strand exchange proteins, Rad51, or its meiosis-specific homolog, Dmc1, to form nucleoprotein filaments (11). These filaments represent the active species that search and pair with an intact template, the sister chromatid or the homologous chromosome depending on whether the filament consists of Rad51 or Dmc1, respectively (12). Pairing results in the formation of a D-loop in a process called single end invasion (13). The invading 3′-end in the D-loop intermediate primes DNA synthesis, which serves to recover DNA content that is lost during resection at the opposite end (14, 15). At this point, either the extended D-loop can dissociate, and the replicated end can anneal with the second end of the DNA break (16), or it can engage directly with the second end of the break (17). The former process is called synthesis-dependent strand annealing and leads to formation of non-crossovers. The latter process forms double Holliday junctions that are mainly resolved as crossovers (16).
Meiotic and mitotic recombination machineries have components with overlapping functions. Examples of overlapping activities are two DNA strand exchange proteins, Rad51 and Dmc1, and two DNA translocases, Rad54 and Tid1. Dmc1 physically interacts with Tid1 as shown using a two-hybrid assay (18). Genetic analyses indicate that Rad51 and Rad54 are utilized primarily for sister chromatid recombination in vegetative and meiotic cells, whereas Dmc1 and Tid1 are involved exclusively in interhomolog recombination during meiosis (19–21). However, direct evidence for this functional coupling is lacking.
In this work, using purified proteins, we biochemically delineate the functional relationship between Dmc1/Rad51 and Tid1/Rad54. We show that Dmc1 binds ssDNA as well as double-stranded DNA (dsDNA), forms helical filaments, and catalyzes DNA strand pairing. Importantly, we demonstrate that, whereas Rad51 is stimulated by Rad54 and Tid1, Dmc1 is specifically stimulated by Tid1. Our data provide biochemical evidence for the hypothesis (22) that Dmc1 and Tid1 interact to promote homologous pairing during meiotic recombination.
EXPERIMENTAL PROCEDURES
Enzymes and Reagents
T4 DNA ligase, T4 polynucleotide kinase, and restriction endonucleases were purchased from New England Biolabs. Proteinase K was purchased from GE Healthcare. [γ-32P]ATP (6000 Ci/mmol) was purchased from PerkinElmer Life Sciences. Thrombin, pyruvate kinase, and phosphoenolpyruvate were purchased from Sigma.
DNA
Two oligodeoxyribonucleotides, a 55-mer and a 100-mer, complementary to nucleotides 2451–2505 and 2451–2550 of the minus strand of pUC19, respectively, were purchased from Sigma Genosys and gel-purified on a 12% denaturing polyacrylamide gel. They were 5′-32P-labeled using T4 polynucleotide kinase and [γ-32P]ATP and purified using MicroSpin G-25 columns (GE Healthcare). Supercoiled pUC19 DNA was purified by non-alkaline lysis followed by cesium chloride density gradient centrifugation (23). Concentrations of the 55-mer, 100-mer, and pUC19 (2686 bp) were determined using molar (nucleotide) extinction coefficients at 260 nm of 9880, 9980, and 6600 m−1 cm−1, respectively. DNA concentrations are expressed in moles of nucleotides (nt), base pairs (bp), or molecules as indicated. Poly(dT) was obtained from Sigma, and its concentration was determined using a molar (nucleotide) extinction coefficient at 260 nm of 8520 m−1 cm−1.
Construction of GST-Dmc1 and GST-Tid1 Expression Plasmids
The full-length Dmc1 and Tid1 genes were amplified by PCR (AccuPrime Pfx, Invitrogen) using yeast genomic DNA (W303) as template and primers AN24 (5′-TTGTGGCTCGAGATATGTCTGTTACAGGAACTGAGATCGATAGTGATACA-3′) and AN25 (5′-CCACAACCCGGGTTAGTCACTTGAATCGGTAATACCTTTTTCACC-3′) for Dmc1, and primers AN20 (5′-TTGTGGCTCGAGATATGCAGATACCGAAATATGAGAACAAGCC-3′) and AN21 (5′-CCACAACCCGGGTCATTGTTCTCTGAGACATATCTCGCCGGGC-3′) for Tid1. The start and stop codons are indicated in bold text. The italicized sequences indicate recognition sequences for XhoI and XmaI. The parameters for PCR amplification were as follows: 95 °C/5 min (activation of Pfx), 95 °C/15 s (strand denaturation), 49 °C/30 s (primer annealing), and 68 °C/1 min for Dmc1 and 3 min for Tid1 (extension, 30 cycles). The amplified PCR products were digested with XhoI and XmaI and cloned in between the XhoI and XmaI sites of the vector (pWDH597; a kind gift from Dr. W. Heyer, University of California, Davis) such that it was in-frame and downstream of a glutathione S-transferase (GST) tag. The resultant recombinant proteins are therefore 63 (26.5-kDa GST and 36.5-kDa Dmc1) and 136.5 kDa (26.5-kDa GST and 110-kDa Tid1). The expression constructs were designed to contain a thrombin cleavage site immediately downstream of the GST tag. The authenticity of the clones was verified by restriction mapping as well as sequencing.
Expression of Recombinant Proteins
GST-Dmc1 and GST-Tid1 fusion proteins (henceforth referred to as Dmc1 and Tid1, respectively) were expressed using an identical protocol. The expression plasmids were introduced individually into yeast expression strain yWDH668 (a kind gift from Dr. W. Heyer, University of California, Davis) using the lithium acetate method (23). For large scale expression, a 12-liter culture was initially grown to an A600 of 1. Protein expression was induced by the addition of galactose to a final concentration of 1%. Cells were harvested 8 h postinduction. Expression of the recombinant proteins was confirmed by Western blotting using anti-GST antibody (Santa Cruz Biotechnology). The total yield of cells starting from a 12-liter culture was 60 and 45 g for GST-Dmc1- and GST-Tid1-expressing cells, respectively. The cells were stored at −80 °C until purification.
Purification of Recombinant Proteins
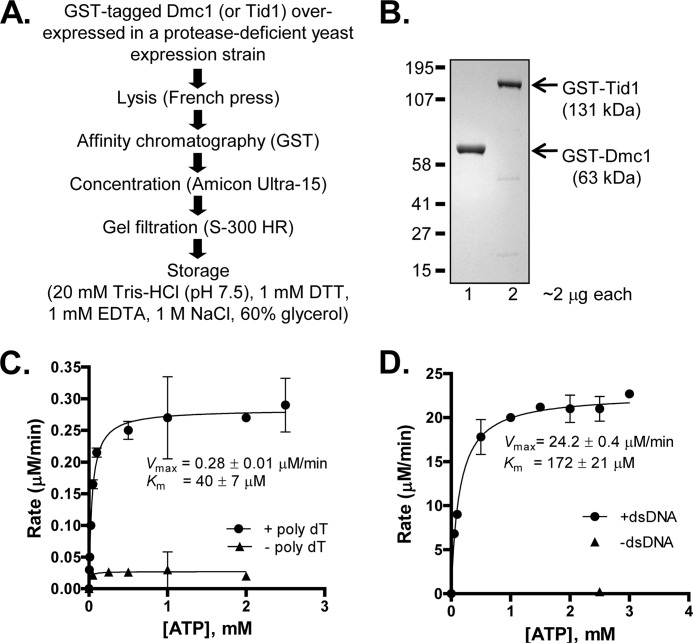
The fusion proteins were purified to homogeneity using a two-column purification scheme (see Fig. 1A) similar to the one used to purify Rad54 (24). All steps were performed at 4 °C. Cells expressing either recombinant protein (∼20 g) were uniformly suspended in Buffer A (20 mm Tris-HCl (pH 7.5), 1 m NaCl, 10% glycerol, 1 mm EDTA, 10 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, and EDTA-free protease inhibitor (one tablet/40 ml of buffer; Roche Applied Science) in a ratio of 4 ml/g of cells. The cells were lysed by three passages through a French press (American Instrument Co.) at 1100 p.s.i. The lysate was clarified by centrifuging at 35,000 rpm for 1 h. The clarified lysate was loaded on a 20-ml GST-PrepFF column (GE Healthcare) equilibrated with Buffer A. Bound protein was eluted using Buffer A + 20 mm glutathione (pH 7.5). The fractions were analyzed for the presence of protein using 12% SDS-polyacrylamide gel electrophoresis (PAGE). The peak fractions were pooled (∼15 ml) and concentrated down to less than ∼1 ml using an Amicon Ultra-15 concentration device. The concentrated protein was loaded on an ∼20-ml S-300 HR column (Amersham Biosciences) equilibrated and eluted using Buffer A. In either case, the protein eluted in two distinct peaks. The first peak coincided with the void volume and consisted of high molecular weight aggregates formed presumably due to contaminating DNA. The second peak was free of DNA and was pooled and concentrated by dialyzing against storage buffer (20 mm Tris-HCl (pH 7.5), 1 m NaCl, 1 mm DTT, 1 mm EDTA, and 60% glycerol), flash frozen using liquid nitrogen, and stored at −80 °C. The purity of the proteins was estimated to be >95% as determined by 12% SDS-PAGE followed by Coomassie Brilliant Blue staining (see Fig. 1B, lanes 1 and 2). The concentration of each protein was determined spectrophotometrically using molar extinction coefficients of 52,800 and 106,800 m−1 cm−1 at 280 nm for Dmc1 and Tid1, respectively. The purified proteins were determined to be free of contaminating ssDNA- and dsDNA-specific nucleases because incubation of an ∼10-fold molar excess of either protein over 5′-end-labeled ssDNA (100-mer) or 3′-end-labeled dsDNA (2.6 kb fragment) for 1 h did not generate degradation products. Rad51 (25), Rad54 (26), and His-tagged Dmc1 (27) were purified as described.
FIGURE 1.
Purified recombinant Dmc1 and Tid1 proteins are active ATPases. A, flow chart showing steps involved in the purification of GST-tagged Dmc1 and Tid1. Both proteins were independently overexpressed and purified using a two-step approach. B, image showing 2 μg of purified Dmc1 (lane 1) and Tid1 (lane 2) resolved by 12% SDS-PAGE. The positions of molecular mass markers are as indicated. C, ssDNA-dependent ATPase activity of Dmc1. The rate of Dmc1 (1 μm)-catalyzed hydrolysis of ATP in the presence as well as absence of ssDNA (poly(dT); 10 μm nt) is plotted as a function of ATP concentration. D, dsDNA-dependent ATPase activity of Tid1. The rate of Tid1 (7 nm)-catalyzed hydrolysis of ATP in the presence as well as absence of dsDNA (pUC19 supercoiled DNA, 0.8 μm bp) is plotted as a function of ATP concentration. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
GST Tag Removal Using Thrombin
GST-tagged Dmc1 and Tid1 were individually treated with thrombin (Sigma; dissolved in 1× phosphate buffered saline (PBS) (pH 7.4) (10 mm sodium phosphate, 2 mm potassium phosphate, 150 mm NaCl, and 3 mm KCl) at a final concentration of 4 units/mg of tagged protein for 12 h at 4 °C. As controls, the tagged proteins were incubated with PBS for the same duration of time. Cleavage was verified by analyzing the thrombin-treated proteins using 10% PAGE followed by Coomassie Brilliant Blue staining. Untagged Rad54 was a kind gift from Dr. Wolf Heyer (University of California, Davis).
ATP Hydrolysis Assay for Dmc1
To determine the kinetic parameters for ATP hydrolysis, Dmc1 (1 μm) was incubated with poly(dT) (10 μm nt) in a buffer containing 20 mm Tris acetate (pH 7.5), 5 mm magnesium acetate or calcium acetate, 1 mm dithiothreitol, 100 μg/ml bovine serum albumin, ATP (varying amounts), and 20 μCi/ml [γ-32P]ATP in a reaction volume of 10 μl at 30 °C. At the indicated times, the reactions were stopped by spotting 0.6-μl aliquots on a polyethyleneimine-modified cellulose TLC plate (EMD Chemicals; 5 × 10 cm). The spots were air-dried and resolved via capillary action using a running buffer consisting of 1 m formic acid and 0.8 m lithium chloride. The buffer front was allowed to rise to the top of the plate (∼5 cm) after which the plate was air-dried and exposed to a storage phosphor screen for ∼30 min. Reaction products were visualized and quantified with a GE Healthcare Storm 860 using ImageQuant version 5.2. The amount of ATP hydrolyzed in μm was derived from the percentage of ATP hydrolysis at each ATP concentration; the rate of ATP hydrolysis (μm/min) was determined by dividing by the incubation time. The graphs were generated using GraphPad Prism version 5, and the kinetic parameters Km and Vmax were determined by fitting the data to the Michaelis-Menten equation. The observed kcat for ssDNA-dependent ATPase activity of Dmc1 was determined by dividing the rate by the molar concentration of the protein used in the reaction.
ATP Hydrolysis Assay for Tid1
The hydrolysis of ATP by Tid1 was monitored using a spectrophotometric assay that couples production of ADP to the oxidation of NADH. To determine the kinetic parameters of Tid1-mediated ATP hydrolysis, Tid1 (7 nm) was incubated with pUC19 supercoiled DNA (0.8 μm bp) in a buffer containing 25 mm Tris acetate (pH 7.5), 0.1 mm dithiothreitol, 10 mm magnesium acetate, varying ATP, 1.5 mm phosphoenolpyruvate, 0.16 mg/ml NADH, 30 units/ml pyruvate kinase, and 30 units/ml lactate dehydrogenase at 30 °C. ATP hydrolysis was monitored continuously using a multicuvette holder connected to HP Agilent 8453. The analysis was described previously (28, 29).
DNA Binding Assay for Dmc1
The fluorescence increase upon Dmc1 binding to M13 etheno-DNA was measured by exciting at 305 nm (bandwidth, 5 nm) and monitoring at 410 nm (bandwidth, 30 nm) using a Shimadzu spectrofluorometer (30). The reaction buffer was 20 mm Tris acetate (pH 7.5), 1 mm DTT, 1 mm ATP, and 5 mm magnesium or calcium acetate. The concentration of M13 etheno-DNA was 1 μm (nt).
DNA Binding Using an Electrophoretic Mobility Shift Assay for Dmc1
Dmc1 (0.2 μm) was incubated with ssDNA (55-mer; 0.1 μm nt) in buffer containing 33 mm Na-HEPES (pH 7.5), 5 mm calcium acetate or magnesium acetate, 1 mm ATP (or analog), 0.1 mm DTT, 100 μg/ml BSA, and varying amounts of NaCl for 1 h at 30 °C. The reaction products were analyzed by 6% PAGE in Tris borate-EDTA buffer (12.5 V/cm; 2 h) at 4 °C. Following electrophoresis, the gels were dried on DE81 paper (Whatman) and exposed to a storage phosphor screen. The reaction products were visualized and quantified with a GE Healthcare Storm 860 using ImageQuant version 5.2.
Atomic Force Microscopy (AFM)
DNA used for AFM was a 1.1-kb linear dsDNA fragment (the Dmc1 open reading frame obtained by PCR amplification of W303 genomic DNA using primers AN23 and AN24 as described earlier). Dmc1 was incubated with DNA in a ratio of 1 μm protein to 3 μm DNA (bp) in buffer containing 25 mm Na-HEPES (pH 7.5), 5 mm calcium acetate or magnesium acetate, and 1 mm nucleotide for 30 min at 30 °C in a total reaction volume of 20 μl. Complexes were adsorbed onto a ruby mica surface modified with polylysine. After 5 min, the surface was washed with 1 ml of distilled water and dried with compressed nitrogen gas. Polylysine-modified mica surfaces were prepared by depositing 25 μl of 10 ng/μl polylysine on freshly cleaved mica for 5 min. The mica was then rinsed with water and air-dried. Protein-DNA complexes were imaged using Tapping Mode (Nanoscope IIIa, Digital Instruments) using BS-Tap300Al silicon AFM probes. Images were captured at scan sizes of 0.6–2 μm and processed by first-order flattening to remove sample tilt. Pitch and handedness were determined by visual inspection and by Fourier analysis of the images.
Fast Fourier Transform Analysis of Dmc1-DNA Complexes
Images were cropped and rotated such that continuous sections of complexes were oriented vertically. Two-dimensional fast Fourier transforms were computed for the images using the Gwyddion version 2.26 software package. In the fast Fourier transform output, there were distinct symmetric intensities at a distance of 99 μm−1 from the origin that corresponded to a physical spacing of 10 nm. The peak to peak distance of the symmetric peaks was measured. The peaks were noted to be rotated counterclockwise relative to the vertical axis, which is consistent with a right-handed structure.
Electron Microscopy (EM)
Dmc1-dsDNA complexes were formed as described for AFM. The complexes were fixed by addition of glutaraldehyde to 0.2% followed by 15-min incubation at 37 °C. Samples were diluted and washed in 5 mm magnesium acetate prior to uranyl acetate staining. Complexes were visualized using a Philips CM12 electron microscope (31).
Joint Molecule Assay
Joint molecules (D-loops) were formed as follows: 5′-32P-labeled 100-mer (0.9 μm nt; 9 nm molecules) was preincubated with DNA strand exchange protein (Dmc1 or Rad51 at 0.3 μm) in a buffer containing 25 mm Tris acetate (pH 7.5), varying concentrations of magnesium acetate or calcium acetate, 20 units/ml pyruvate kinase, 3 mm phosphoenolpyruvate, 1 mm DTT, 1 mm ATP, and 100 μg/ml BSA for 5 min at 30 °C. The reaction was supplemented with Tid1 or Rad54 protein (0.3 μm) followed by immediate addition of pUC19 supercoiled DNA (8 μm bp; 3 nm molecules). Reactions were incubated at 30 °C for 10 min and stopped by the addition of termination buffer (final concentration, 2% SDS, 3 μg/μl proteinase K, and 50 mm EDTA). After 30 min, reaction products were analyzed by electrophoresis in 1% agarose (4.5 V/cm for ∼1.5 h). Following electrophoresis, the gels were dried on DE81 paper (Whatman) and exposed to a storage phosphor screen. The reaction products were visualized and quantified with a GE Healthcare Storm 860 using ImageQuant version 5.2. The yield of D-loops was expressed as a percentage of the limiting supercoiled DNA. Data were analyzed using GraphPad Prism version 5.
Pulldown Assays
His-tagged Dmc1 and either Tid1 or Rad54 (1 μg each) were incubated in binding buffer (20 mm Na-HEPES (pH 7.5), 250 mm NaCl, 5 mm MgCl2, 0.1 mm DTT, and 0.05% Triton X-100) containing 20 mm imidazole at 30 °C for 15 min. Ni2+-NTA magnetic beads (Qiagen) were added to a concentration of 1% and incubated at 30 °C for 30 min with constant mixing. The beads were isolated using a magnet and washed (once with 50 μl and twice with 100 μl of binding buffer containing 100 mm imidazole). Beads were boiled in SDS-PAGE loading buffer. The load and bound fractions were analyzed by 12% SDS-PAGE and Coomassie Brilliant Blue staining.
RESULTS
Purified Recombinant Dmc1 and Tid1 Proteins Are Active ATPases
We purified Dmc1 and Tid1 as GST fusion proteins (Fig. 1, A and B). This N-terminal GST-tagged Dmc1 is indeed an ssDNA-dependent ATPase (Fig. 1C) whose Km for ATP and Vmax values are 40 ± 7 μm and 0.28 ± 0.01 μm/min, respectively. The kcat was calculated to be 0.30 ± 0.01 min−1, a value that is comparable with that reported for the His-tagged Dmc1 protein (27). We demonstrated previously that N-terminal GST-tagged Tid1 actively translocates on dsDNA in an ATP-dependent manner (32). Here, we examined the kinetic parameters for ATP hydrolysis (Fig. 1D) and determined that Tid1 is a dsDNA-dependent ATPase with Km for ATP and Vmax values of 172 ± 21 μm and 24.2 ± 0.4 μm/min, respectively. The kcat was determined to be 3500 ± 60 min−1, which is comparable with the published value (33). We therefore conclude that the recombinant proteins are biochemically active with regard to ATP hydrolysis.
To confirm that the presence of the tag does not adversely affect the biochemical activity of these proteins, we compared the ATPase activities of the tagged and untagged forms of the two proteins (supplemental Fig. S1A). The kcat values from a time course of ssDNA-dependent ATPase activity at 2 μm protein are similar: 0.39 ± 0.02 and 0.41 ± 0.02 min−1 for tagged and untagged Dmc1, respectively (supplemental Fig. S1B). In supplemental Fig. S1C, we compared dsDNA-dependent ATP hydrolysis by tagged and untagged Tid1 as a function of protein concentration. Here as well, the kcat values were identical (3550 ± 300 and 3500 ± 350 min−1 for tagged and untagged Tid1, respectively). We therefore conclude that the GST tags do not affect the ATPase activity of either protein.
Dmc1-ssDNA Complexes Are Stabilized in Ca2+
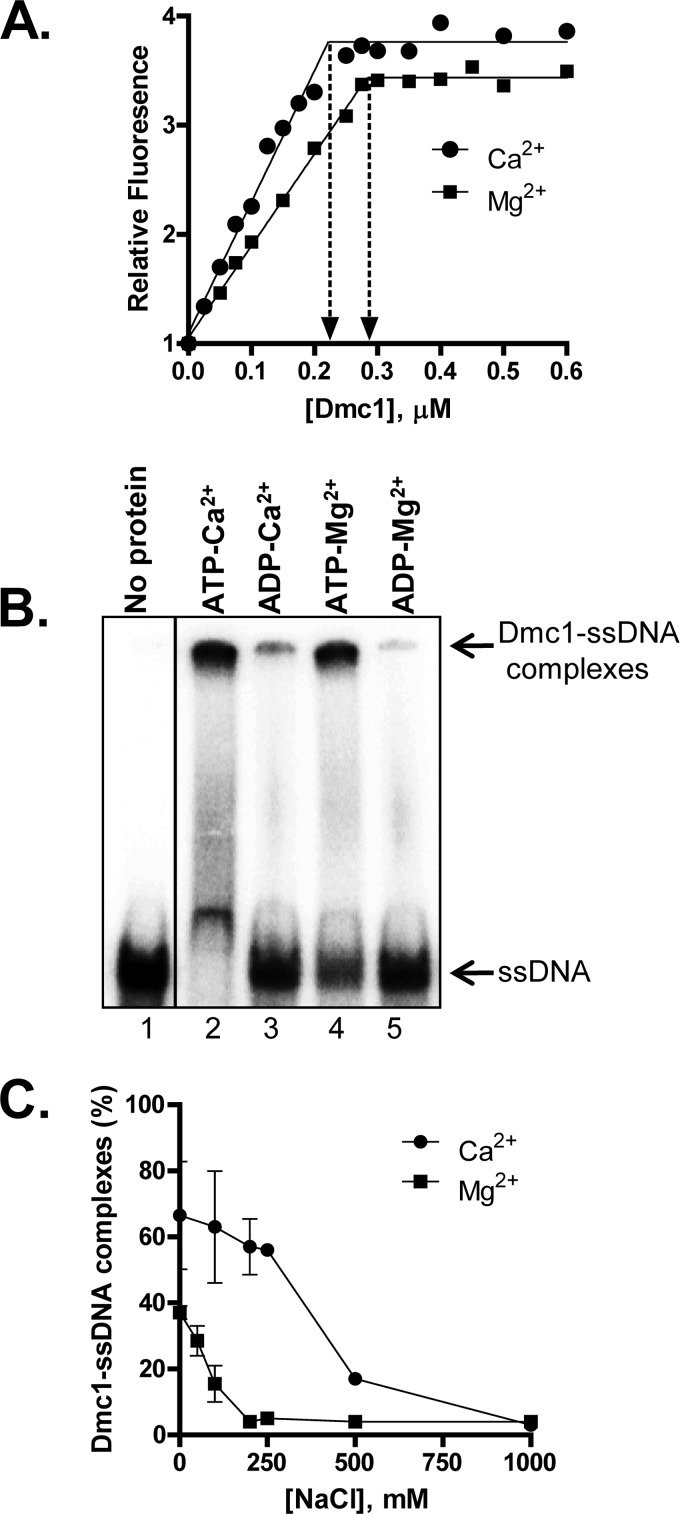
The ssDNA-dependent ATPase activity of Dmc1 (Fig. 1C) established that the protein binds ssDNA. Previous reports established that yeast, as well as human, Dmc1 forms stable filaments only in the presence of calcium (34, 35). We therefore examined the ability of Dmc1 to form complexes with ssDNA in the presence of magnesium acetate or calcium acetate (henceforth referred to as Mg2+ and Ca2+, respectively). DNA binding experiments using etheno-DNA showed that Dmc1-ssDNA filament formation exhibited comparable binding stoichiometries of 3.6 ± 0.1 nt/Dmc1 in Mg2+ and 4.3 ± 0.2 nt/Dmc1 in Ca2+ (Fig. 2A).
FIGURE 2.
Dmc1-ssDNA complexes are stabilized in Ca2+ by blocking ATP hydrolysis. A, binding of Dmc1 to etheno-DNA. The increase in fluorescence of etheno-DNA (1 μm) upon Dmc1 binding is plotted as a function of Dmc1 concentration. The y axis values are relative to the signal in the absence of Dmc1, which is assigned a value of 1. The site size was calculated by fitting the data to a two-segment line using GraphPad Prism version 5. B, formation of Dmc1-ssDNA complexes as a function of NTP and divalent cation. Dmc1 (0.2 μm)-ssDNA (55-mer; 0.1 μm nt) complexes were formed in the presence of the indicated nucleotide and divalent cation using standard conditions. C, the percentage of Dmc1-ssDNA complexes formed upon Dmc1 binding to 55-mer ssDNA at varying NaCl concentrations. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
To confirm these results, we also tested the ability of Dmc1 to bind ssDNA using a 55-mer oligonucleotide and an electrophoretic mobility shift assay (Fig. 2B). Here as well, protein-DNA complexes were identified in the presence of ATP and either divalent cation, but the extent of binding was ∼2-fold lower in the presence of Mg2+, suggesting less stable complex formation in Mg2+ (Fig. 2, B and C, compare values at 0 mm NaCl concentration). In the presence of ADP, complexes were not detected regardless of the divalent cation (Fig. 2B). To validate the difference in the stability of the Dmc1-ssDNA complexes formed in ATP with Ca2+ versus Mg2+, we challenged complexes formed in the presence of either cation with increasing amounts of NaCl. An increase in the salt concentration of the buffer disrupts protein-DNA interactions; for the same protein-DNA complexes, the weaker complexes are more susceptible to dissociation (36, 37). Such salt titration experiments confirmed that Dmc1-ssDNA complexes formed in Ca2+ are more resistant to the disruptive effect of NaCl than when formed in Mg2+, with the midpoint for the salt titrations being 250 and 100 mm NaCl for Ca2+ and Mg2+, respectively (Fig. 2C). We therefore conclude that although Dmc1 binds ssDNA in the presence of either cation, Dmc1-ssDNA filaments formed in Mg2+ are less stable than those formed in Ca2+.
AFM and EM Analyses of Dmc1-DNA Complexes
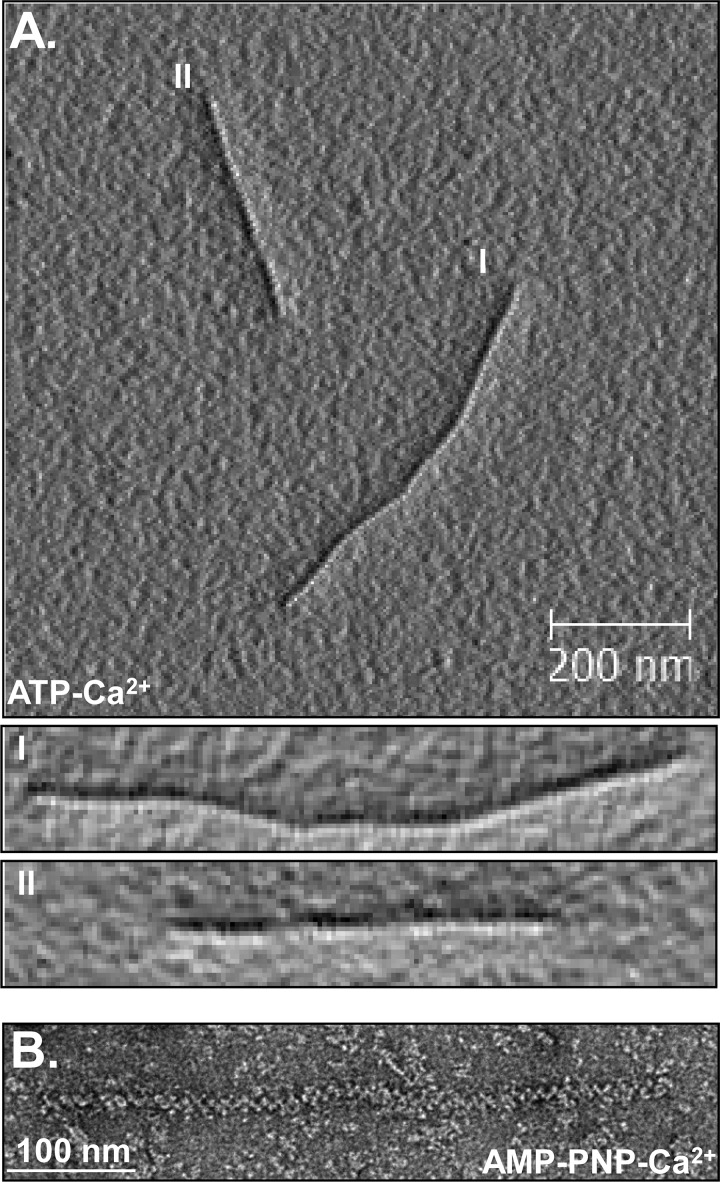
We next visualized the complexes of Dmc1 formed on dsDNA by AFM and EM. AFM images showed that in the presence Ca2+, Dmc1 formed complete filaments and extended the DNA by 1.5 ± 0.1-fold (Fig. 3A, supplemental Fig. S2A, and Table 1). The filaments were right-handed with a pitch of 101 ± 5 Å, which is similar to the values for RadA (109 and 100 Å by AFM and EM, respectively) (38, 39) and RAD51 (99 Å) (40) nucleoprotein filaments. EM analysis showed that the pitch of Dmc1-dsDNA complexes formed in AMP-PNP, a non-hydrolyzable analog of ATP (39), and Ca2+ is 109 ± 2 Å (Fig. 3B), which is comparable with the pitch obtained from AFM. Overall, the pitch values calculated using either method are equivalent to the numbers reported previously (41). AFM images also showed that, in contrast to filaments formed in Ca2+, Dmc1-dsDNA filaments formed in the presence of Mg2+ appeared unstable and incomplete (supplemental Fig. S2A). EM images showed a similar difference in the stability of filament formation: whereas filaments were evident in the presence of either cation, the filaments in Mg2+ were diffuse and incomplete (supplemental Fig. S2B).
FIGURE 3.
AFM and EM images of Dmc1-dsDNA filaments. A, AFM image showing Dmc1-dsDNA complexes in ATP-Ca2+. I and II show magnified images of the two complexes. B, EM image showing Dmc1-dsDNA complexes in the presence of AMP-PNP-Ca2+. The pitches of the complexes in A and B are 101 ± 5 (n = 7) and 109 ± 2.3 Å (n = 6), respectively.
TABLE 1.
Length and pitch measurement for Dmc1-dsDNA filament images obtained using AFM
The length analysis was performed on images such as those shown in supplemental Fig. S2A. The pitch of Dmc1-dsDNA filaments was calculated from images such as those shown in Fig. 3A. N.A. and N.D. are not applicable and not determined, respectively, and n is the number of molecules measured.
| Length | Pitch | |
|---|---|---|
| nm | Å | |
| Naked dsDNA | 355 ± 10 (n = 12) | N.A. |
| Dmc1-dsDNA (ATP-Ca2+) | 519 ± 40 (n = 9) | 101 ± 5 (n = 7) |
| Dmc1-dsDNA (AMP-PNP-Mg2+) | 480 ± 76 (n = 12) | N.D. |
Dmc1 Does Not Hydrolyze ATP in Ca2+-containing Buffer
We wondered whether this contrasting behavior of the filaments in Ca2+ and Mg2+ can be explained by the inhibition of ATP hydrolysis in the presence of Ca2+ because ATP hydrolysis promotes protein dissociation by producing the ADP-bound forms of DNA strand exchange proteins, which are known to display lower affinity for DNA (29). We therefore compared the ability of Dmc1-ssDNA nucleoprotein filaments to hydrolyze ATP in the presence of Ca2+ and Mg2+. As we anticipated, Dmc1 failed to hydrolyze ATP in the presence of Ca2+ (supplemental Fig. S3), which would maintain the protein-DNA complex in a stable form. To verify whether inhibition of ATP hydrolysis leads to stabilization of the Dmc1-DNA filament, we also analyzed AFM images of Dmc1-DNA complexes formed in the presence of Mg2+ and AMP-PNP. Indeed, stable Dmc1-DNA filaments were formed in the presence of Mg2+ when ATP was replaced with AMP-PNP with the filaments being extended to an extent comparable with that seen with ATP and Ca2+ (1.35 ± 0.2-fold; supplemental Fig. S2A and Table 1). These DNA binding and activity studies establish that Ca2+-induced inhibition of ATP hydrolysis results in the formation of stable Dmc1-DNA filaments.
Dmc1 Forms Joint Molecules in Presence of Ca2+ but Not Mg2+
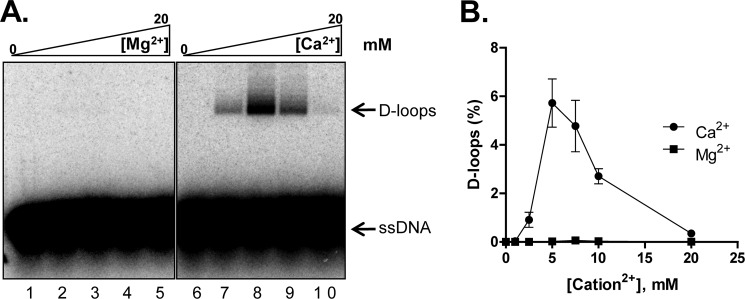
Because the nucleoprotein filament is the active species that catalyzes DNA strand invasion during homologous recombination, we expected the stable filaments formed in the presence of Ca2+ to be competent in joint molecule formation. We therefore tested the ability of Dmc1 to form D-loops as a function of Ca2+ and Mg2+. D-loop formation occurred in the presence of Ca2+ but not Mg2+, with the efficiency being highest at 5 mm Ca2+ (Fig. 4, A and B). This behavior of Dmc1 is similar to human RAD51 (42). D-loop formation by the tagged and untagged Dmc1 at the optimal Ca2+ concentration was comparable (supplemental Fig. S4, A and B), demonstrating that the GST tag does not influence Dmc1 activity.
FIGURE 4.
Dmc1 forms joint molecules in presence of Ca2+ but not Mg2+. A, gel showing D-loop formation catalyzed by Dmc1 as a function of Mg2+ or Ca2+, respectively. B, graphical representation of data from A. The positions of free ssDNA and D-loops are indicated. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
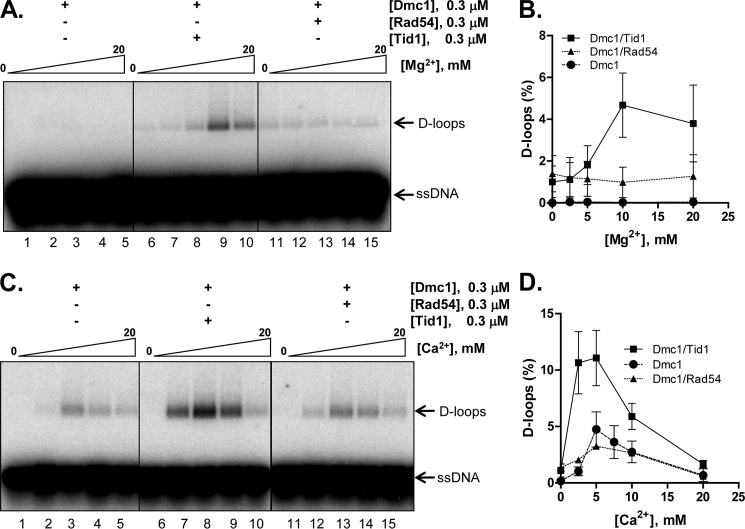
Dmc1 Is Specifically Stimulated by Tid1
Dmc1 and Tid1 work together during meiosis to establish crossovers (18). To determine whether Tid1 stimulates DNA pairing by Dmc1, we compared the effect of Tid1 and Rad54 in D-loop formation by Dmc1. We first studied the effect of Tid1 in the presence of varying concentrations of Mg2+. As established before (Fig. 4), Dmc1 does not function in reactions containing Mg2+. Interestingly, however, Tid1 enabled Dmc1-mediated D-loop formation in the presence of Mg2+, with the optimum concentration being 10 mm Mg2+ (Fig. 5, A and B). In comparison, Rad54 failed to stimulate Dmc1, suggesting a specific functional interaction between Dmc1 and Tid1. We next performed a similar analysis in the presence of Ca2+ instead of Mg2+. As seen earlier in Fig. 4, Dmc1 formed D-loops in the presence of Ca2+. Upon addition of Tid1, the efficiency of Dmc1-mediated D-loop formation increased ∼2–3-fold with maximum stimulation observed at 5 mm Ca2+ (Fig. 5, C and D). Importantly, untagged Dmc1 was equally stimulated by either tagged or untagged Tid1, establishing that stimulation is not affected by the presence of the GST tags (supplemental Fig. S4, C and D). Rad54 failed to stimulate Dmc1 in Ca2+ as well (Fig. 5, C and D), and experiments with untagged Rad54 showed that it stimulated untagged Dmc1 to an extent similar to that of the tagged Rad54, which is less than that observed with either tagged or untagged Tid1 (supplemental Fig. S4, E and F). These findings establish that the specificities of stimulation observed for these four proteins are unaltered by the presence or absence of the GST tags.
FIGURE 5.
Dmc1 is specifically stimulated by Tid1. A and C, gels showing D-loop formation catalyzed by the indicated sets of proteins as a function of Mg2+ or Ca2+, respectively. B and D, graphical representation of data from A and C. The positions of free ssDNA and D-loops are indicated. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
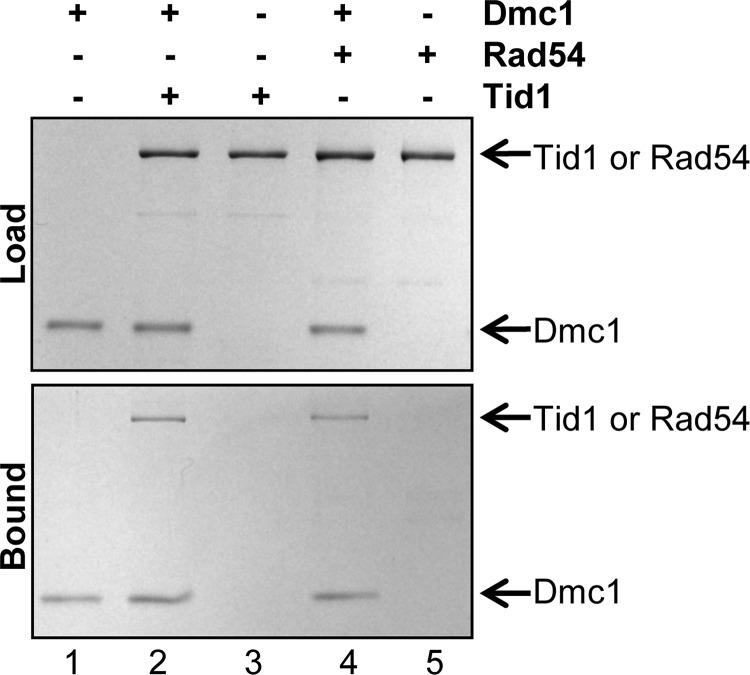
Dmc1 Interacts Physically with Tid1 as Well as with Rad54
Tid1 was identified as a Dmc1-interacting protein in a two-hybrid screen, and genetic evidence showed that Dmc1 and Tid1 interacted physically as well as functionally (18). To determine whether the specific stimulation of Dmc1 by Tid1 in joint molecule formation (Fig. 5) could be explained by a direct interaction, we purified Dmc1 as a His-tagged protein as described (27). We performed pulldown experiments using Ni2+-NTA magnetic beads, exploiting the presence of the His6 tag on the N-terminal end of Dmc1 to determine whether Tid1 and Rad54 interact with Dmc1. The data show that Tid1 is pulled down only when Dmc1 was bound to the beads (Fig. 6, lanes 2 and 3), demonstrating that the two proteins physically interact. Interestingly, Dmc1 interacted with Rad54 as well (Fig. 6, lanes 4 and 5). The observation that Dmc1 interacts with both of the translocases indicates that the functionally specific stimulation of Dmc1 by Tid1 is not simply a consequence of a direct physical interaction between the two proteins.
FIGURE 6.
Dmc1 interacts physically with Tid1 as well as with Rad54. Pulldown experiments were performed with Dmc1 (N-terminal His6 tag) and Tid1 or Rad54 using Ni2+-NTA magnetic beads as follows: lanes 1, 3 and 5, Dmc1, Tid1, and Rad54 alone, respectively; lane 2, Dmc1 and Tid1; lane 4, Dmc1 and Rad54. The bound fractions were analyzed by gel electrophoresis followed by Coomassie Blue staining. The panels labeled “load” and “bound” show the starting material and amount of protein bound to the beads, respectively. The positions of Dmc1, Rad54, and Tid1 are indicated.
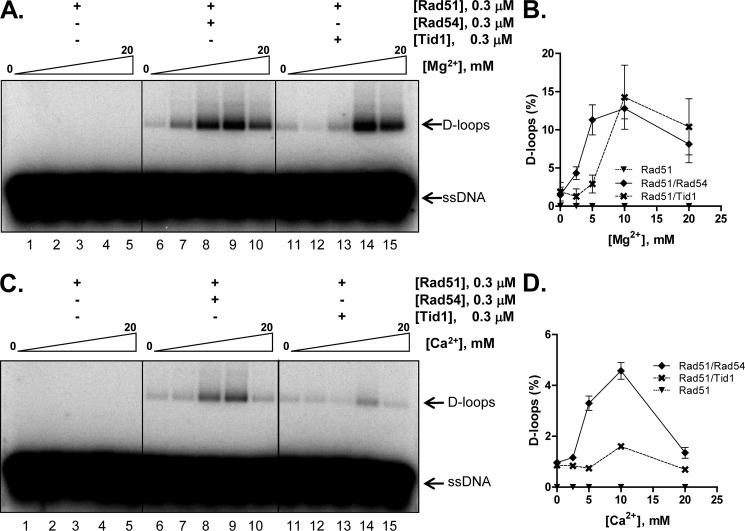
Rad51 Can Be Stimulated by Rad54 as Well as by Tid1
Having established the effect of Tid1 and Rad54 on Dmc1, we next examined the role of Rad54 and Tid1 in Rad51-mediated joint molecule formation. Both Rad54 and Tid1 were reported to stimulate the activity of Rad51 in the presence of Mg2+ (33, 43, 44). Consistent with those reports, we also observed stimulation of Rad51-mediated joint molecule formation by Rad54, with the optimum concentration being 5–10 mm Mg2+ (Fig. 7, A and B). Tid1 also stimulated Rad51, but a higher Mg2+ concentration was needed (Fig. 7, A and B). Therefore, in the presence of Mg2+, Rad51 is stimulated by both Rad54 and Tid1.
FIGURE 7.
D-loop formation by Rad51 can be stimulated by either Rad54 or Tid1 in the presence of Mg2+ but only by Rad54 in the presence of Ca2+. A and B, Rad51 is stimulated by Rad54 as well as by Tid1. A, gel showing D-loop reactions catalyzed by the indicated sets of proteins as a function of Mg2+. C and D, Rad51 is specifically stimulated by Rad54 in Ca2+. C, gel showing D-loop reactions catalyzed by the indicated sets of proteins as a function of Ca2+. B and D, graphical representation of data from A and C. The positions of free ssDNA and D-loops are indicated. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
Rad51 Is Specifically Stimulated by Rad54 in Ca2+
Unlike Dmc1, yeast Rad51 cannot form joint molecules in the presence of Ca2+ (Fig. 7C, lanes 1–5). To determine whether either Rad54 or Tid1 can stimulate Rad51 activity in the presence of Ca2+, we tested their abilities to activate Rad51-mediated D-loop formation at varying concentrations of Ca2+. Rad54 did indeed stimulate Rad51 at an optimum Ca2+ concentration of 10 mm (Fig. 7, C and D), but the stimulation was ∼⅓ of the yield seen with Mg2+ (Fig. 7, A and B). However, in the presence of Ca2+, Tid1 did not significantly stimulate Rad51 (Fig. 7, C and D). Similar results were obtained using untagged Rad54 and Tid1, thereby establishing that for DNA pairing reactions mediated by Rad51 as well, the tags do not dictate the specificity of the interaction (supplemental Fig. S4, E and F). These data show that, in the presence of Ca2+, Rad51 is preferentially stimulated by Rad54 but not by Tid1.
The ability of Rad54 and Tid1 to stimulate DNA strand exchange proteins in Ca2+ suggested that these translocases can function as ATPases in Ca2+ because the ATPase-dead mutants of either protein do not stimulate DNA strand exchange (32, 40). Indeed, Tid1 and Rad54 hydrolyzed ATP in Ca2+-containing buffer to an extent comparable with that with Mg2+-containing buffer (supplemental Fig. S5, A and B). To ascertain that the activity seen in the presence of Ca2+ was not due to contaminating Mg2+, we measured the activity of RecA in the same buffers. As expected (29), RecA was inhibited by the Ca2+ buffer (supplemental Fig. S5C).
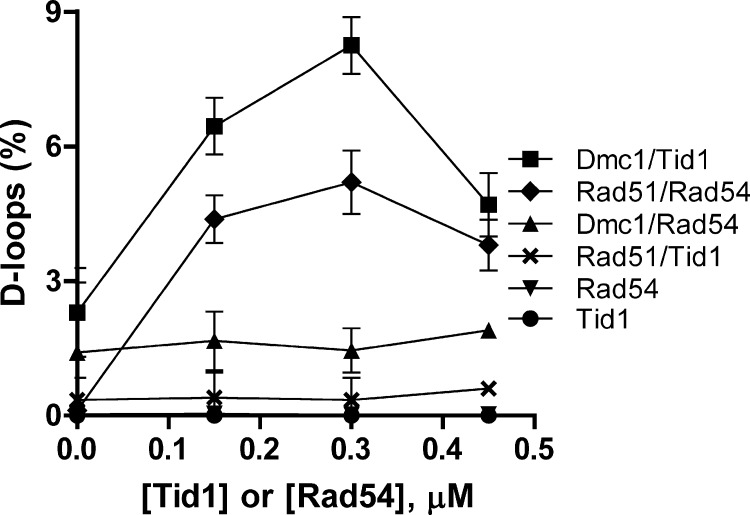
The Specific Stimulation of Dmc1 by Tid1 and Rad51 by Rad54 Is Revealed in Ca2+-containing Reactions
To confirm the aforementioned findings regarding the specificity of stimulation, we performed titrations with Tid1 and Rad54 at fixed concentrations of Dmc1 and Rad51 (Fig. 8). The data indicated that specificity holds over a wide range of translocase concentrations. In the presence of Mg2+, Rad51 and Rad54 had been shown to produce the maximum amount of D-loops when present at equimolar concentrations (26). Here we show that in the presence of Ca2+ the two proteins also demonstrate an optimal yield at a 1:1 ratio (Fig. 8, diamonds). Similarly, Dmc1 and Tid1 yielded maximum products when present at equimolar amounts (squares). The non-cognate pairs (Dmc1-Rad54 and Rad51-Tid1) yielded minimal amounts of joint molecule products. Our results thus establish that Dmc1 and Rad51 faithfully couple with Tid1 and Rad54, respectively, albeit with the need for Ca2+.
FIGURE 8.
The specific stimulation of Dmc1 by Tid1 and Rad51 by Rad54 is revealed in Ca2+-containing reactions. D-loop reactions were performed in 5 mm Ca2+. The concentration of Dmc1 and Rad51 was 0.3 μm. The concentrations of Tid1 and Rad54 were varied from 0 to 0.45 μm. Error bars indicate S.D. from at least three independent experiments and are smaller than the symbols when not evident.
DISCUSSION
In this study, we presented the purification and characterization of yeast Dmc1 and Tid1 proteins. We showed that the GST tag used for purification did not hinder the innate ssDNA- and dsDNA-dependent activities of either Dmc1 or Tid1, respectively. Furthermore, we established that the tagged versions of these proteins demonstrate identical specificities in their stimulation of joint molecule formation when compared with their untagged counterparts. We demonstrated that Dmc1 forms complete and stable nucleoprotein filaments in the presence of Ca2+, that Dmc1 mediates DNA strand invasion, and that depending on conditions this reaction is stimulated specifically and more efficiently by Tid1 in comparison to Rad54. In contrast, joint molecule formation by Rad51 can be stimulated by either Rad54 or Tid1 when the presumed physiologically meaningful divalent ion, Mg2+, is used, but a specific requirement for Rad54 is revealed when Ca2+ is substituted for Mg2+. Our results thereby provide biochemical evidence for the preferential functional pairwise interactions between Dmc1 and Tid1 and between Rad51 and Rad54.
Our results showed that Dmc1 alone can mediate DNA strand invasion although only in the presence of Ca2+. Imaging and activity analysis demonstrated that the Dmc1-DNA filaments formed in the presence of Ca2+ are stable. Previous reports suggested that Ca2+ converts the inactive ring form of Dmc1 into dispersed monomers that allow archetypal right-handed nucleoprotein formation (34). We showed that Ca2+ facilitates Dmc1-DNA filament formation by both increasing the stability of the Dmc1-DNA complex and blocking the ATPase activity of Dmc1. Ca2+ also stimulates joint molecule formation by human DMC1 and RAD51 by stabilizing the nucleoprotein complex, although there is an important mechanistic difference in the behavior of these two proteins with regard to the effect of Ca2+. In the case of RAD51, Ca2+ maintains an active presynaptic filament by blocking ATP hydrolysis and thereby preventing the self-inhibition that accompanies accumulation of ADP within the filament. In contrast, for DMC1, Ca2+ does block ATP hydrolysis, but this is not the basis for stimulation because a similar stimulation by Ca2+ is observed in the presence of the non-hydrolyzable AMP-PNP; thus, Ca2+ apparently induces a conformational change in the filament assembled on ssDNA that enables more effective DNA strand exchange (35, 42). For yeast Dmc1, the modulation by Ca2+ is similar to human DMC1 in that Ca2+ also stimulates joint molecule formation slightly when AMP-PNP is used (supplemental Fig. S6), although stimulation by Ca2+ is greater when ATP is used, suggesting that inhibition of ATP hydrolysis has a larger impact. Analysis of RAD51-ssDNA filaments by linear dichroism established that the complexes formed in Ca2+, but not in Mg2+, assume an ordered conformation that contributes to stabilizing protein-DNA interaction (45). Ca2+ may have a similar effect on Dmc1 filaments.
The role of Ca2+ in stabilizing protein-DNA interactions is not unprecedented: it was established more than two decades ago that the ATPase activity of RecA in the presence of Ca2+ is 4% of that observed in Mg2+, resulting in higher affinity of the protein for ssDNA (29). The stabilization of the nucleoprotein filament by blocking ATP hydrolysis is a strategy effectively used by the cell as well: ATP hydrolysis by human RAD51 is reduced by BRC repeats 1–4 of the mediator protein BRCA2 (46, 47) and by BRCA2 itself (48), and ATP hydrolysis by Caenorhabditis elegans RAD-51 is blocked by BRC-2 (49); in all cases, the result is a stable nucleoprotein filament.
Whereas the stabilized Dmc1-DNA nucleoprotein filament formed in Ca2+ showed potent DNA strand invasion activity, filaments formed in Mg2+ appeared inactive (Fig. 4). In the presence of Mg2+, Dmc1 hydrolyzes ATP to produce the inactive and less stable ADP-Dmc1-ssDNA complex, which is unable to pair DNA (Fig. 4). Interestingly, stable filament formation (supplemental Fig. S2A) and DNA pairing (27) occur in Mg2+ when ATP is replaced with a non-hydrolyzable analog (AMP-PNP), indicating that blocking dissociation of Dmc1 from ssDNA is a component of the mechanism of Ca2+-dependent stimulation of Dmc1 activity. This conclusion in turn implies that the activation of Dmc1 by Tid1 in Mg2+ buffers results from a stabilization of the Dmc1 nucleoprotein filament. This stabilization is specific to Tid1 as Rad54 provides negligible stimulation under conditions where Dmc1 can hydrolyze ATP (Fig. 5B). We speculate that the stimulation may be attributed to the enhanced stability of Dmc1-ssDNA in the presence of Tid1, similar to both the stimulation and stabilization of the Rad51-ssDNA complex afforded by Rad54 (34).
Given that Ca2+ plays an important role during the early stages of meiosis (50), it is tempting to hypothesize that Ca2+ is a universal modulator during meiotic DNA break repair. But the cellular concentration of Ca2+ in vivo during this phase is in the nanomolar range (51), making it unlikely to stimulate homologous pairing in the cell. It is nevertheless possible that the effects of Ca2+ on DNA pairing observed in vitro are a mimic of the situation in vivo, perhaps mediated by proteins that specifically modulate the activity of Dmc1 or stabilize the Dmc1-ssDNA complex.
Although Tid1 and Rad54 can be partially redundant in vivo, genetic evidence suggests that each also has autonomous functions mediated through functional interactions with Dmc1 and Rad51, respectively (19–21, 52). During mitosis, unlike rad54 mutants, tid1 mutants are not sensitive to methyl methanesulfonate (20), and tid1 mutants are not defective for recombination except in diploids (20, 21). In meiosis, tid1 mutants are defective in recombination, but the rad54 mutants are not (21). We established that joint molecule formation by Dmc1 is stimulated by Tid1 but not by Rad54. Our observation correlates with physical analysis of crossover formation in vivo that shows that formation of recombinants is not dependent on Rad54 (21). DNA breaks formed during meiosis are repaired via two distinct pathways: one that depends on Rad51 alone and another that requires Rad51 and Dmc1 (53). The first one is termed the “Rad51-only” pathway, and the repair products generated by this pathway are non-crossovers. The second pathway is termed the “Dmc1-dependent” pathway and is essential for crossover formation and subsequent chromosome segregation. Interestingly, Rad51 is also required for the Dmc1-dependent pathway. This is evident in a rad51 single mutant wherein the intermediates show an 8-fold decrease in the ratio of interhomolog to intersister joint molecules (54). In the presence of both pathways, it is necessary to down-regulate the Rad51-only pathway to ensure that at least a subset of the breaks are channeled toward crossover formation (55, 56). It is believed that the bias on crossover versus non-crossover formation is established either during filament formation or by controlling the DNA pairing activity (1). Regarding that notion, our study shows that Tid1 specifically stimulates the innate ability of Dmc1 to mediate DNA pairing, thereby tilting the bias toward crossover formation.
At the biochemical level, single end invasion reactions catalyzed by Rad51 or Dmc1 are similar: both proteins form ATP-dependent helical filaments on DNA and perform DNA strand exchange (41). The comparable in vitro activities of Dmc1 and Rad51 support the view that differences in the meiotic function of Rad51 and Dmc1 are more likely to result from the influence of distinct sets of accessory proteins. The ssDNA can be utilized either by the Rad51-only or Dmc1-dependent pathway with Rad51 and Dmc1 nucleoprotein filaments being stabilized by Rad54 and Tid1, respectively. There is a notion that Rad51 and Dmc1 assemble on opposite ends of DNA breaks due to the differential regulation of the two nucleoprotein complexes (13, 57). During meiosis, specific factors ensure that, whereas Dmc1 nucleoprotein filaments pair with the homolog, Rad51 activity is suppressed. Indeed, the meiosis-specific factors Mek1, Red1, and Hop1 form a complex that is responsible in part for creating a barrier to sister chromatid repair (58). In addition, germ line-specific Hed1 downplays Rad51 activity by interfering with the recruitment of Rad54, which is indispensable for Rad51-mediated recombination (43, 59). In addition to blocking Rad51 activity, the cell also encodes factors that stimulate Dmc1 activity. The meiosis-specific Mei5-Sae3 heterodimer recruits Dmc1 (60) and acts as a mediator to stimulate Dmc1 pairing activity in the presence of replication protein A (61). Whereas joint molecules formed by Rad51 are channeled toward the synthesis-dependent strand annealing pathway and generate non-crossovers, joint molecules formed by Dmc1 lead to the formation of double Holliday junctions and are primarily resolved into crossover products. Indeed, biochemical investigation of human counterparts established that joint molecules formed by DMC1 are more resistant to the disruptive activity of RAD54 and Bloom syndrome helicase than those formed by RAD51 (62). Elucidation of these control mechanisms in yeast will require continued biochemical investigation of these regulatory components.
Supplementary Material
Acknowledgment
We thank all members of the Kowalczykowski laboratory for helpful discussions on this work and for comments and insights on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62653 (to S. C. K.).

This article contains supplemental Figs. S1–S6.
- ssDNA
- single-stranded DNA
- nt
- nucleotides
- AFM
- atomic force microscopy
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1. Ehmsen K. T., Heyer W. D. (2008) Biochemistry of meiotic recombination: formation, processing, and resolution of recombination intermediates. Genome Dyn. Stab. 3, 91–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roeder G. S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600–2621 [DOI] [PubMed] [Google Scholar]
- 3. Hassold T., Hunt P. (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 [DOI] [PubMed] [Google Scholar]
- 4. Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S. (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 [DOI] [PubMed] [Google Scholar]
- 5. Romanienko P. J., Camerini-Otero R. D. (2000) The mouse spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 [DOI] [PubMed] [Google Scholar]
- 6. Heyer W. D., Ehmsen K. T., Liu J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keeney S., Neale M. J. (2006) Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 34, 523–525 [DOI] [PubMed] [Google Scholar]
- 8. Mimitou E. P., Symington L. S. (2009) Nucleases and helicases take center stage in homologous recombination. Trends Biochem. Sci. 34, 264–272 [DOI] [PubMed] [Google Scholar]
- 9. Sun H., Treco D., Schultes N. P., Szostak J. W. (1989) Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338, 87–90 [DOI] [PubMed] [Google Scholar]
- 10. Cao L., Alani E., Kleckner N. (1990) A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089–1101 [DOI] [PubMed] [Google Scholar]
- 11. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 12. Hunter N. (2007) in Molecular Genetics of Recombination (Aguilera A., Rothstein R., eds) pp. 381–442, Springer-Verlag, Berlin [Google Scholar]
- 13. Hunter N., Kleckner N. (2001) The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 [DOI] [PubMed] [Google Scholar]
- 14. McIlwraith M. J., West S. C. (2008) DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol. Cell 29, 510–516 [DOI] [PubMed] [Google Scholar]
- 15. Li X., Stith C. M., Burgers P. M., Heyer W. D. (2009) PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell 36, 704–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Allers T., Lichten M. (2001) Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8, 225–231 [DOI] [PubMed] [Google Scholar]
- 17. Schwacha A., Kleckner N. (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791 [DOI] [PubMed] [Google Scholar]
- 18. Dresser M. E., Ewing D. J., Conrad M. N., Dominguez A. M., Barstead R., Jiang H., Kodadek T. (1997) DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147, 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arbel A., Zenvirth D., Simchen G. (1999) Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 18, 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klein H. L. (1997) RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shinohara M., Shita-Yamaguchi E., Buerstedde J. M., Shinagawa H., Ogawa H., Shinohara A. (1997) Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147, 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shinohara M., Sakai K., Shinohara A., Bishop D. K. (2003) Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics 163, 1273–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Second Edition, pp. 92–96 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Kiianitsa K., Solinger J. A., Heyer W. D. (2002) Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J. Biol. Chem. 277, 46205–46215 [DOI] [PubMed] [Google Scholar]
- 25. Zaitseva E. M., Zaitsev E. N., Kowalczykowski S. C. (1999) The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 274, 2907–2915 [DOI] [PubMed] [Google Scholar]
- 26. Mazin A. V., Bornarth C. J., Solinger J. A., Heyer W. D., Kowalczykowski S. C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell 6, 583–592 [DOI] [PubMed] [Google Scholar]
- 27. Hong E. L., Shinohara A., Bishop D. K. (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276, 41906–41912 [DOI] [PubMed] [Google Scholar]
- 28. Roman L. J., Kowalczykowski S. C. (1989) Characterization of the adenosinetriphosphatase activity of the Escherichia coli RecBCD enzyme: relationship of ATP hydrolysis to the unwinding of duplex DNA. Biochemistry 28, 2873–2881 [DOI] [PubMed] [Google Scholar]
- 29. Menetski J. P., Varghese A., Kowalczykowski S. C. (1988) Properties of the high-affinity single-stranded DNA binding state of the Escherichia coli recA protein. Biochemistry 27, 1205–1212 [DOI] [PubMed] [Google Scholar]
- 30. Menetski J. P., Kowalczykowski S. C. (1985) Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181, 281–295 [DOI] [PubMed] [Google Scholar]
- 31. Sogo J., Stasiak A., DeBernardin W., Losa R., Koller T. (1987) in Electron Microscopy in Molecular Biology (Sommerville J., Scheer U., eds) pp. 61–79, IRL Press, Oxford [Google Scholar]
- 32. Nimonkar A. V., Amitani I., Baskin R. J., Kowalczykowski S. C. (2007) Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J. Biol. Chem. 282, 30776–30784 [DOI] [PubMed] [Google Scholar]
- 33. Petukhova G., Sung P., Klein H. (2000) Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee M. H., Chang Y. C., Hong E. L., Grubb J., Chang C. S., Bishop D. K., Wang T. F. (2005) Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 280, 40980–40984 [DOI] [PubMed] [Google Scholar]
- 35. Bugreev D. V., Golub E. I., Stasiak A. Z., Stasiak A., Mazin A. V. (2005) Activation of human meiosis-specific recombinase Dmc1 by Ca2+. J. Biol. Chem. 280, 26886–26895 [DOI] [PubMed] [Google Scholar]
- 36. Record M. T., Jr., Lohman M. L., De Haseth P. (1976) Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 107, 145–158 [DOI] [PubMed] [Google Scholar]
- 37. Menetski J. P., Varghese A., Kowalczykowski S. C. (1992) The physical and enzymatic properties of Escherichia coli recA protein display anion-specific inhibition. J. Biol. Chem. 267, 10400–10404 [PubMed] [Google Scholar]
- 38. Seitz E. M., Brockman J. P., Sandler S. J., Clark A. J., Kowalczykowski S. C. (1998) RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 12, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X., Jacobs S. A., West S. C., Ogawa T., Egelman E. H. (2001) Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang S., Yu X., Seitz E. M., Kowalczykowski S. C., Egelman E. H. (2001) Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J. Mol. Biol. 314, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 41. Sheridan S. D., Yu X., Roth R., Heuser J. E., Sehorn M. G., Sung P., Egelman E. H., Bishop D. K. (2008) A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36, 4057–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bugreev D. V., Mazin A. V. (2004) Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petukhova G., Stratton S., Sung P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393, 91–94 [DOI] [PubMed] [Google Scholar]
- 44. Mazin A. V., Alexeev A. A., Kowalczykowski S. C. (2003) A novel function of Rad54 protein: stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278, 14029–14036 [DOI] [PubMed] [Google Scholar]
- 45. Fornander L. H., Frykholm K., Reymer A., Renodon-Cornière A., Takahashi M., Nordén B. (2012) Ca2+ improves organization of single-stranded DNA bases in human Rad51 filament, explaining stimulatory effect on gene recombination. Nucleic Acids Res. 40, 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carreira A., Hilario J., Amitani I., Baskin R. J., Shivji M. K., Venkitaraman A. R., Kowalczykowski S. C. (2009) The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136, 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carreira A., Kowalczykowski S. C. (2011) Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 108, 10448–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jensen R. B., Carreira A., Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petalcorin M. I., Sandall J., Wigley D. B., Boulton S. J. (2006) CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J. Mol. Biol. 361, 231–242 [DOI] [PubMed] [Google Scholar]
- 50. Whitaker M. (2006) Calcium at fertilization and in early development. Physiol. Rev. 86, 25–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Keith C. H., Maxfield F. R., Shelanski M. L. (1985) Intracellular free calcium levels are reduced in mitotic Pt K2 epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 82, 800–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmuckli-Maurer J., Heyer W. D. (2000) Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma 109, 86–93 [DOI] [PubMed] [Google Scholar]
- 53. Tsubouchi H., Roeder G. S. (2003) The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5, 915–925 [DOI] [PubMed] [Google Scholar]
- 54. Schwacha A., Kleckner N. (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123–1135 [DOI] [PubMed] [Google Scholar]
- 55. Kaback D. B., Barber D., Mahon J., Lamb J., You J. (1999) Chromosome size-dependent control of meiotic reciprocal recombination in Saccharomyces cerevisiae: the role of crossover interference. Genetics 152, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsubouchi H., Roeder G. S. (2006) Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20, 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shinohara M., Gasior S. L., Bishop D. K., Shinohara A. (2000) Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 97, 10814–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N. M. (2005) Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16, 5804–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Busygina V., Sehorn M. G., Shi I. Y., Tsubouchi H., Roeder G. S., Sung P. (2008) Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22, 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsubouchi H., Roeder G. S. (2004) The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics 168, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ferrari S. R., Grubb J., Bishop D. K. (2009) The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J. Biol. Chem. 284, 11766–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bugreev D. V., Pezza R. J., Mazina O. M., Voloshin O. N., Camerini-Otero R. D., Mazin A. V. (2011) The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat. Struct. Mol. Biol. 18, 56–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.