Background: MITA is an adapter protein critically involved in virus-triggered type I IFN induction and cellular antiviral response.
Results: The E3 ligase TRIM32 targets MITA for K63-linked ubiquitination and knockdown of TRIM32 inhibits virus-triggered type I IFN induction.
Conclusion: TRIM32-mediated K63-linked ubiquitination of MITA is important for cellular antiviral response.
Significance: These findings provide insights on the mechanisms of regulation of cellular antiviral response.
Keywords: Innate Immunity, Interferon, Signal Transduction, Ubiquitin Ligase, Virus
Abstract
Viral infection activates several transcription factors including NF-κB and IRF3, which collaborate to induce type I interferons (IFNs) and innate antiviral response. MITA (also called STING) is a critical adaptor protein that links virus-sensing receptors to IRF3 activation upon infection by both RNA and DNA pathogens. Here we show that the E3 ubiquitin ligase tripartite motif protein 32 (TRIM32) ubiquitinated MITA and dramatically enhanced MITA-mediated induction of IFN-β. Overexpression of TRIM32 potentiated virus-triggered IFNB1 expression and cellular antiviral response. Consistently, knockdown of TRIM32 had opposite effects. TRIM32 interacted with MITA, and was located at the mitochondria and endoplasmic reticulum. TRIM32 targeted MITA for K63-linked ubiquitination at K20/150/224/236 through its E3 ubiquitin ligase activity, which promoted the interaction of MITA with TBK1. These findings suggest that TRIM32 is an important regulatory protein for innate immunity against both RNA and DNA viruses by targeting MITA for K63-linked ubiquitination and downstream activation.
Introduction
Viral infection triggers a series of cellular events that lead to induction of type I interferons (IFNs) and proinflammatory cytokines (1, 2). Type I IFNs activate the JAK-STAT signal transduction pathways, leading to transcriptional induction of a wide range of downstream antiviral genes and innate antiviral response (3–5).
Pattern recognition receptors (PRRs)2 detect pathogen-associated molecular patterns (PAMPs) of microbes to initiate innate immune response. Viral nucleic acids, including viral genomic RNA/DNA and replicating intermediates, act as typic PAMPs (6). In most cell types, cytosolic viral RNA is recognized by RIG-I-like receptors (RLRs), including RIG-I, MDA5, and LGP2 (2). Upon sensing viral RNAs through their C-terminal RNA helicase domains, RIG-I and MDA5 undergo conformational changes and are recruited to the downstream adaptor protein VISA (also called MAVS, IPS-1, and Cardif) through their respective CARD domains (7, 8). VISA is located at the outer membrane of mitochondria and acts as a central platform for assembly of a complex that mediates both the NF-κB and IRF3 activation pathways after viral infection. TRAF6 and the IKK complex are responsible for VISA-mediated NF-κB activation, whereas TRAF3 and the noncanonical IKK family members TBK1 and IKKϵ function downstream of VISA to activate IRF3 (9).
Although considerable progress has been made on the mechanisms of type I IFN induction and antiviral innate immunity in responding to RNA viruses, how DNA pathogens trigger innate antiviral response is less understood. So far, multiple cytosolic sensors for viral or microbial DNA have been identified, including DAI (aso known as ZBP1 or DLM-1), polymerase III, AIM2, IFI16, DHX36, DHX9, and DDX41 (10–15). However, whether these DNA sensors act in a cell-context specific manner or play redundant roles is still unclear.
MITA, also known as STING/ERIS/MPYS, has been identified as a mitochondrion- and ER-associated membrane protein that is critically involved in type I IFN induction and antiviral innate immunity in responding to both RNA and DNA virus infection under certain circumstances (16–20). In some cell lines, knockdown of MITA inhibits type I IFN induction upon RNA virus infection. MEFs lacking MITA are defective in VSV- and SeV-triggered type I IFN production or STAT6-mediated induction of a set of chemokines, suggesting that MITA plays an pivotal role in antiviral innate immune response to RNA viruses (16–18, 21). In addition, certain cell types derived from MITA-deficient mice fail to induce type I IFN production in response to dsDNA and infection with herpes simplex virus 1 (HSV-1). MITA-deficient mice are also susceptible to HSV-1 infection, suggesting that MITA is an indispensable component of viral DNA-mediated innate immune response (20). Recently, it has been demonstrated that MITA acts as a direct sensor of cyclic dinucleotides which is known as a pathogen-secreted second messenger (22). These studies together reveal a critical role for MITA in innate immunity triggered by cytosolic viral or microbial nucleic acids.
Virus-triggered type I IFN induction and antiviral response are heavily regulated by ubiquitination of the signaling components. Since MITA plays a critical role in virus-triggered type I IFN induction and antiviral response, we screened for E3 ubiquitin ligases that target MITA. We identified tripartite motif protein 32 (TRIM32) as an E3 ubiquitin ligase that targets MITA for K63-linked ubiquitination at multiple lysine residues. Furthermore, our findings suggest that TRIM32-mediated K63-linked ubiquitination of MITA is an important step in virus-triggered IFN induction and cellular antiviral innate immunity.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Recombinant TNFα, IL-1β, and IFN-γ (R&D Systems); mouse monoclonal antibodies against FLAG (Sigma), HA (Covance), β-actin (Sigma), AIF and KDEL (Santa Cruz Biotechnology); rabbit polyclonal antibodies against ubiquitin (Santa Cruz Biotechnology), IRF3 and phospho-IRF3 (Santa Cruz Biotechnology), were purchased from the indicated manufacturers. SeV, VSV, GFP-NDV, HSV-1, anti-RIG-I, anti-cIAP1, and anti-MITA sera were previously described (16, 23). Mouse anti-TRIM32 was raised against recombinant human full-length TRIM32.
Constructs
NF-κB, ISRE, IRF1, and IFN-β promoter luciferase reporter plasmids, mammalian expression plasmids for HA-, Flag-, or GFP-tagged MITA and its mutants, and RIG-I, TBK1 were previously described (7, 16, 24, 25). The pCMV-C-HA-ERIS/MITA was provided by Dr. Zhengfan Jiang (Peking University). Mammalian expression plasmids for human HA-, Flag-, or Cherry-tagged TRIM32 and its mutants were constructed by standard molecular biology techniques.
Expression Cloning
The cDNA expression clones encoding 352 ubiquitin-related enzymes were obtained from the Ubiquitin-GFC Transfection Array purchased from Origene (Cat. #UBGB19601). The clones were transfected together with MITA into 293 cells and ubiquitination of MITA was examined by coimmunoprecipitation and immunoblots.
Transfection and Reporter Assays
293 cells (1 × 105) were seeded on 24-well dishes and transfected the following day by standard calcium phosphate precipitation method. Empty control plasmid was added to ensure that each transfection receives the same amount of total DNA. To normalize for transfection efficiency, 0.01 μg of pRL-TK (Renilla luciferase) reporter plasmid was added to each transfection. Approximately 18 h after transfection, luciferase assays were performed using a dual-specific luciferase assay kit (Promega).
Coimmunoprecipitation, Immunoblot Analysis, Re-immunoprecipitation, and Native PAGE and VSV Plaque Assays
These experiments were performed as described (7, 16, 24, 25).
Virus Manipulation
Viral infection was performed when cells were 70% confluent. The culture medium was replaced by serum-free DMEM, and then NDV-GFP was added into the medium at various multiplicities of infection (MOI) according to the specific experiments. After 1 h, the medium was removed, and the cells were fed with DMEM containing 10% FBS. NDV-GFP replication in 293 cells was visualized by fluorescent microscopy.
In Vitro Ubiquitination Assays
The tested proteins were expressed with a TnT Quick-coupled Transcription/Translation Systems kit (Promega) following instructions of the manufacturer. Ubiquitination was analyzed with an ubiquitination kit (Enzo Life Science) following protocols recommended by the manufacturer.
RNAi Experiments
Double-stranded oligonucleotides corresponding to the target sequences were cloned into the pSuper.Retro RNAi plasmid (oligoengine Inc.). The following sequences were targeted for human TRIM32 cDNA. #1: GAAGTTGAGAAGTCCAATA; #2: CAGTCAAGGTGAAGTACTA; #3: CAGTTAACGTGGAAGATTC. A pSuper.retro RNAi plasmid targeting GFP was used as control for all RNAi-related experiments.
Real-time PCR
Total RNA was isolated from cells using Trizol reagent (TAKARA, Japan) and subjected to real-time PCR analysis to measure expression of mRNA. Gene-specific primer sequences were as follows: IFNB1: TTGTTGAGAACCTCCTGGCT (forward), TGACTATGGTCCAGGCACAG (reverse); RNATES: GGCAGCCCTCGCTGTCATCC (forward), GCAGCAGGGTGTGGTGTCCG (reverse); RIG-I: ACGCAGCCTGCAAGCCTTCC (forward); TGTGGCAGCCTCCATTGGGC (reverse); TNFα: GCCGCATCGCCGTCTCCTAC (forward), CCTCAGCCCCCTCTGGGGTC (reverse); IL-1β: GCAGGCCGCGTCAGTTGTTG (forward), CCCGGAGCGTGCAGTTCAGT (reverse); GAPDH: GAGTCAACGGATTTGGTCGT (forward), GACAAGCTTCCCGTTCTCAG (reverse).
Subcellular Fractionation
The 293 cells (5 × 107) infected with SeV or left uninfected for the indicated times were washed with PBS and lysed by douncing 40 times in 2 ml of homogenization buffer (10 mm Tris-HCl, pH7.4, 2 mm MgCl2, 10 mm KCl, 250 mm sucrose). The homogenate was centrifuged at 500 × g for 10 min for two times, and the pellet (P5) was saved as crude nuclei. The supernatant (S5) was centrifuged at 5,000 × g for 10 min to precipitate crude mitochondria (P5K). The supernatant (S5K) was further centrifuged at 50,000 × g for 60 min to generate S50K and P50K.
Immunofluorescent Confocal Microscopy
The transfected cells were incubated with the MitoTracker Red or ER Tracker Green (Molecular Probes) for 30 min. The cells were then fixed with 4% paraformaldehyde for 10 min and observed with an Olympus confocal microscope under a ×100 oil objective.
RNAi-transduced Stable THP-1 Cells
The 293 cells were transfected with two packaging plasmids pGag-Pol and pVSV-G and the GFP-RNAi control or TRIM32-RNAi retroviral plasmid by calcium phosphate precipitation. Cells were washed 12 h after transfection and new medium without antibiotics was added for 24 h. The recombinant virus-containing medium was filtered and used to infect THP-1 cells in the presence of polybrene (4 μg/ml). The infected THP-1 cells were selected with puromycin (0.5 μg/ml) for 2 weeks before additional experiments were performed.
RESULTS
Modulation of MITA Ubiquitination and Function by TRIM32
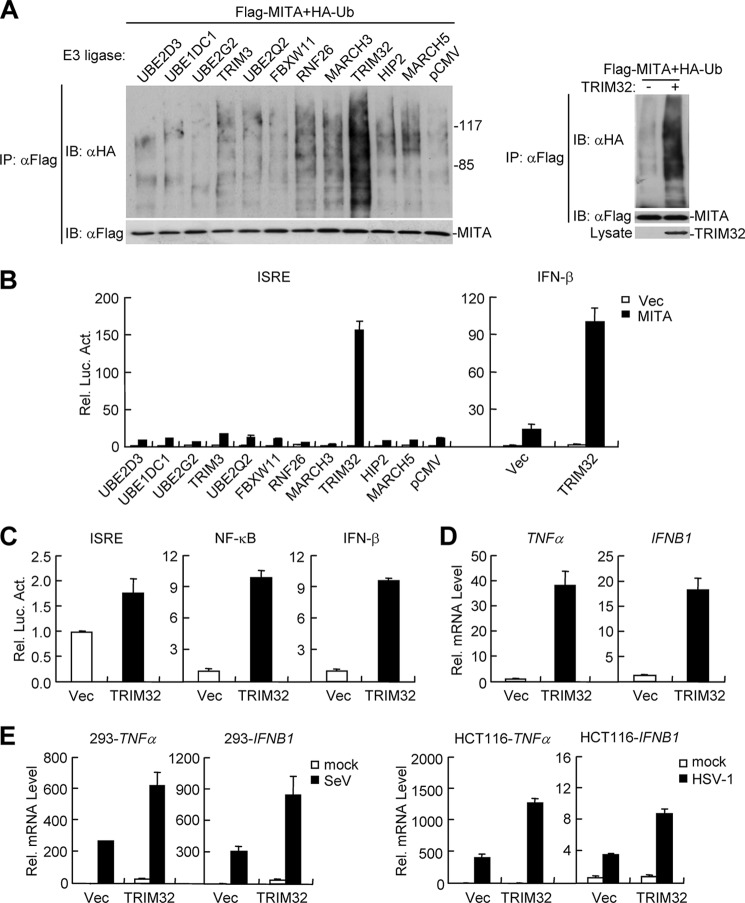
Ubiquitination has emerged as a critical post-translational regulatory mechanism for virus-triggered type I IFN induction pathways. Since MITA is an important adapter protein in these pathways, we attempted to unambigously identify E3 ubiquitin ligases that regulate MITA ubiquitination and function. To do this, we screened a cDNA array containing 352 expression clones of ubiquitin-related enzymes by ubiquitination assays with MITA as a substrate. In these assays, each of the clones was co-transfected with MITA into 293 cells, and then ubiquitination of MITA was examined by coimmunoprecipitation and immunoblot. The clones that could cause MITA ubiquitination were further tested for their ability to modulate MITA-mediated ISRE activation. These assays indentified TRIM32 as an E3 ligase that could cause MITA ubiquitination (Fig. 1A). In similar assays, TRIM32 did not ubiquitinate RIG-I, MDA5, and VISA (supplemental Fig. S1). In reporter assays, TRIM32 potently enhanced MITA-mediated activation of ISRE and the IFN-β promoter (Fig. 1B). In fact, overexpression of TRIM32 alone could weakly activate ISRE and NF-κB, as well as the IFN-β promoter (Fig. 1C). Real-time PCR experiments further confirmed that overexpression of TRIM32 could induce transcription of IFNB1 gene and the NF-κB targeted downstream gene TNFα (Fig. 1D). To determine whether TRIM32 plays a role in virus-induced expression of downstream genes, we examined its effects on Sendai virus (SeV, a negative-sense RNA virus)- and Herpes Simplex Virus 1 (HSV-1, a DNA virus)-triggered transcription of TNFα and IFNB1 genes. The results indicated that overexpression potentiated SeV- and HSV-1-induced transcription of TNFα and IFNB1 genes (Fig. 1E).
FIGURE 1.
TRIM32 plays a role in virus-triggered induction of IFN-β and TNFα. A, screens for E3 ubiquitin ligases that cause MITA ubiquitination. Left panels: the 293 cells (4 × 105) were transfected with Flag-MITA (2 μg) along with HA-ubiquitin (0.2 μg) and the indicated E3 ligases (0.2 μg each). Cell lysates were immunoprecipitated with anti-Flag and the immunoprecipitates were analyzed by immunoblots with anti-HA or anti-Flag. Right panels: the 293 cells were transfected with the indicated plasmids. Cell lysates were denatured by adding 1% SDS and heating for 10 min. The denatured lysates were diluted for immunoprecipitation and immunoblot analysis as described above. B, effects of the indicated E3 ligases on MITA-mediated activation of ISRE and the IFN-β promoter. The 293 cells (1 × 105) were transfected with MITA (0.1 μg) and the indicated E3 ligases (0.1 μg) along with ISRE or the IFN-β promoter reporter plasmid (0.1 μg). Luciferase assays were performed 22 h after transfection. Graphs show mean ± S.D., n = 3. C, overexpression of TRIM32 results in activation of NF-κB and the IFN-β promoter. The 293 cells (1 × 105) were transfected with the indicated reporter (0.1 μg) and TRIM32 (0.1 μg) plasmids. Luciferase assays were performed 22 h after transfection. Graphs show mean ± S.D., n = 3. D, overexpression of TRIM32 leads to transcription of TNFα and IFNB1 genes. The 293 cells (2 × 105) were transfected with the indicated expression plasmids (0.1 μg each). Real-time PCR was performed 20 h after transfection. E, TRIM32 potentiates SeV- and HSV-1-induced transcription of TNFα and IFNB1 genes. For the left histographs, the 293 cells (2 × 105) were transfected with the indicated expression plasmids (0.1 μg each). 18 h after transfection, cells were left untreated or infected with SeV for 12 h before real-time PCR were performed. For the right histograph, HCT116 cells (2 × 105) were transfected with the indicated expression plasmids (0.1 μg each). 18 h after transfection, cells were left untreated or infected with HSV-1 for 24 h before real-time PCR was performed.
TRIM32 Is Required for Innate Immune Response to Both Cytosolic RNA and DNA
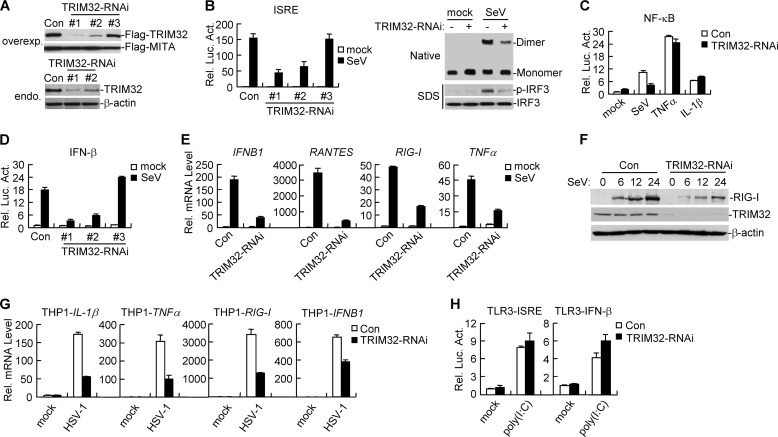
Because overexpression of TRIM32 potentiated virus-triggered IFN-β induction, we next determined whether endogenous TRIM32 regulates virus-triggered IFN-β induction. We constructed three human TRIM32-RNAi plasmids. Immunoblot analysis indicated that the #1 and #2 RNAi plasmids could markedly inhibit the expression of transfected and endogenous TRIM32 in 293 cells, whereas the #3 RNAi plasmid had little effect on TRIM32 expression (Fig. 2A). We transfected the TRIM32-RNAi vectors into 293 cells and performed reporter assays. As shown in Fig. 2B, knockdown of TRIM32 inhibited SeV-induced ISRE activation. The degree of inhibition was correlated with the efficiencies of knockdown of TRIM32 expression by each RNAi vector (Fig. 2, A and B). We selected TRIM32-RNAi-#1 plasmid for additional experiments described below and similar results were obtained with the #2 RNAi plasmid. IRF3, a critical player in the induction of type I IFNs, undergoes phosphorylation, dimerization, and nuclear translocation upon viral infection (26). As shown in Fig. 2B, knockdown of TRIM32 markedly inhibited SeV-induced phosphorylation and dimerization of IRF3. Knockdown of TRIM32 also inhibited SeV- but not TNFα- or IL-1β-induced NF-κB activation (Fig. 2C), suggesting that TRIM32 is specifically involved in SeV-induced NF-κB activation pathway. Consistently, knockdown of TRIM32 markedly inhibited SeV-induced activation of the IFN-β promoter (Fig. 2D). Real-time PCR experiments indicated that knockdown of TRIM32 inhibited SeV-induced transcription of downstream genes such as IFNB1, RANTES, and RIG-I and TNFα in 293 cells (Fig. 2E). Immunoblot analysis indicated that knockdown of TRIM32 inhibited SeV-induced expression of RIG-I protein, which has been demonstrated as a virus-inducible RNA sensor (Fig. 2F). Since MITA is indispensable for induction of IFN-β by non-CpG intracellular DNA species, we examined the effect of TRIM32 knockdown on HSV-1-induced expression of IFN-β in THP-1 cells which is sensitive to HSV-1 infection. The results indicated that knockdown of TRIM32 inhibited HSV-1-induced transcription of downstream genes including IL-1β, TNFα, RIG-I, and IFNB1 (Fig. 2G). Consistantly, knockdown of TRIM32 dramatically inhibited cytoplasmic poly(I:C)- and poly(dA:dT)-induced activation of ISRE and the IFN-β promoter in reporter assays (supplemental Fig. S2). In contrast, reporter assays indicated that knockdown of TRIM32 could not inhibit extracellular poly(I:C)-induced and TLR3-mediated activation of ISRE and the IFN-β promoter in TLR3-stable cells (Fig. 2H). These results suggest that TRIM32 is specifically involved in cytoplasmic viral nucleic acid sensors, but not membrane TLR3-induced signaling pathways.
FIGURE 2.
Effects of TRIM32 knockdown on virus-triggered induction of downstream genes. A, effects of TRIM32-RNAi plasmids on the expression of transfected and endogenous TRIM32. In the upper panels, the 293 cells (2 × 105) were transfected with expression plasmids for Flag-TRIM32 and Flag-MITA (0.1 μg each), and the indicated RNAi plasmids (1 μg each). At 24 h after transfection, cell lysates were analyzed by immunoblot with anti-Flag. In the lower panels, the 293 cells (1 × 107) were transfected with control or the indicated TRIM32-RNAi plasmids (8 μg each) for 24 h. Cell lysates were immunoprecipitated with anti-TRIM32, and the immunoprecipitates were analyzed by immunoblots with anti-TRIM32. The lysates were also analyzed by immunoblot with anti-β-actin as control. B, effects of TRIM32-RNAi on SeV-induced ISRE activation in 293 cells. In the left panel, the 293 cells (1 × 105) were transfected with the indicated RNAi plasmids (0.5 μg each) and ISRE reporter plasmid (0.1 μg). Twenty-four hours after transfection, cells were left untreated or infected with SeV for 12 h before luciferase assays were performed. Graphs show mean ± S.D., n = 3. In the right panels, the 293 cells (2 × 105) were transfected with the indicated RNAi plasmids (2 μg each). Twelve hours after transfection, cells were selected with puromycin (1 μg/ml) for 24 h, then infected with SeV or left uninfected for 8 h. Cell lysates were separated by native (upper panel) and SDS (bottom two panels) PAGE and analyzed with the indicated antibodies. C, effects of TRIM32-RNAi on SeV-, TNFα-, and IL-1β-induced NF-κB activation in 293 cells. Reporter assays were performed similarly as in B except that TNFα (10 ng/ml) and IL-1β (10 ng/ml) were used as stimuli. D, effects of TRIM32-RNAi on SeV-induced activation of the IFN-β promoter in 293 cells. Reporter assays were similarly performed as in B. E, effects of TRIM32-RNAi on SeV-induced transcription of IFNB1, RANTES, RIG-I, and TNFα genes. 293 cells (2 × 105) were transfected with the indicated RNAi plasmids (2 μg each). 12 h after transfection, cells were selected with puromycin (1 μg/ml) for 24 h, then infected with SeV or left uninfected for 12 h before real-time PCR was performed. F, knockdown of TRIM32 inhibited SeV-induced expression of RIG-I. The 293 cells (2 × 105) were transfected with TRIM32-RNAi plasmid for 12 h, selected with puromycin (1 μg/ml) for 24 h, then infected with SeV or left uninfected for the indicated time points. Cell lysates were analyzed by immunoblots with the indicated antibodies. G, effects of TRIM32-RNAi on HSV-1-induced transcription of downstream genes in THP-1 cells. THP-1 cells were transduced with GFP-RNAi or TRIM32-RNAi by retrovirus-mediated gene transfer. The cells were then either untreated or infected with HSV-1 for 24 h before real-time PCR experiments were performed. H, effects of TRIM32-RNAi on poly(I:C)-induced activation of ISRE and the IFN-β promoter in TLR3 cell lines. TLR3-stable cells (2 × 105) were transfected with TRIM32-RNAi plasmid (0.5 μg each) along with the indicated reporter plasmids (0.1 μg). 24 h after transfection, cells were left untreated or treated with poly(I:C) (20 μg/ml) in the medium for 12 h before luciferase assays were performed. Graphs show mean ± S.D., n = 3.
TRIM32 Interacts with MITA
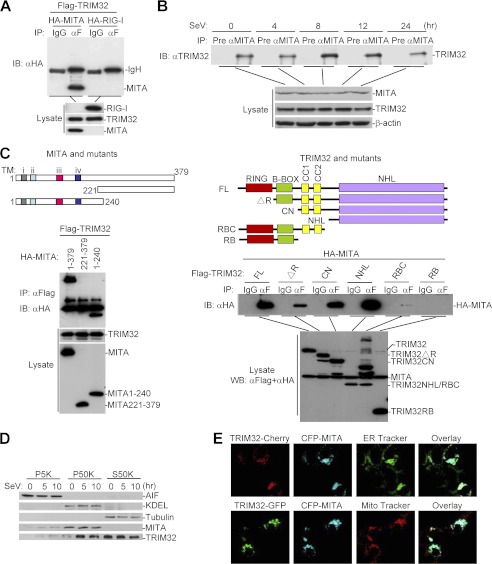
Since TRIM32 causes ubiquitination of MITA and is important for virus-induced IFN-β induction, we examined whether TRIM32 is physically associated with MITA. Coimmunoprecipitation experiments indicated that TRIM32 interacted with MITA in mammalian overexpression system (Fig. 3A). In untransfected cells, endogenous TRIM32 constitutively interacted with MITA and SeV infection enhanced this interaction at 8 h post-infection (Fig. 3B). Domain mapping experiments indicated that the N-terminal transmembrane domain-containing fragment of MITA and the C-terminal NHL domain of TRIM32 were important for their interaction (Fig. 3C). Subcellular fractionation experiments indicated that TRIM32 was located at mitochondria, ER and cytosol, and SeV infection caused translocation of a fraction of TRIM32 from cytosol to the ER and mitochondria (Fig. 3D). Confocal microscopy further confirmed that TRIM32 was colocalized with MITA at both the mitochondria and ER in 293 cells (Fig. 3E).
FIGURE 3.
TRIM32 interacts and colocalizes with MITA. A, TRIM32 interacts with MITA in mammalian overexpression system. 293 cells (1 × 107) were transfected with the indicated plasmids (5 μg each). Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. B, endogenous association between TRIM32 and MITA. The 293 cells (3 × 107) were left uninfected or infected with SeV for the indicated time points before coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. C, domain mapping of the interaction between TRIM32 and MITA. Upper panels: a schematic presentation of MITA, TRIM32 and their mutants. Lower panels: the 293 cells (1 × 107) were transfected with the indicated plasmids (5 μg each), followed by coimmunoprecipitation and immunoblot analysis with the indicated antibodies. D, subcellular distribution of TRIM32. The 293 cells (5 × 107) were left untreated or infected with SeV for the indicated times. The cellular fractions were analyzed by immunoblots with the indicated antibodies. E, TRIM32 and MITA colocalize to the mitochondria and ER in 293 cells. The 293 cells were transfected with the indicated plasmids. 20 h after transfection, cells were stained with Mito-Tracker Red or ER-Tracker Green for 30 min. The cells were fixed with 4% paraformaldehyde and subjected for confocal microscopy.
TRIM32 Targets MITA for K63-linked Ubiquitination at K20/150/224/236
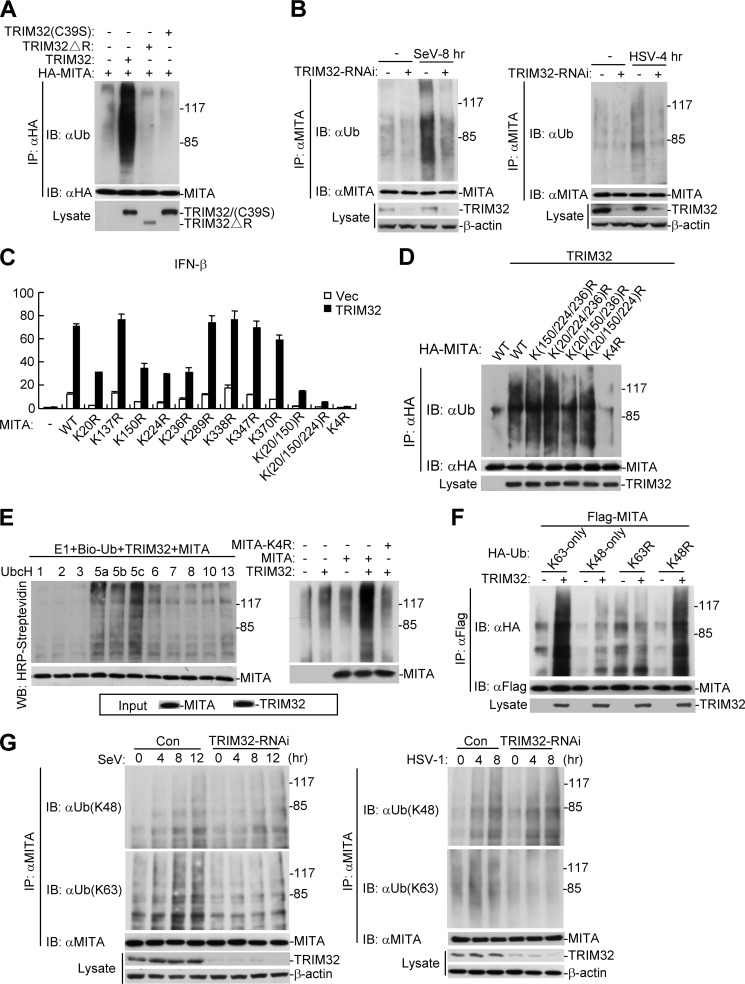
TRIM32 has been considered as an E3 ubiquitin ligase due to the presence of its RING finger domain. Therefore, we examined whether TRIM32 is an E3 ubiquitin ligase for MITA. As shown in Fig. 4A, wild-type TRIM32 but not TRIM32-ΔR or TRIM32(C39S), two enzymatic inactive mutants, could ubiquitinate MITA in transient transfection and coimmunoprecipitation experiments (Fig. 4A). Consistently, knockdown of TRIM32 attenuated SeV- and poly(dA:dT)-induced ubiquitination of overexpressed MITA (supplemental Fig. S3), or SeV- and HSV-1-induced ubiquitination of endogenous MITA (Fig. 4B). Taken together, these data suggest that TRIM32 plays an important role in ubiquitination of MITA. To identify residues of MITA that are targeted by TRIM32, we mutated individually all the lysine residues of MITA to arginine. As shown in Fig. 4C, mutation of K20, K150, K224, or K236 to arginine reduced the ability of MITA to activate the IFN-β promoter, whereas mutation of all the four lysines to arginines simultaneously abolished the ability of MITA to activate the IFN-β promoter and its ability to potentiate TRIM32-mediated activation the IFN-β promoter (Fig. 4C). Consistently, mutation of each of the four lysine residues of MITA did not markedly impair its ubiquitination by TRIM32, whereas simultaneous mutation of all the four lysine residues (K20, K150, K224, and K236) abolished its ubiquitination by TRIM32 (Fig. 4D). To test whether TRIM32 can directly ubiquitinate MITA, we performed an in vitro ubiquitination assay. The results indicated that TRIM32 could directly ubiquitinate MITA but not the MITA-K(20/150/224/236)R mutant (designated as MITA-K4R) (Fig. 4E), suggesting that the four lysine residues of MITA are targeted by TRIM32. Utilizing K63-only, K48-only, K63R, and K48R mutant ubiquitin, we found that MITA was modified by K63-linked ubiquitin chains (Fig. 4F). Furthermore, knockdown of TRIM32 inhibited both SeV- and HSV-1-induced K63-linked but not K48-linked ubiquitination of endogenous MITA (Fig. 4G). Taken together, these data suggest that TRIM32 mediates K63-linked ubiquitination of MITA under physiological condition or upon viral infection.
FIGURE 4.
TRIM32 targets MITA for K63-linked ubiquitination at K20/150/224/236. A, overexpression of wild-type but not the mutant TRIM32 promotes ubiquitination of MITA. The 293 cells (1 × 107) were transfected with MITA (5 μg) together with TRIM32 or its mutants (2 μg). 24 h after transfection, coimmunoprecipitation was performed with anti-HA. The immunoprecipitates were denatured and reimmunoprecitated with anti-HA and then analyzed by immunoblot with anti-ubiquitin (upper panel). The expression levels of the proteins were examined by immunoblots with the indicated antibodies (lower panels). B, knockdown of TRIM32 attenuated virus-induced ubiquitination of endogenous MITA. The 293 (left panel) or HeLa (right panel) cells were transduced with GFP-RNAi or TRIM32-RNAi by retrovirus-mediated gene transfer and selected with puromycin to establish stable cell lines. The cells (3 × 107) were then left uninfected or infected with SeV or HSV-1 respectively for the indicated times before coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. C, effects of MITA mutation on activation of the IFN-β promoter. The 293 cells (1 × 107) were transfected with TRIM32 and the indicated MITA mutants along with the IFN-β promoter reporter plasmid. Luciferase assays were performed 24 h after transfection. Graphs show mean ± S.D., n = 3. D, TRIM32 targets MITA for ubiquitination at K20/150/224/236. 293 cell (1 × 107) were transfected with TRIM32 (3 μg) and the indicated MITA mutant plasmids (6 μg). Ubiquitination experiments were carried out similarly as in A. E, TRIM32 ubiquitinates MITA but not MITA-K4R in vitro. TRIM32 and MITA were translated in vitro, and biotin-ubiquitin, E1 and the indicated E2s were added for ubiquitination assays. Ubiquitin-conjugated proteins were detected by immunoblot with HRP-streptavidin. The input levels of the translated proteins were detected by immunoblots. F, TRIM32 targetes MITA for K63-linked ubiquitination. The 293 cells (2 × 106) were transfected with MITA (2 μg) along with the indicated plasmids (0.2 μg). 24 h after transfection, immunoprecipitation, and immunoblot analysis were performed with the indicated antibodies. G, knockdown of TRIM32 inhibits virus-induced K63-linked but not K48-linked ubiquitination of endogenous MITA. Experiments were carried out similar to B except that different antibodies were used for the immunoblot.
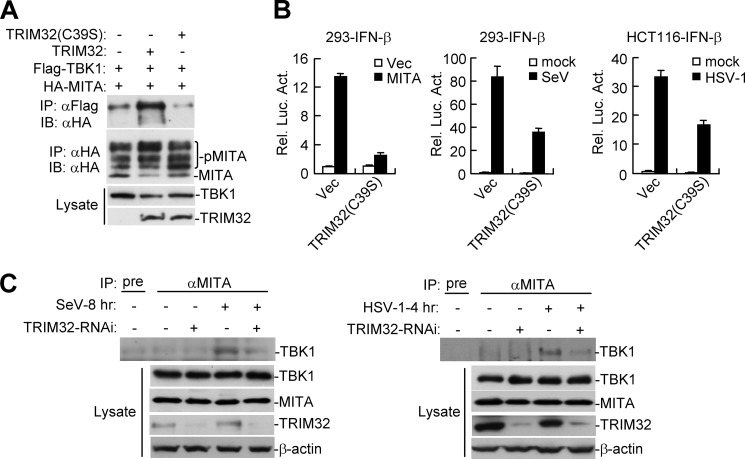
TRIM32 Is Required for the Interaction between MITA and TBK1
To further explore the mechanism on how TRIM32 exerts its effects on virus-triggered IFN induction pathways, we examined whether TRIM32 has any effects on interaction between MITA and TBK1. Transient transfection and coimmunoprecipitation experiments indicated that TRIM32, but not TRIM32(C39S), could markedly enhance the interaction between MITA and TBK1 (Fig. 5A). Consistent with a role of TRIM32 in K63-linked ubiquitination of MITA, the enzyme-inactive mutant TRIM32(C39S) markedly inhibited MITA-mediated, as well as SeV- or HSV-1-induced activation of the IFN-β promoter (Fig. 5B), confirming that the ubiquitination activity of TRIM32 is required for its role in virus-triggered IFN induction pathways. Moveover, knockdown of TRIM32 could reduce the endogenous interaction between MITA and TBK1 upon viral infection (Fig. 5C). Collectively, the data suggest that TRIM32-mediated ubiquitination of MITA is required for downstream TBK1 activation and IFN-β induction.
FIGURE 5.
TRIM32 is required for the interaction between MITA and TBK1. A, effects of TRIM32 and TRIM32(C39S) on interaction between MITA and TBK1. The 293 cells (1 × 107) were transfected with the indicated plasmids for 22 h, and then coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. B, effects of TRIM32(C39S) on MITA-, SeV-, and HSV-1-induced activation of the IFN-β promoter. The 293 or HCT116 cells were transfected with the indicated expression and reporter plasmids. The cells were left untreated, or infected with SeV (12 h) or HSV-1 (24 h) before reporter assays were performed. C, effects of knockdown of TRIM32 on the endogenous interaction between MITA and TBK1. The control or TRIM32 knockdown 293 (left panels) or HeLa (right panels) cells were left uninfected with infected with SeV or HSV-1 as indicated. Coimmunoprecipitation and immunoblots were performed with the indicated antibodies.
TRIM32 Is Required for Cellular Antiviral Response
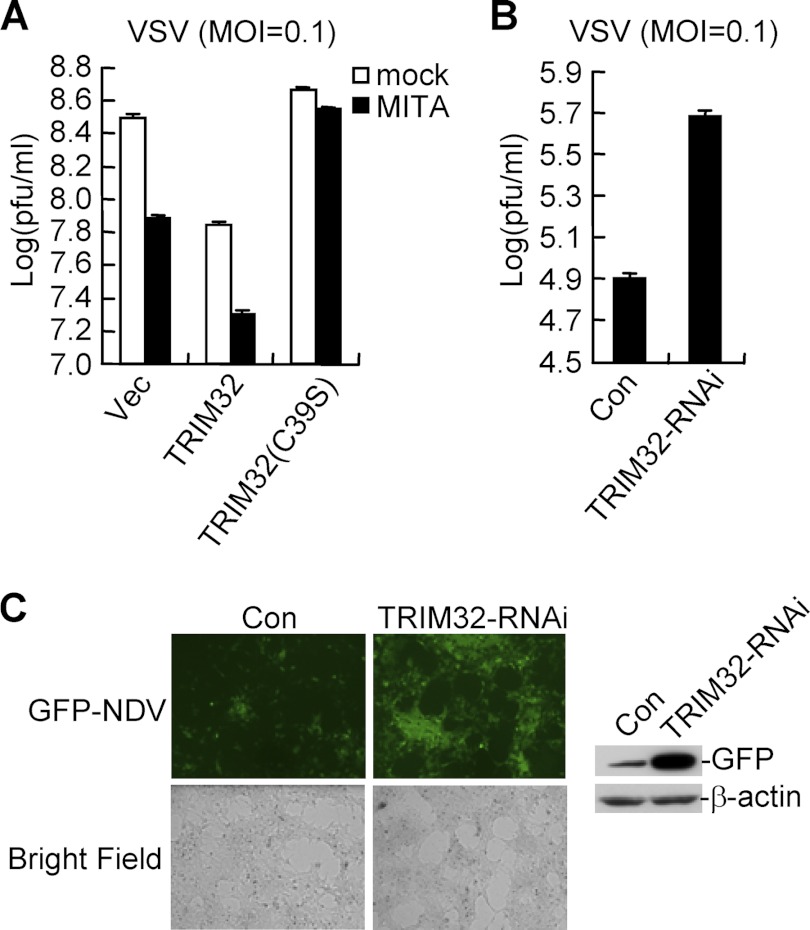
Since TRIM32 is involved in K63-linked ubiquitination of MITA and virus-triggered induction of IFN-β, we examined its roles in cellular antiviral response. In these experiments, we used Vesicular Stomatitis Virus (VSV), a negative-strand RNA virus suitable for plaque assays. Overexpression of TRIM32 but not TRIM32(C39S) could inhibit VSV replication by itself, and enhance MITA-mediated inhibition of VSV replication (Fig. 6A). Conversely, knockdown of TRIM32 potentiated VSV replication (Fig. 6B). Similarly, knockdown of TRIM32 also enhanced Newcastle Disease Virus (NDV) replication (Fig. 6C). These results indicated that TRIM32 is required for efficient cellular antiviral response.
FIGURE 6.
Roles of TRIM32 in cellular antiviral response. A, effects of TRIM32 and its mutant on VSV replication. The 293 cells (1 × 105) were transfected with the indicated expression plasmids (0.5 μg each). 24 h after transfection, cells were infected with VSV (MOI = 0.1). The supernatants were harvested 24 h after infection for standard plaque assays. Graphs show mean ± S.D., n = 3. B, effects of TRIM32-RNAi on VSV replication. Plaque assays were similarly performed as in A except that a TRIM32-RNAi plasmid (#1) (1 μg) was transfected and cells were infected with VSV (MOI = 0.1) for 16 h. Graphs show mean ± S.D., n = 3. C, effects of TRIM32-RNAi on NDV replication. The 293 cells were transfected with TRIM32-RNAi plasmid (1 μg each). 24 h later, cells were infected with NDV-GFP (MOI = 0.1) for 36 h and imaged by microscopy or analyzed by immunoblots with the indicated antibodies.
DISCUSSION
MITA is an important adapter in both viral RNA- and DNA-triggered induction of type I IFNs and innate antiviral immunity. In this study, we identified TRIM32 as a new E3 ubiquitin ligase that targets MITA. Overexpression of TRIM32 potentiated induction of IFN-β and other downstream genes triggered by MITA overexpression or infection with both RNA and DNA viruses, whereas knockdown of TRIM32 had opposite effects. These results reveal a critical role for TRIM32 in innate immune response against various types of viruses.
Previous studies have demonstrated that MITA is required for cytoplasmic poly(I:C)- and poly(dA:dT)-induced type I IFN signaling and on the other hand is dispensable for TLR3 signaling in human cells (16, 18). Consistently, we found that TRIM32 was indispensable for cytoplasmic poly(I:C)- and poly(dA:dT)-induced signaling but had no marked roles in TLR3-mediated signaling pathways. These observations further support our conclusion that TRIM32 is a positive regulator of innate antiviral response by targeting MITA.
TRIM32 has E3 ubiquitin ligase activity due to its N-terminal RING finger domain (27). In vitro and in vivo experiments indicated that wild-type TRIM32, but not a truncation mutant lack of RING finger domain or an enzymatic inactive point mutant, caused ubiquitination of MITA. Knockdown of TRIM32 inhibited basal or virus-induced K63-linked ubiquitination of MITA. Site-directed mutagenesis indicated that TRIM32 ubiquitinated MITA at lysines 20/150/224/236. Functionally, overexpression of wild-type TRIM32 potentiated MITA-mediated, and SeV- and HSV-1-triggered IFN-β induction, whereas an enzymatic inactive TRIM32 mutant acted as a dominant negative mutant to inhibit virus-triggered IFN-β induction. These findings suggest that K63-linked ubiquitination of MITA by TRIM32 plays a critical role in virus-triggered IFN-β induction pathways. Interestingly, previous studies have demonstrated that TRIM32 is an E3 ubiquitin ligase which targets proteins for degradation (28–32), our study for the first time demonstrated that TRIM32 could target a protein for K63-linked ubiquitination and promote its activation.
Previously, two studies have identified K150 of MITA as a residue modified by either K48- or K63-linked ubiquitination. It has been shown that the E3 ubiquitin ligase RNF5 causes K48-linked ubiquitination of MITA at K150 upon infection with RNA viruses, leading to proteasome-mediated degradation of MITA and negative regulation of virus-triggered induction of IFN-β. Another study demonstrates that the E3 ubiquitin ligase TRIM56 mediates K63-linked ubiquitination of MITA at K150, which potentiates IFN-β induction (33, 34). In the current study, we found that TRIM32 catalized K63-linked ubiquitination of MITA at residues 20, 150, 224, and 236. In addition, as shown in supplemental Fig. S3, experiments with K63-only or K48-only ubiquitin and MITA(K150R) mutant indicated that SeV-induced K63-linked ubiquitination occurred at 8 and 12 h post-infection, whereas SeV-induced K48-linked ubiquitination occurs at 8 but not 12 h post-infection. These results together suggest that ubiquitination and functions of MITA are differentially regulated by different E3 ligases, linkage-specific ubiquitins as well as viral infection duration. Nevertheless, our findings provide insights on the mechanisms of regulation of cellular antiviral response.
Supplementary Material
Acknowledgment
We thank Dr. Zhengfan Jiang for reagent.
This work was supported by Grants from the National Science Foundation of China (31130020, 30921001), and the Chinese Ministry of Science and Technology (2012CB910201, 2010DFA31100).

This article contains supplemental Figs. S1–S3.
- PRR
- pattern recognition receptors
- PAMP
- pathogen-associated molecular pattern
- TRIM32
- tripartite motif protein 32.
REFERENCES
- 1. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 2. Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 3. Durbin J. E., Fernandez-Sesma A., Lee C. K., Rao T. D., Frey A. B., Moran T. M., Vukmanovic S., García-Sastre A., Levy D. E. (2000) Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 164, 4220–4228 [DOI] [PubMed] [Google Scholar]
- 4. Levy D. E., García-Sastre A. (2001) The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12, 143–156 [DOI] [PubMed] [Google Scholar]
- 5. Levy D. E., Marié I. J. (2004) RIGging an antiviral defense–it's in the CARDs. Nat Immunol 5, 699–701 [DOI] [PubMed] [Google Scholar]
- 6. Baccala R., Gonzalez-Quintial R., Lawson B. R., Stern M. E., Kono D. H., Beutler B., Theofilopoulos A. N. (2009) Sensors of the innate immune system: their mode of action. Nat. Rev. Rheumatol. 5, 448–456 [DOI] [PubMed] [Google Scholar]
- 7. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 8. O'Neill L. A., Bowie A. G. (2010) Sensing and signaling in antiviral innate immunity. Curr. Biol. 20, R328–333 [DOI] [PubMed] [Google Scholar]
- 9. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) IKKϵ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 10. Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 11. Chiu Y. H., Macmillan J. B., Chen Z. J. (2009) RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rathinam V. A., Jiang Z., Waggoner S. N., Sharma S., Cole L. E., Waggoner L., Vanaja S. K., Monks B. G., Ganesan S., Latz E., Hornung V., Vogel S. N., Szomolanyi-Tsuda E., Fitzgerald K. A. (2010) The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., Sirois C. M., Jin T., Latz E., Xiao T. S., Fitzgerald K. A., Paludan S. R., Bowie A. G. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., Bover L., Plumas J., Chaperot L., Qin J., Liu Y. J. (2010) Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 107, 15181–15186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y. J. (2011) The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 12, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H. B. (2008) The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 17. Ishikawa H., Barber G. N. (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin L., Hill K. K., Filak H., Mogan J., Knowles H., Zhang B., Perraud A. L., Cambier J. C., Lenz L. L. (2011) MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J. Immunol. 187, 2595–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishikawa H., Ma Z., Barber G. N. (2009) STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen H., Sun H., You F., Sun W., Zhou X., Chen L., Yang J., Wang Y., Tang H., Guan Y., Xia W., Gu J., Ishikawa H., Gutman D., Barber G., Qin Z., Jiang Z. (2011) Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147, 436–446 [DOI] [PubMed] [Google Scholar]
- 22. Burdette D. L., Monroe K. M., Sotelo-Troha K., Iwig J. S., Eckert B., Hyodo M., Hayakawa Y., Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao A. P., Li S., Zhong B., Li Y., Yan J., Li Q., Teng C., Shu H. B. (2010) Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-β (IFN-β) and cellular antiviral response. J. Biol. Chem. 285, 9470–9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diao F., Li S., Tian Y., Zhang M., Xu L. G., Zhang Y., Wang R. P., Chen D., Zhai Z., Zhong B., Tien P., Shu H. B. (2007) Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc. Natl. Acad. Sci. U.S.A. 104, 11706–11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J., Liu T., Xu L. G., Chen D., Zhai Z., Shu H. B. (2005) SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 24, 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Honda K., Takaoka A., Taniguchi T. (2006) Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 [DOI] [PubMed] [Google Scholar]
- 27. Sato T., Okumura F., Kano S., Kondo T., Ariga T., Hatakeyama S. (2011) TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J. Cell Sci. 124, 3492–3502 [DOI] [PubMed] [Google Scholar]
- 28. Locke M., Tinsley C. L., Benson M. A., Blake D. J. (2009) TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum. Mol. Genet. 18, 2344–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kudryashova E., Kudryashov D., Kramerova I., Spencer M. J. (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 354, 413–424 [DOI] [PubMed] [Google Scholar]
- 30. Albor A., El-Hizawi S., Horn E. J., Laederich M., Frosk P., Wrogemann K., Kulesz-Martin M. (2006) The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J. Biol. Chem. 281, 25850–25866 [DOI] [PubMed] [Google Scholar]
- 31. Kano S., Miyajima N., Fukuda S., Hatakeyama S. (2008) Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 68, 5572–5580 [DOI] [PubMed] [Google Scholar]
- 32. Schwamborn J. C., Berezikov E., Knoblich J. A. (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., Wang Y. Y., Zhang X. L., Shu H. B. (2009) The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30, 397–407 [DOI] [PubMed] [Google Scholar]
- 34. Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S. (2010) The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33, 765–776 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.