Abstract
Background
Carriers of a germline mutation in a DNA mismatch repair (MMR) gene—that is, persons with Lynch syndrome—have substantially high risks of colorectal (CRC), endometrial, and several other cancers. The proportion of carriers who have de novo mutations (not inherited from either parent) is not known. This study reports a case series of de novo mutations in MMR genes and estimates the frequency of de novo mutation in MMR genes using the Colon Cancer Family Registry.
Methods
Screening for germline MLH1, MSH2, MSH6, and PMS2 mutations was performed for all incident CRC cases recruited from cancer registries (population based probands) displaying microsatellite instability (MSI) or loss of expression of MMR genes by immunohistochemistry (IHC) and probands with CRC in multi-case families recruited from clinics (clinic based probands), regardless of MSI or IHC status. All relatives of probands with a pathogenic mutation who donated a blood sample underwent testing for the mutation identified in the proband.
Results
Of 261 probands (202 clinic based, 59 population based) with MMR gene mutations for whom it was possible to determine the origin of the mutation, six (2.3%, 95% CI 0.9% to 5.0%) were confirmed as de novo, and the remaining 255 (97.7%, 95% CI 95.0% to 99.1%) were inherited. Of the de novo mutation carriers, three were clinic based probands (1.5%, 95% CI 0.3% to 4.5%) and three were population based probands (5.1%, 95% CI 1.2% to 14.5%). Two were in MLH1, three in MSH2, and one in MSH6.
Conclusion
De novo MMR gene mutations are uncommon causes of Lynch syndrome.
INTRODUCTION
Lynch syndrome, historically known as hereditary non-polyposis colorectal cancer syndrome (MIM 114500), describes a spectrum of malignancies caused by a germline mutation in one of four DNA mismatch repair (MMR) genes; MLH1, MSH2, MSH6 or PMS2.1 MMR genes produce proteins that detect and repair DNA mismatches occurring during cell replication or after damage to DNA.2 Carriers of a mutation in an MMR gene are at increased risk of colorectal, endometrial, and other cancers.3–5 Lynch syndrome accounts for 1–3% of all colorectal cancers (CRCs)6,7 and 10–15% of CRCs diagnosed before the age of 50 years.8 One estimate is that about 1 in 3100 people inherit a germline mutation in MLH1 or MSH2,9 and given that mutations in MSH6 and PMS2 account for about one sixth of all MMR mutations,10,11 then approximately 1 in 2700 people are MMR mutation carriers. However, the prevalence might be as high as 1 in 360.12
A de novo germline mutation is a constitutional mutation carried by a person in all their nucleated cells, and absent in both parents. It is thought to occur in a parental germ cell. Each gene has its own characteristic rate of de novo mutation.13–19 This has yet to be defined for the MMR genes. To our knowledge, there have been only four reports of single cases with de novo mutations in MMR genes: two cases for MLH1 (exon 18: c.2101C/T;→p.Gln701X20 and exon 8: c.666dupA; p.Ser225X21) and two cases for MSH2 (exon 2: c.344delA22 and exon 13: c.2006-2A/G;→p.Pro670fs→676X23). We now report the first case series of de novo mutations in MMR genes from the Colon Cancer Family Registry and have estimated the frequency of de novo mutations in MMR genes.
METHODS
The study sample was from the Colon Cancer Family Registry, an international resource for studies on the aetiology of CRC that was initiated in 1997 and is described in detail elsewhere24 and at http://epi.grants.cancer.gov/CFR/. Briefly, the Colon Cancer Family Registry recruited families through six administrative centres: the University of Melbourne consortium (Melbourne, Victoria, Australia), the Mayo Clinic (Rochester, Minnesota, USA), the Fred Hutchinson Cancer Research Center (Seattle, Washington State, USA), the University of Hawaii (Honolulu, Hawaii, USA), the University of Southern California Consortium (Los Angeles, California, USA), and Cancer Care Ontario (Toronto, Canada). Families were recruited via recently diagnosed CRC cases ascertained through population complete cancer registries (population based probands) or persons from multi-case families referred to family cancer clinics (clinic based probands) ascertained by the University of Melbourne consortium (through family cancer clinics in Adelaide, Perth, Sydney, Brisbane and Melbourne, Australia and Auckland, New Zealand), University of Southern California Consortium (through the Cleveland Clinic), and the Mayo Clinic. Written informed consent was obtained from all study participants, and the study protocol was approved at each Colon Cancer Family Registry centre.
Data collection
At recruitment, baseline information on demographics, personal characteristics, personal and family history of cancer, cancer screening history, history of polyps, polypectomy and other surgery was obtained from all participants. Participants were followed-up approximately 5 years and 10 years after baseline to update all the information. Reported cancer diagnoses and age at diagnoses were confirmed, where possible, using pathology reports, medical records, corroboration among relatives, cancer registry reports, and/or death certificates. Blood samples and tumour tissues were collected for genetic testing. The present study was based on all available baseline and follow-up data.
Definition
A pathogenic mutation was defined as a variant that was predicted to result in a stop codon, a frameshift mutation, a large insertion or deletion, or a missense mutation previously reported within the scientific literature to be pathogenic. An inherited mutation was defined as a mutation carried by a proband that was tested for and found in any relative (excluding probands' offspring). A de novo mutation was defined as a mutation carried by a proband that was tested for but not found in either parent.
Mutation screening and testing
Screening for germline mutations in MLH1, MSH2, MSH6, and PMS2 was performed for: all population based probands who had a colorectal tumour displaying evidence of impaired MMR function as indicated by either microsatellite instability (MSI), or by lack of MMR protein expression by immunohistochemistry (IHC); and the proband with CRC from each clinic based family regardless of MSI or MMR protein expression status. Mutation testing was performed by Sanger sequencing or denaturing high pressure liquid chromatography, followed by confirmatory DNA sequencing. Large insertion and deletion mutations were detected by multiplex ligation dependent probe amplification according to the manufacturer's instructions (MRC Holland, Amsterdam, The Netherlands).4,24,25 All participants who donated a blood sample, and who were relatives of probands with a pathogenic mutation, underwent mutation analysis for the same mutation identified in the proband.
Paternity and maternity testing
Non-paternity and non-maternity testing was performed at two centres, both employing the use of microsatellite markers; nine markers for one test (Australia) and 12 for the other (Mayo Clinic). Microsatellite markers were amplified by PCR (conditions available upon request). The PCR products were then pooled along with an internal molecular weight standard and analysed on Applied Biosystems Genetic Analyzer (ABI 3100 for Australia and ABI 3130xl for Mayo).
Statistical analysis
The proportion of MMR mutations that were de novo (frequency of de novo mutations in MMR genes) was estimated by dividing the number of de novo mutations by the sum of de novo and inherited mutations. Ninety-five per cent confidence intervals (95% CIs) for the proportion estimates were calculated using the method of Agresti and Coull.26,27 The estimated mean age of onset of CRC for de novo mutation carriers was compared with that for inherited mutation carriers using Student t test.
RESULTS
Of a total of 471 probands with MMR gene mutations, 261 mutations could be classified as either de novo or inherited. For 210 probands, the origin of the mutation could not be determined because of non-availability of blood from either parents or any other relatives. Of 261 probands with MMR mutations, six (2.3%, 95% CI 0.9% to 5.0%) could be classified as de novo mutations and 255 (97.7%, 95% CI 95.0% to 99.1%) as inherited (table 1). Of the de novo mutation carriers, three were from 202 clinic based probands (1.5%, 95% CI 0.3% to 4.5%) and three were from 59 population based probands (5.1%, 95% CI 1.2% to 14.5%). Two de novo mutations were detected in the 102 MLH1 mutation carriers (2.0%, 95% CI 0.1% to 7.3%), three in the 132 MSH2 mutation carriers (2.3%, 95% CI 0.5% to 6.8%), one in the 22 MSH6 mutation carriers (4.5%, 95% CI 0.1% to 23.5%), and none in the five PMS2 mutation carriers (one-sided 95% CI 0.0% to 45.1%).
Table 1.
Summary of mismatch repair gene mutation carriers from the Colon Cancer Family Registry
| Number of probands |
|||
|---|---|---|---|
| Carrier | Combined (n = 471) | Population based (n = 153) | Clinic based (n = 318) |
| Inherited | 255 | 56 | 199 |
| Mother | 48 | 15 | 33 |
| Father | 33 | 11 | 22 |
| Other relative | 174 | 30 | 144 |
| Undetermined | 210 | 94 | 116 |
| De novo | 6 | 3 | 3 |
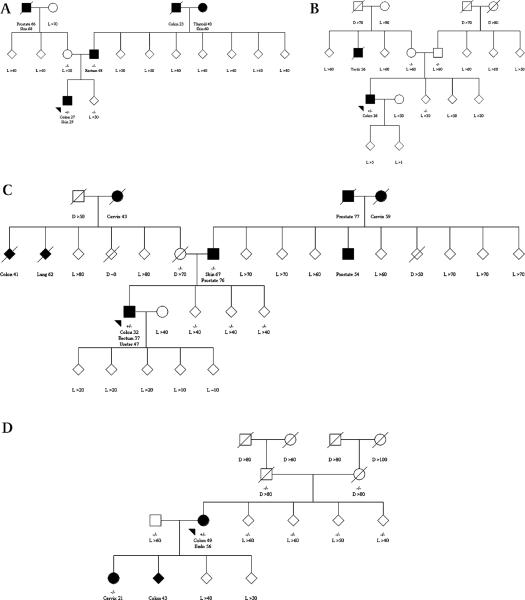
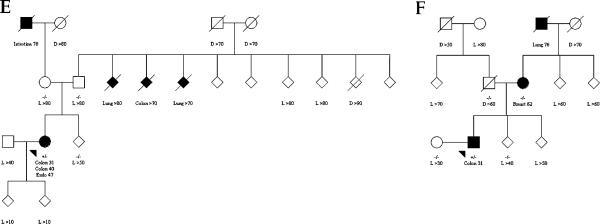
Three de novo mutation carriers were recruited from family cancer clinics in Perth and Brisbane and via the Victorian Cancer Registry, Australia, and three were recruited from the Mayo Clinic and via the Minnesota Cancer Surveillance System, USA (table 2). Two (probands B and F) had no family history of CRC or other Lynch syndrome associated cancer, and one (proband D) had no family history of any cancer in the parents, grandparents, and siblings (figure 1).
Table 2.
Mutation,cancer diagnosis and ascertainment of the probands with de novo mutations in mismatch repair genes
| Proband | Gene | Exon | Mutation description | MMR protein loss | MSI | Age at CRC (year) | TNM stage | Source | Family history of CRC (age) |
|---|---|---|---|---|---|---|---|---|---|
| A | MLH1 | 1–19 | Deletion | MLH1, PMS2 | – | 27 | T3N1M0 | Population | Father (48), grandfather (23) |
| B | MLH1 | 13 | c.1459C→T p.Arg487X | MLH1, PMS2, MSH6 | – | 36 | T3N0M0 | Clinic | Nil |
| C | MSH2 | 8 | Deletion | MSH2, MSH6 | High | 32 | T3N0M0 | Population | Aunt/Uncle (41) |
| D | MSH2 | 5 | c.942+3→T p.Val265_Gln314del | MSH2, MSH6 | – | 49 | T3N1M9 | Clinic | Son (43) |
| E | MSH2 | 6 | c.1034G→A p.Trp345X | MSH2, MSH6 | – | 31 | T3N0M9 | Clinic | Aunt/Uuncle (70), Grandfather (78) |
| F | MSH6 | 4 | c.1422_1423delGCinsAT p.Gln475X | MSH6 | High | 31 | T3N0M9 | Population | Nil |
CRC, colorectal cancer; MMR, mismatch repair; MSI, microsatellite instability.
Figure 1.
(A–F) Status of mismatch repair gene mutation and cancer diagnosis for family members of de novo germline mismatch repair gene mutation carriers. Square for male, circle for female, diamond for unidentified sex, shaded for cancer-affected (cancer diagnoses are named with corresponding age), unshaded for cancer-unaffected, arrow for proband, +/− for carrier, −/− for non-carrier, D for age of death in years, L for last known age in years. To increase confidentiality, the sex of all relatives is provided as unidentified, except for those who were parents or when diagnosed with sex specific cancers.
The mean age at CRC diagnosis for de novo mutation carriers (34.3 years, SD 7.7 years) was 8.5 (95% CI 0.3 to 16.7) years younger than that for inherited mutation carriers (42.8 years, SD 10.1 years) (p=0.04).
Genotyping of the probands and parents demonstrated allele segregation across all microsatellite markers tested (nine for the Australian families and 12 for the Mayo families) over multiple chromosomal regions. These data provide convincing evidence that each of the six probands with de novo mutations were biologically related to their parents.
DISCUSSION
We have reported six cases of de novo germline mutation in MMR genes (two MLH1, three MSH2, and one MSH6) from the Colon Cancer Family Registry. This is the first report, to our knowledge, on a case series of de novo germline mutation in MMR genes.
All specific de novo mutations except one that we observed in this study were different from the previously reported de novo mutations in MLH1 or MSH2.20–23 The MSH2 mutation in exon 5: c.942+3A→T p. Val265_Gln314del that we observed in proband 4 was the same mutation reported by Desai et al28 in which the 942+3 mutation was recognised as being relatively common in Lynch syndrome series around the world. They estimated that this type of de novo mutation accounted for 11% of all known pathogenic MSH2 mutations. Because haplotype sharing across families was not found, the authors concluded that this mutation occurred de novo with a relatively high frequency. They hypothesised that it may arise as a consequence of misalignment of DNA strands at replication. Indeed, this mutation was found to be one of the six de novo mutation carriers in our series.
If a disease-causing mutation found in the proband cannot be detected in the DNA of either parent, three possible explanations need to be considered: non-paternity/non-maternity, germline mosaicism in a parent, or that the mutation is de novo. Non-paternity/non-maternity could be confidently excluded as a possible explanation in each of the six de novo mutation probands. However, we did not have the opportunity to test multiple tissues in our patients and their parents for the mutation and, therefore, could not rule out the possibility that these mutations were early postzygotic mutations or due to parental germline mosaicism.
We have estimated the proportion of MMR gene mutation carriers that have de novo mutations to be between 1 in 100 (0.9%) and 1 in 20 (5.0%). This is comparable to previous small studies which estimated proportions of 1% (1/114)23 or 2% (1/47).21 De novo germline mutations in MMR genes, therefore, appear to be rare in contrast to other hereditary CRC syndromes: 11–25% in the adenomatous polyposis coli (APC) gene mutations that cause familial adenomatous polyposis (MIM 175100),13–16 25–50% in the LKB1/STK11 gene mutations that cause Peutz-Jeghers syndrome (MIM 175200),17,18 and ~25% in BMPR1A and SMAD4 mutations that cause juvenile polyposis syndrome (MIM 174900).18,19
We observed that the average age of onset for CRC in the de novo carriers was younger than for the non-de novo carriers. Overreliance on family history of cancer—for example, the Amsterdam criteria29—to guide MMR gene mutation testing will contribute to the under-diagnosis of Lynch syndrome resulting from de novo mutations. None of the six de novo mutation carriers had a family history that met the Amsterdam criteria. A proposed alternative approach25 uses a tumour based strategy that has been demonstrated to identify persons with Lynch syndrome more efficiently. All the CRC tumours from the six probands with de novo mutations lacked the MMR protein corresponding to the mutated gene. Recognition of the presence of de novo mutations in three different MMR genes and a relatively high frequency of de novo mutations in population based probands supports the application of tumour testing for loss of MMR function to identify people without a family history, but at high risk of having a pathogenic MMR gene mutation.
Acknowledgements
The authors thank all study participants of the Colon Cancer Family Registry and staff for their contributions to this project.
Funding This work was supported by the National Cancer Institute, National Institutes of Health under RFA #CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry and Principal Investigators of Australasian Colorectal Cancer Family Registry (U01 CA097735) and Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the CFRs, nor does mention of trade names, commercial products, or organisations imply endorsement by the US Government or the CFR. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
Footnotes
Competing interests None.
Ethics approval Written informed consent was obtained from all study participants, and the study protocol was approved at each Colon Cancer Family Registry center.
Contributors AKW—study concept and design; acquisition of data; statistical analysis and interpretation of data; drafting and final approval of the manuscript. MAJ—statistical analysis and interpretation of data; drafting and final approval of the manuscript. DDB—genetic testing and interpretation of data; drafting and final approval of the manuscript. MC—genetic testing and interpretation of data; drafting and final approval of the manuscript. JPY—genetic testing and interpretation of data; drafting and final approval of the manuscript. GGG—case ascertainment; analysis and interpretation of data; drafting and final approval of the manuscript. JG—case ascertainment; analysis and interpretation of data; drafting and final approval of the manuscript. BAL—case ascertainment; analysis and interpretation of data; drafting and final approval of the manuscript. JLH—statistical analysis and interpretation of data; drafting and final approval of the manuscript. SNT—genetic testing and interpretation of data; drafting and final approval of the manuscript. NML—study concept and design; case ascertainment; analysis and interpretation of data; drafting and final approval of the manuscript.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Vasen HF. Clinical diagnosis and management of hereditary colorectal cancer syndromes. J Clin Oncol. 2000;18:81S–92S. [PubMed] [Google Scholar]
- 2.Muller A, Fishel R. Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC) Cancer Invest. 2002;20:102–9. doi: 10.1081/cnv-120000371. [DOI] [PubMed] [Google Scholar]
- 3.Baglietto L, Lindor NM, Dowty JG, White DM, Wagner A, Gomez Garcia EB, Vriends AH, Cartwright NR, Barnetson RA, Farrington SM, Tenesa A, Hampel H, Buchanan D, Arnold S, Young J, Walsh MD, Jass J, Macrae F, Antill Y, Winship IM, Giles GG, Goldblatt J, Parry S, Suthers G, Leggett B, Butz M, Aronson M, Poynter JN, Baron JA, Le Marchand L, Haile R, Gallinger S, Hopper JL, Potter J, de la Chapelle A, Vasen HF, Dunlop MG, Thibodeau SN, Jenkins MA, Dutch Lynch Syndrome Study Group Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, Lindblom A, Lagerstedt K, Thibodeau SN, Lindor NM, Young J, Winship I, Dowty JG, White DM, Hopper JL, Baglietto L, Jenkins MA, de la Chapelle A. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–28. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, Watson P, Gruber SB, Euhus D, Kinzler KW. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aaltonen LA, Sankila R, Mecklin JP, Jarvinen H, Pukkala E, Peltomaki P, de la Chapelle A. A novel approach to estimate the proportion of hereditary non-polyposis colorectal cancer of total colorectal cancer burden. Cancer Detect Prev. 1994;18:57–63. [PubMed] [Google Scholar]
- 7.Burt RW, DiSario JA, Cannon-Albright L. Genetics of colon cancer: impact of inheritance on colon cancer risk. Annu Rev Med. 1995;46:371–9. doi: 10.1146/annurev.med.46.1.371. [DOI] [PubMed] [Google Scholar]
- 8.Hopper JL. Application of genetics to the prevention of colorectal cancer. Recent Results Cancer Res. 2005;166:17–33. doi: 10.1007/3-540-26980-0_2. [DOI] [PubMed] [Google Scholar]
- 9.Dunlop MG, Farrington SM, Nicholl I, Aaltonen L, Petersen G, Porteous M, Carothers A. Population carrier frequency of hMSH2 and hMLH1 mutations. Br J Cancer. 2000;83:1643–5. doi: 10.1054/bjoc.2000.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampel H. Genetic testing for hereditary colorectal cancer. Surg Oncol Clin N Am. 2009;18:687–703. doi: 10.1016/j.soc.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woods MO, Younghusband HB, Parfrey PS, Gallinger S, McLaughlin J, Dicks E, Stuckless S, Pollett A, Bapat B, Mrkonjic M, de la Chapelle A, Clendenning M, Thibodeau SN, Simms M, Dohey A, Williams P, Robb D, Searle C, Green JS, Green RC. The genetic basis of colorectal cancer in a population-based incident cohort with a high rate of familial disease. Gut. 2010;59:1369–77. doi: 10.1136/gut.2010.208462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampel H, de la Chapelle A. The search for unaffected individuals with Lynch syndrome: do the ends justify the means? Cancer Prev Res (Phila) 2011;4:1–5. doi: 10.1158/1940-6207.CAPR-10-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ripa R, Bisgaard ML, Bulow S, Nielsen FC. De novo mutations in familial adenomatous polyposis (FAP) Eur J Hum Genet. 2002;10:631–7. doi: 10.1038/sj.ejhg.5200853. [DOI] [PubMed] [Google Scholar]
- 14.Rustin RB, Jagelman DG, McGannon E, Fazio VW, Lavery IC, Weakley FL. Spontaneous mutation in familial adenomatous polyposis. Dis Colon Rectum. 1990;33:52–5. doi: 10.1007/BF02053203. [DOI] [PubMed] [Google Scholar]
- 15.Aretz S, Uhlhaas S, Caspari R, Mangold E, Pagenstecher C, Propping P, Friedl W. Frequency and parental origin of de novo APC mutations in familial adenomatous polyposis. Eur J Hum Genet. 2004;12:52–8. doi: 10.1038/sj.ejhg.5201088. [DOI] [PubMed] [Google Scholar]
- 16.Bisgaard ML, Fenger K, Bulow S, Niebuhr E, Mohr J. Familial adenomatous polyposis (FAP): frequency, penetrance, and mutation rate. Hum Mutat. 1994;3:121–5. doi: 10.1002/humu.1380030206. [DOI] [PubMed] [Google Scholar]
- 17.Westerman AM, Entius MM, Boor PP, Koole R, de Baar E, Offerhaus GJ, Lubinski J, Lindhout D, Halley DJ, de Rooij FW, Wilson JH. Novel mutations in the LKB1/STK11 gene in Dutch Peutz-Jeghers families. Hum Mutat. 1999;13:476–81. doi: 10.1002/(SICI)1098-1004(1999)13:6<476::AID-HUMU7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Schreibman IR, Baker M, Amos C, McGarrity TJ. The hamartomatous polyposis syndromes: a clinical and molecular review. Am J Gastroenterol. 2005;100:476–90. doi: 10.1111/j.1572-0241.2005.40237.x. [DOI] [PubMed] [Google Scholar]
- 19.Haidle JL, Howe JR. Juvenile polyposis syndrome. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. Gene Reviews. University of Washington, Seattle; Seattle, WA: 1993. [PubMed] [Google Scholar]
- 20.Stulp RP, Vos YJ, Mol B, Karrenbeld A, de Raad M, van der Mijle HJ, Sijmons RH. First report of a de novo germline mutation in the MLH1 gene. World J Gastroenterol. 2006;12:809–11. doi: 10.3748/wjg.v12.i5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plasilova M, Zhang J, Okhowat R, Marra G, Mettler M, Mueller H, Heinimann K. A de novo MLH1 germ line mutation in a 31-year-old colorectal cancer patient. Genes Chromosomes Cancer. 2006;45:1106–10. doi: 10.1002/gcc.20374. [DOI] [PubMed] [Google Scholar]
- 22.Kraus C, Kastl S, Günther K, Klessinger S, Hohenberger W, Ballhausen WG. A proven de novo germline mutation in HNPCC. J Med Genet. 1999;36:919–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Morak M, Laner A, Scholz M, Madorf T, Holinski-Feder E. Report on de-novo mutation in the MSH2 gene as a rare event in hereditary non-polyposis colorectal cancer. Eur J Gastroenterol Hepatol. 2008;20:1101–5. doi: 10.1097/MEG.0b013e328305e185. [DOI] [PubMed] [Google Scholar]
- 24.Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, Hall D, Hopper JL, Jass J, Le Marchand L, Limburg P, Lindor N, Potter JD, Templeton AS, Thibodeau S, Seminara D, the Colon Cancer Family R Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–43. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 25.Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, Tesoriero AA, Smith LD, Jennings K, Grubb G, Royce SG, Walsh MD, Barker MA, Young JP, Jass JR, St John DJ, Macrae FA, Giles GG, Hopper JL. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J Clin Oncol. 2005;23:6524–32. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- 26.Agresti A, Coull B. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52:119–26. [Google Scholar]
- 27.Brown L, Cai T, DasGupta A. Interval estimation for a binomial proportion. Stat Sci. 2001;16:101–33. [Google Scholar]
- 28.Desai DC, Lockman JC, Chadwick RB, Gao X, Percesepe A, Evans DG, Miyaki M, Yuen ST, Radice P, Maher ER, Wright FA, de La Chapelle A. Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet. 2000;37:646–52. doi: 10.1136/jmg.37.9.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–6. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]