Abstract
Recent studies have examined the influence on patterns of human genetic variation of a variety of cultural practices. In India, centuries-old marriage customs have introduced extensive social structuring into the contemporary population, potentially with significant consequences for genetic variation. Social stratification in India is evident as social classes that are defined by endogamous groups known as castes. Within a caste, there exist endogamous groups known as gols (marriage circles), each of which comprises a small number of exogamous gotra (lineages). Thus, while consanguinity is strictly avoided and some randomness in mate selection occurs within the gol, gene flow is limited with populations outside the gol. Gujarati Patels practice this form of “exogamic endogamy.” We have analyzed genetic variation in one such group of Gujarati Patels, the Chha Gaam Patels (CGP), who comprise individuals from six villages. Population structure analysis of 1,200 autosomal loci offers support for the existence of distinctive multilocus genotypes in the CGP with respect to both non-Gujaratis and other Gujaratis, and indicates that CGP individuals are genetically very similar. Analysis of Y-chromosomal and mitochondrial haplotypes provides support for both patrilocal and patrilineal practices within the gol, and a low-level of female gene flow into the gol. Our study illustrates how the practice of gol endogamy has introduced fine-scale genetic structure into the population of India, and contributes more generally to an understanding of the way in which marriage practices affect patterns of genetic variation.
Keywords: Endogamy, Gene Flow, HVS1, India, Y-chromosomal
Among the factors shaping present-day patterns of human genetic variation, culturally-driven marital practices provide a key instance of an interaction between social and genetic processes. For example, culturally-learned mating preferences have been implicated in the evolution of hair, eye, and skin coloration phenotypes during human evolution (Aoki, 2002; Frost, 2006; Laland et al., 2010). Additionally, the consequences of patrilocal and matrilocal practices on local gene flow have been shown to influence neutral patterns of genetic variation in various populations (Cordaux et al., 2004; Kayser et al., 2006; Nasidze et al., 2006; Oota et al., 2001). One region of particular interest has been India, where many recent studies have considered population structure in terms of cultural, geographic, and linguistic groupings that affect marital practices (Bamshad et al., 2001; Basu et al., 2003; Cordaux et al., 2004; Indian Genome Variation Consortium, 2008; Kashyap et al., 2006; Kivisild et al., 2003; Reich et al., 2009; Rosenberg et al., 2006; Roychoudhury et al., 2001; Sahoo et al., 2006; Sengupta et al., 2006; Thangaraj et al., 2008; Thanseem et al., 2006; Tripathi et al., 2008; Watkins et al., 2008).
The contemporary population of India is estimated to consist of at least 1,028 million people (Census of India, 2001) and is culturally stratified as tribals, who constitute 8.2% of the total population, and non-tribals. Over 87% of the non-tribal population belongs to the Hindu religion (Census of India, 2001), within which strict traditions have introduced a complex arrangement of hereditary groups and extensive social structure within the population. It has been estimated that across India, these traditions have created 50,000 to 60,000 essentially endogamous groups (Gadgil et al., 1998). These groups can be understood in a framework with three different levels (Fig. 1), or orders, of population structure that are similar across diverse regions of India and that are commonly known as the caste system (Shah, 1982; Shah, 1998).
Figure 1.
An overview of caste divisions in the Hindu population of India. The hierarchy among the top three Leva Kanbi Patidar gols was taken from Thakkar (Thakkar, 1999). We do not show the fourth and last order of caste divisions called tads (Shah, 1982), endogamous groups composed of a few families who are all members of the same gol. To the best of our knowledge, these “fourth-order” divisions do not exist among the Leva Kanbi Patidar.
Divisions of the first-order separate the population into broad hereditary groups, often termed jātis or castes (Shah, 1982; Shah, 1998), which are loosely associated with a traditional job function. These castes are distinguished from each other by traditions that prohibit the sharing of food and marital (bride) transactions, and they therefore constitute distinct endogamous groups (Shah, 1982; Shah, 1998). It has been estimated that India contains ~3,000 of these major castes (Bittles, 2002), examples of which include the Rajput (rulers and warriors), Brahmin (teachers, scholars, and priests), Vania (also termed Bania; traders), and Kanbi (also termed Kunbi; agriculturalists) (Shah, 1982; Shah, 1998). There also exist an estimated ~1,000 scheduled castes (formerly known as Dalits or untouchables) (Gadgil et al., 1998), which constitute ~17.6% of the non-tribal population of India (Census of India, 2001). In traditional Indian society, the castes themselves each exist among one of five Varnas or broad social classes (Brahmin, Kshatriya, Vaishya, Shudra, and Panchama) (Flood, 1996). However, it has been noted that in modern India, it is impossible to place all castes into the five-tier Varna system (Shah, 1998), and therefore, we do not consider Varna as a caste division here.
Divisions of the second-order separate each caste into what are sometimes referred to as sub-castes, endogamous groups that are distinguished by traditions that prohibit marital (bride) transactions, but that permit the sharing of food. The number of divisions varies between the different castes (Shah, 1982; Shah, 1998); in the state of Gujarat, while 80 such second-order divisions exist among the Brahmins, only five exist among the Kanbi (Fig. 1) (Shah, 1982).
Divisions of the third-order separate many second-order divisions into groups known as ekdās (units) or gols (marriage circles) (Shah, 1982; Shah, 1998). In Gujarat, each gol is composed of a number of gotra (lineages or clans) living in certain gaam (villages) that are generally geographically close to each other. Among Gujarati Patels, a village is the equivalent of a gotra. Each individual within a gol identifies as belonging to his or her father’s village (patrilocality), and property and surnames are inherited through the paternal line (patrilineality). Gols are in essence endogamous groups that are distinguished from one another by strict traditions restricting marital (bride) transactions, so that most marriages occur between individuals from the same gol (Pocock 1972). Within a gol, there is a strict practice of exogamy, whereby an individual cannot marry anyone from his or her village, as that would be considered marrying kin (Pocock, 1972). Thus, while consanguinity is avoided and some randomness in mate selection occurs through the practice of village exogamy within a gol, there is a degree of gene flow restriction among gols owing to the practice of endogamy within the gol.
These traditions have partitioned a vast portion of the population of India into tens of thousands of endogamous gols. However, they have also allowed for a small amount of gene flow between gols within the same second-order division through the practice of hypergamy (Ghurye, 1969; Nath, 1973), in which a bride marries a groom from a family whose gol has a higher status. There exist hierarchies both within each gol, where families and gotra vary in their status (Patel and Rutten, 1999), as well as between gols, which also vary in standing. Marriages occur much less frequently between individuals from different gols than between individuals within the same gol, primarily because of the financial burden placed upon the bride’s family by the higher dowry that would need to be paid for such a marriage compared to a within-gol marriage, and by the fine imposed by the bride’s gol for marrying outside it (Nath, 1973; Patel and Rutten, 1999; Pocock, 1957). The increase in status that would be gained by the bride’s family by such a marriage (Pocock, 1957), and the financial gain that it offers to the groom’s family through the higher dowry that may offset those dowries paid when arranging the marriages of their own daughters, make desirable such marriages between brides from families of high status in a gol of lower standing and grooms from families of low status in a gol of higher standing. Among the motivating forces for the creation of ekdās or gols was the prevention of hypergamy (Clark, 1983). This was particularly so for the gols of higher standing (Das, 1973), who were concerned that if some family among them gave in to the temptation of profitable alliance with a lower-status family, the gates of entry into a higher societal status would open for the lower sections, potentially leaving a shortage of grooms for their own daughters.
In this study, we investigate the effect of restricted marital practices in India on patterns of genetic variation. We have studied the Patels of the Chha Gaam Patels (Circle of Six Villages; “CGP” henceforth) (Patel and Rutten, 1999), a division of the third-order located in the Charotar region of the western Indian state of Gujarat. All Patels, or Patidars (Patel and Rutten, 1999), were until 1931 known as the Leva Kanbi and Kadva Kanbi (Pocock, 1972; Pocock, 1993), the former having higher standing (Nath, 1973). Historically, the Leva Kanbi and Kadva Kanbi were endogamous groups (Nath, 1973; Pocock, 1972) located primarily in Gujarat, most notably within the Charotar region (Nath, 1973), although also in the states of Rajasthan, Maharashtra, Uttar Pradesh and Madhya Pradesh.
Originally, within the Patidar, there were no classes, gols, or restrictions on marriages. Then, approximately 500 years ago, inequality in the movement of brides arose among the Patidar, primarily driven by the economic differences between Patidar living in cities such as Ahmedabad, Gujarat, and Patidar who remained in the villages. In 1869 CE, Nadiad, Sojitra, Vaso and fifteen other villages created their own gol. Over subsequent years, some fifty gols were formed among the Patidar, with the inclusion and exclusion of villages from the original gol of 18 villages leading to the creation of the modern CGP. This gol comprises the villages of Bhadran, Dharmaj, Karamsad, Nadiad, Sojitra, and Vaso (Fig. 2), all of which are populated by Patels who were formerly Leva Kanbi (Fig. 1). The six villages of the CGP are large in size, relatively wealthy, and urbanized, and are therefore considered to be the highest-status villages among the Patels (Fig. 1) (Patel and Rutten, 1999; Pocock, 1993; Thakkar, 1999; Vyas et al., 1958). Thus, under the rules of hypergamy, while males of the CGP can marry a female from any village, a female of the CGP can only marry a male from the CGP (Patel and Rutten, 1999).
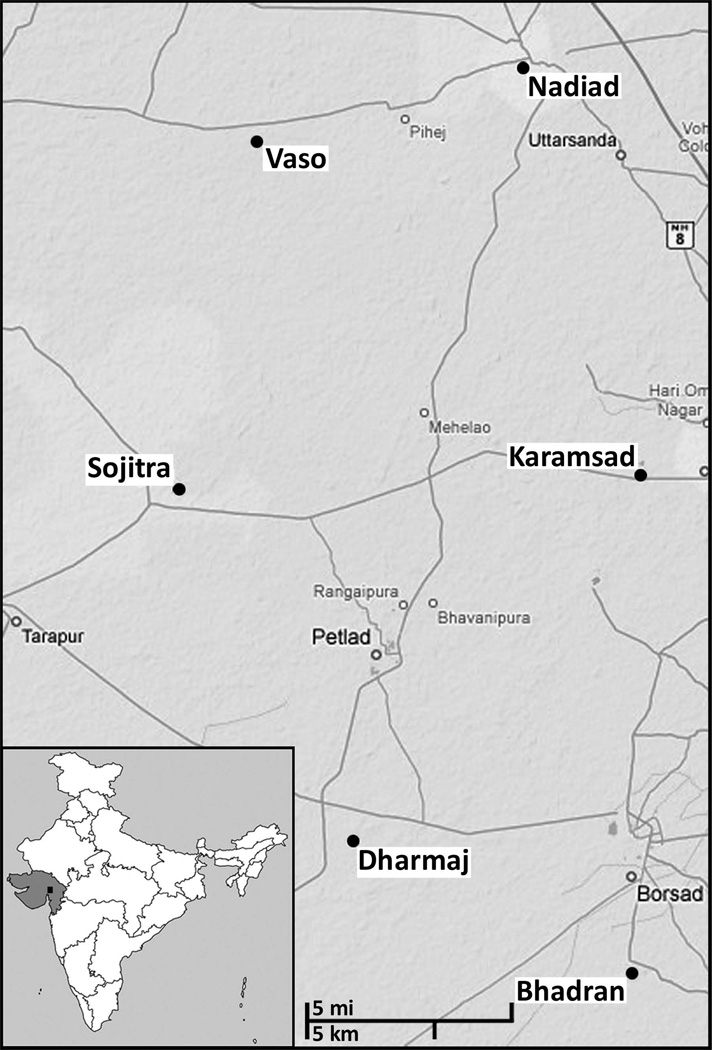
Figure 2.
Map of the six CGP villages. The map was obtained from Google Maps (http://maps.google.com/), retrieved on February 24, 2009. Inset, a map of India with the state of Gujarat shaded in grey. A black rectangle indicates the location of the six CGP villages.
We have analyzed 1,200 autosomal loci together with 26 Y-chromosomal markers and the HVS1 region of the mitochondrial genome in 194 individuals of the CGP. We analyze these data in conjunction with similar data on 31 Gujaratis from outside the CGP (“other Gujaratis” henceforth) and autosomal data from 382 individuals from 14 other population subgroups defined by language from across India (Rosenberg et al., 2006), to determine how the restricted marital practices of the caste system have influenced patterns of genetic variation in this distinctive exogamous-endogamic group. We show that while the practice of gotra exogamy has made the descendants from the six villages genetically very similar, the practice of gol endogamy has made these descendants genetically distinguishable both from non-Gujaratis and from other Gujaratis. We further show that while the practice of patrilocality has made Y-chromosome haplotypes highly specific to particular villages, mitochondrial haplotypes are by contrast highly variable and show no village-specific patterns. This high variability in mitochondrial haplotypes can be explained in part by the migration of females from outside the CGP into the six villages through the practice of gol hypergamy. These results shed light on the genetic history of the CGP, and provide further evidence that the restricted marital practices of the caste system have introduced fine-scale genetic structure into the Hindu population of India.
MATERIALS AND METHODS
Samples
We used a previously described Asian Indian sample (Rosenberg et al., 2006), which consists of India-born individuals sampled in the United States whose four grandparents each spoke the same language and originated from the same state in India. Of the 279 Gujarati individuals previously reported by Rosenberg et al. (Rosenberg et al., 2006), we restricted our analysis to a set of 249 individuals for whom the village of origin was known. Of these 249 Gujarati individuals, 218 emigrated from one of the six CGP villages (Fig. 2). The 31 Gujarati individuals who were not from one of the CGP villages emigrated from one of 25 villages, with not more than 3 individuals emigrating from any given village; of these 25 villages, over two-thirds are located within a ~20-mile radius of the CGP villages.
For the population-structure analysis, 382 unrelated individuals from 14 other groups defined by language from across India were included: Assamese (25), Bengali (27), Hindi (28), Kannada (24), Kashmiri (25), Konkani (42), Malayalam (25), Marathi (26), Marwari (25), Oriya (26), Parsi (25), Punjabi (28), Tamil (29), and Telugu (27). These individuals are the same individuals previously considered by Rosenberg et al. (Rosenberg et al., 2006).
To search systematically for relative pairs in the data set, we followed the methods of Rosenberg (Rosenberg, 2006). Among the 249 Gujarati individuals, on the basis of pairwise allele-sharing and the program RELPAIR (v.2.0.1) (Boehnke and Cox, 1997; Epstein et al., 2000) 71 pairs of individuals were inferred to be related at a level closer than first cousins: 12 parent/offspring pairs, 20 full sibling pairs, 2 half sibling pairs, 34 avuncular pairs, 2 grandparent-grandchild pairs, and 1 monozygotic pair. While it is possible that the monozygotic pair does indeed represent twins, it is perhaps more likely that they are duplicate samples.
In each relative pair, one member was removed to produce a data set in which no two individuals were related at a level closer than first cousins. To minimize the number of individuals removed, individuals present in two or more relative pairs were preferentially omitted. In situations where either individual in a relative pair could be removed, the individual with the higher level of missing data was removed. After the exclusion of related individuals, many of which were related to multiple individuals in the initial data set, our final data set consisted of 225 unrelated Gujarati individuals (Table 1).
Table 1.
Sample size and mean heterozygosity.
| Number of individuals | Heterozygosity | ||||
|---|---|---|---|---|---|
| Village | Total | Males | Females | Average across loci |
Standard deviation across loci |
| Bhadran | 30 | 20 | 10 | 0.723 | 0.093 |
| Dharmaj | 31 | 23 | 8 | 0.724 | 0.093 |
| Karamsad | 29 | 20 | 9 | 0.724 | 0.093 |
| Nadiad | 47 | 32 | 15 | 0.724 | 0.092 |
| Sojitra | 36 | 21 | 15 | 0.723 | 0.096 |
| Vaso | 21 | 17 | 4 | 0.728 | 0.093 |
| Gujaratis from outside of CGP | 31 | 16 | 15 | 0.731 | 0.090 |
| 14 other Indian subpopulations | 382 | 236 | 146 | 0.724–0.735 | 0.089–0.098 |
Genotype data
Each individual had been previously genotyped for 1,200 polymorphisms spread across all 22 autosomes (Rosenberg et al,. 2006): 471 insertion/deletion (indel) polymorphisms and 729 microsatellites. The microsatellites were drawn from Marshfield Screening Sets 13 and 52 (Ghebranious et al., 2003), and the insertion/deletion markers were drawn from Marshfield Screening Set 100 (Weber et al., 2002).
We successfully genotyped 26 Y-chromosomal microsatellites (DYS19, DYS385a/b, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS425, DYS426, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS441, DYS442, DYS446, DYS462, Y-GATA-A7.1, Y-GATA-A7.2, Y-GATA-A10, Y-GATA-C4, Y-GATA-H4) in 140 of the 149 male Gujaratis in our data set, using the procedures of Hall et al. (Hall and Ballantyne, 2003) and Hanson et al. (Hanson et al,. 2006). Of these 140 individuals, 124 were from the CGP villages: 19 from Bhadran, 21 from Dharmaj, 19 from Karamsad, 29 from Nadiad, 21 from Sojitra, and 15 from Vaso. The remaining 16 individuals were from villages outside the CGP.
We successfully sequenced both strands of the HVS1 region of the mitochondrial genome in 138 of the 149 males, using primers L15996 and H16401 from Vigilant et al. (Vigilant et al., 1989) and standard dye-terminator sequencing on an ABI3100 genetic analyzer (Applied Biosystems, Foster City, CA). Of these 138 individuals, 122 were from the CGP villages: 19 from Bhadran, 21 from Dharmaj, 18 from Karamsad, 28 from Nadiad, 21 from Sojitra, and 15 from Vaso. The remaining 16 individuals were from villages outside the CGP. Reads for each of the 138 individuals were mapped to the revised Cambridge reference mitochondrial sequence (Anderson et al., 1981; Andrews et al., 1999) (NC012920.1) to identify variable sites. Sites showing evidence of more than one peak (heteroplasmic positions) were scored as the non-reference base.
The autosomal microsatellite and indel genotype data, Y-chromosome microsatellite genotype data, and the genotype data for the 26 variable sites we identified in our mitochondrial HVS1 sequences are available for download from http://rosenberglab.stanford.edu/datasets.html. The consensus mtDNA HVS1 sequence for each of the 138 successfully sequenced individuals can be obtained from the NCBI GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/genbank) using the accession IDs provided in Table S1.
Autosomal analysis
The level of genetic variation in each of the CGP villages, in other Gujaratis, and in each of the 14 other groups defined by language from across India, was evaluated using expected heterozygosity, averaged across the 729 autosomal microsatellite markers, as calculated with a sample size-corrected estimator (Nei, 1987). Levels of genetic variation in the six CGP villages were compared to those in the 15 other groups using a Wilcoxon signed-rank test performed using wilcox.test in R (v.2.11.1) (R Development Core Team, 2008).
Pairwise genetic distances between the six CGP villages, and between the six CGP villages and other Gujaratis, were estimated using FST (Weir and Cockerham, 1984), as calculated using Arlequin (Excoffier et al., 2005) (v.3.5.1.2) on the 729 microsatellites. The significance of the FST estimates under the null hypothesis of no differences between groups was evaluated using the permutation test implemented in Arlequin, which permutes individuals between groups, with 10,000 permutations. As it is possible for the unbiased estimate of FST used here to assume negative values, which do not have a biological interpretation, we set negative estimates of FST to 0. Pairwise genetic distances between the six CGP villages were compared to those between each of the CGP villages and other Gujaratis using a Wilcoxon signed rank test.
Neighbor-joining trees (Saitou and Nei, 1987) were obtained based on pairwise autosomal allele-sharing distance among groups measured using all 1,200 autosomal markers. A greedy-consensus (Bryant, 2003) neighbor-joining tree of all groups in the data set was constructed using the neighbor, consense, and drawtree programs in the phylip package (Felsenstein, 2008) from 1,000 bootstrap resamples across loci generated using microsat (Minch et al., 1998).
Classic multi-dimensional scaling (MDS) analyses (Cailliez, 1983; Gower, 1966) were performed using cmdscale in R on pairwise allele-sharing distance matrices constructed using all 1,200 autosomal markers. In the two-dimensional MDS plot, the pairwise distances between individuals from the six different CGP villages were evaluated using a permutation-based test of the average linkage distance L0 (Le Roux and Rouanet, 2004; Timm, 2002), as described by Kopelman et al. (Kopelman et al., 2009), using 10,000 permutations. To assess the overall significance of the separation of individuals by village affiliation in the MDS plot, we used a Fisher’s combined probability test (Fisher, 1925) to combine probabilities across the 15 pairwise village comparisons, weighting each comparison by the sum of the sample sizes of the two populations included in the pair. This calculation was performed using combine.test from the survcomp package (Schroder et al., 2011) in R.
Y-chromosomal and mitochondrial analysis
Relationships between haplotypes were determined using the program NETWORK v.4.516 or v.4.600. For the Y-chromosomal network, DYS385a/b was excluded because it is not possible to identify the a and b loci separately, and DYS425 was excluded because it showed 2–4 peaks in eight of the individuals, indicating the possible presence of two or more alleles. DYS389a/b was included, with the repeat count at DYS389b obtained by subtracting the DYS389I count from the DYS389II count. Y-GATA-A7.1 showed duplications in two individuals (neither from within the six villages), and in each of these individuals, one arbitrarily-chosen allele was used. After down-weighting the two loci with highest variance (DYS389b and DYS446) by a factor of five to reduce reticulations, a median-joining network was constructed (Bandelt et al. 1999). Estimates of the time to the most recent common ancestor (TMRCA) and its standard deviation were estimated for individual clusters using the ρ statistic implemented in NETWORK, assuming a mutation rate of 6.9×10−4 per microsatellite per 25 years (Zhivotovsky et al., 2004). Clusters were defined as groups of six or more individuals linked by 0–3 mutational steps. The most frequent haplotype was assumed to be ancestral, or for the Bhadran-1 cluster, the most central with respect to the rest of the cluster. The ancestral haplotypes in the Sojitra and Nadiad-2 clusters lay just two mutational steps apart, and village of origin was considered in assigning individuals to one or the other cluster. For a more relaxed cluster definition applied in some analyses, groups of three or more individuals linked by 0–4 mutational steps were assigned to the same cluster. For mtDNA, a median-joining network was constructed using equal weights for all 26 variable positions (m.16051A>G, m.16069C>T, m.16071C>T, m.16086T>C, m.16092T>C, m.16093T>S, m.16111C>T, m.16114C>T, m.16126T>C, m.16129G>A, m.16145G>A, m.16153G>A, m.16154T>C, m.16162A>G, m.16171A>T, m.16172T>C, m.16179C>T, m.16180A>G, m.16181A>G, m.16182A>C, m.16183A>S, m.16184C>A, m.16185C>T, m.16188C>T, m.16189T>C, and m.16190C>T).
RESULTS
Signatures of gol endogamy in the CGP
The practice of endogamy restricts mate selection to within a pre-defined group of individuals, thereby genetically isolating this group from others. If gol endogamy has been practiced by the CGP since its formation, we would expect to find reduced genetic variation within the gol, and increased genetic distances in comparisons that involve CGP villages and populations from outside the gol.
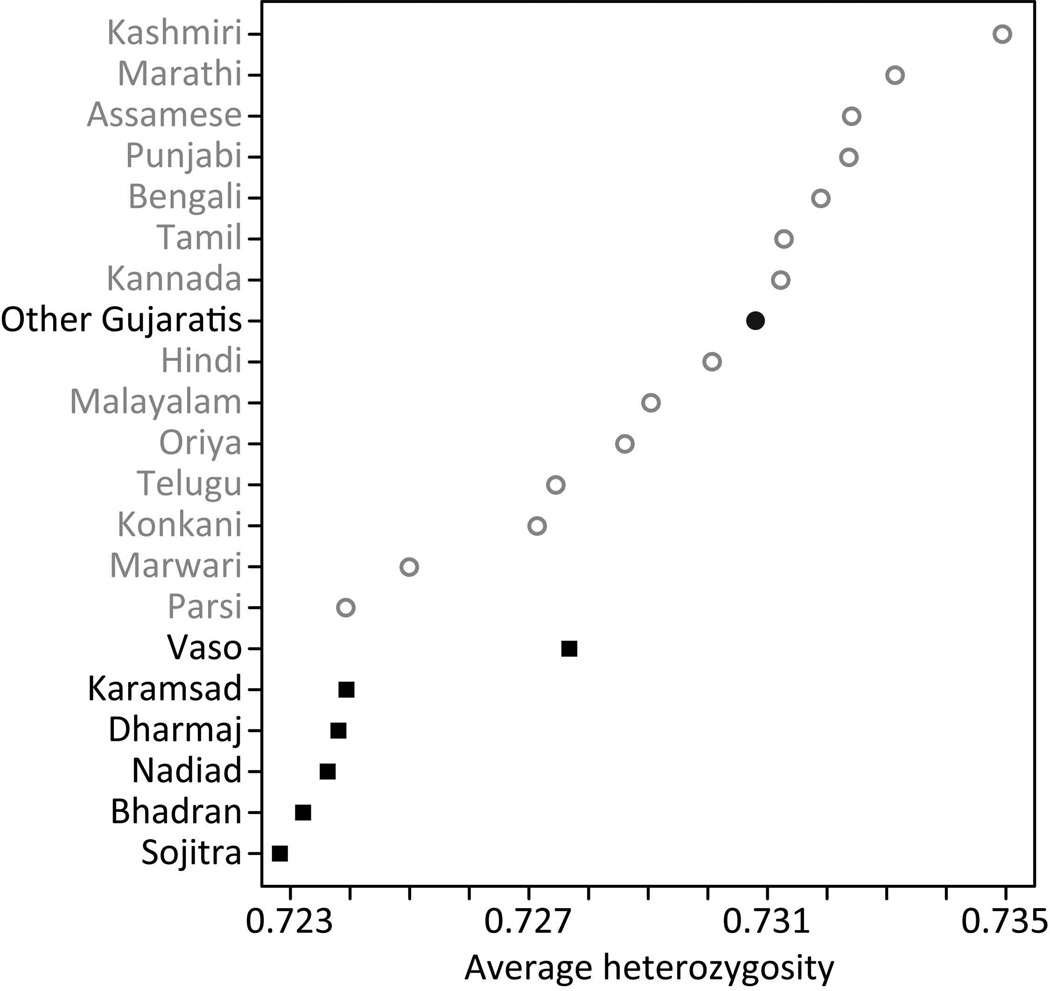
The level of autosomal genetic variation in each of the six CGP villages, as measured by microsatellite heterozygosity, is compatible with the expected reduction of this genetic variation statistic with the practice of endogamy by the CGP, ranging from 0.723 to 0.728 across the six villages, and lying below 0.724 for five of the six (three rounded up to 0.724; Table 1). If we compare the levels of genetic variation in the six CGP villages with those in the 15 groups from outside the gol (0.724 to 0.735), we find genetic variation in the CGP to be significantly lower than in the groups from outside the gol (Fig. 3; P=3.50×10−4, Wilcoxon rank sum test).
Figure 3.
Average microsatellite heterozygosity in each of the six CGP villages (black squares), in other Gujaratis (black circle), and the 14 other language groups (hollow grey circles). Average heterozygosities are provided in Table 1.
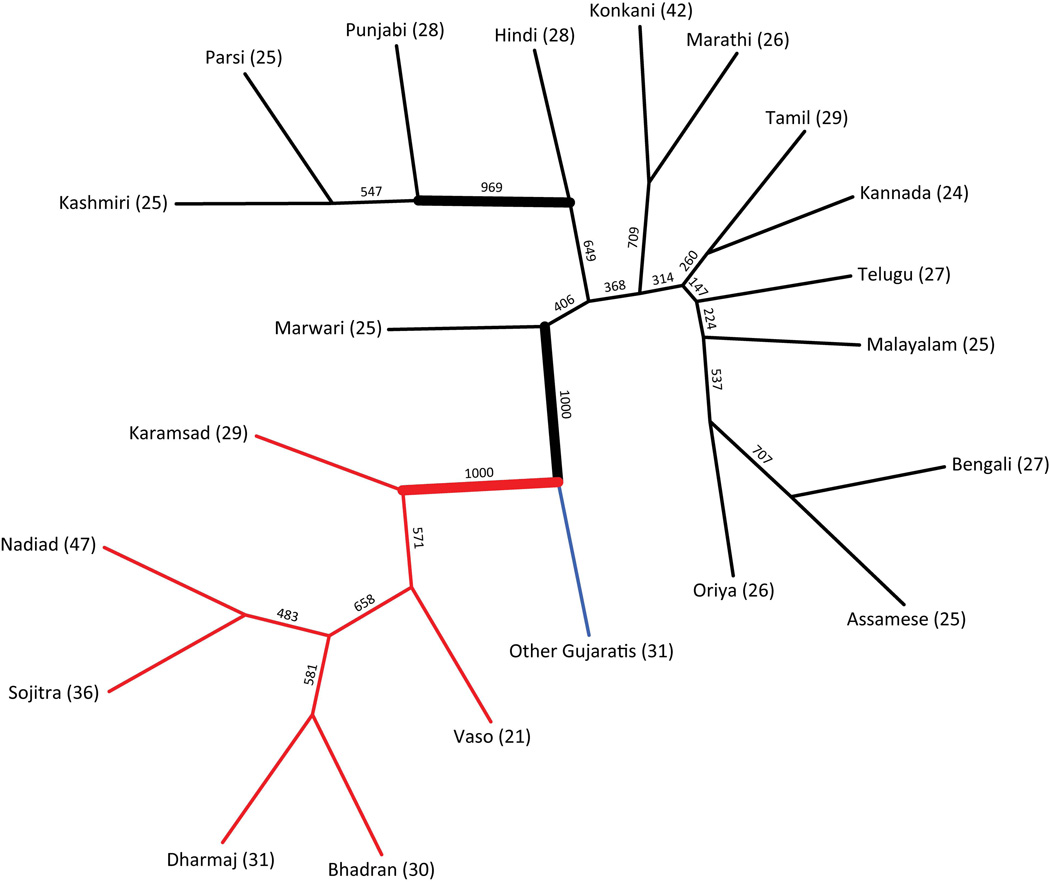
Analysis of population structure in the full sample of individuals using the neighbor-joining clustering algorithm provides 100% bootstrap support for a grouping of the Gujaratis and 100% bootstrap support for a grouping of the CGP within the Gujarati clade (Fig. 4). This latter observation is compatible with the significantly smaller pairwise microsatellite FST values (Table 2) observed in comparisons between pairs of CGP villages than in comparisons between each of the CGP villages and other Gujaratis (P=2.65×10−4, Wilcoxon rank sum test). In agreement with the pattern observed for FST values calculated using both males and females together, FST values calculated separately for males and females were significantly higher in comparisons between each of the CGP villages and other Gujaratis than in comparisons between pairs of CGP villages (Table 3; P=1.29×10−4 and P=0.005, respectively, Wilcoxon rank sum test).
Figure 4.
Consensus neighbor-joining tree estimated from 1,000 bootstraps using 194 individuals from the six CGP villages (red branches), 31 Gujarati individuals not from the CGP villages (blue branch), 382 individuals representing 14 additional Indian language groups (black branches), and 1,200 autosomal markers. The thick edges have at least 95% bootstrap support. The number on each internal branch is the bootstrap support for that branch. The sample size for a group is given in parentheses following the group name.
Table 2.
FST between pairs of CGP villages.
| Village | Bhadran | Dharmaj | Karamsad | Nadiad | Sojitra | Vaso | Other Gujaratis |
|---|---|---|---|---|---|---|---|
| Bhadran | 0.092 | 0.114 | 0.114 | 0.127 | 0.147 | 0.277** | |
| Dharmaj | 0.156 | 0.108 | 0.129 | 0.162 | 0.258** | ||
| Karamsad | 0.104 | 0.046 | 0.150 | 0.234** | |||
| Nadiad | 0.100 | 0.111 | 0.191** | ||||
| Sojitra | 0.101 | 0.241** | |||||
| Vaso | 0.310** |
All FST values (upper triangle) have been multiplied by 100. For the six CGP villages, underlined values indicate the smallest pairwise FST for individual villages and bold values indicate the largest pairwise FST.
P<0.01.
Table 3.
FST between pairs of CGP villages separately for males and females.
| Village | Bhadran | Dharmaj | Karamsad | Nadiad | Sojitra | Vaso | Other Gujaratis |
|---|---|---|---|---|---|---|---|
| Bhadran | - | 0.122 | 0.226 | 0.116 | 0.296* | 0.251 | 0.406** |
| Dharmaj | 0.337 | - | 0.186 | 0.088 | 0.179 | 0.266 | 0.339* |
| Karamsad | 0.034 | 0.373 | - | 0.167 | 0.095 | 0.221 | 0.303* |
| Nadiad | 0.323 | 0.282 | 0.237 | - | 0.191 | 0.208 | 0.243 |
| Sojitra | 0.177 | 0.151 | 0.194 | 0.148 | - | 0.241 | 0.384** |
| Vaso | 0.318 | 0.133 | 0.637 | 0.238 | 0.178 | - | 0.343* |
| Other Gujaratis | 0.483 | 0.412 | 0.333 | 0.340 | 0.301 | 0.499 | - |
All FST values shown have been multiplied by 100, and are given separately for the 149 male individuals (upper triangle) and the 76 female individuals (lower triangle). For the six CGP villages, underlined values indicate the smallest pairwise FST for individual villages and bold values indicate the largest pairwise FST.
P<0.05.
P<0.01.
In summary, members of the gol are genetically distinguishable both from other groups from across India and from other Gujaratis, consistent with restricted gene flow into the CGP as a consequence of the practice of endogamy.
Signature of gotra exogamy and gotra hypergamy in the CGP
The practice of gotra exogamy, which prohibits marriage between individuals from the same gotra, would be expected to maintain gene flow between gotra in the CGP, making individuals in all of the gotra genetically similar. We might then expect to not observe significant separation of individuals by village affiliation in an autosomal MDS plot. However, the practice of gotra hypergamy, in which a female marries a male from a family or gotra of greater standing, might be expected to selectively promote gene flow between certain pairs of villages and thereby reduce the level of genetic differentiation between such pairs. We might then expect significant separation between some pairs of villages but not others.
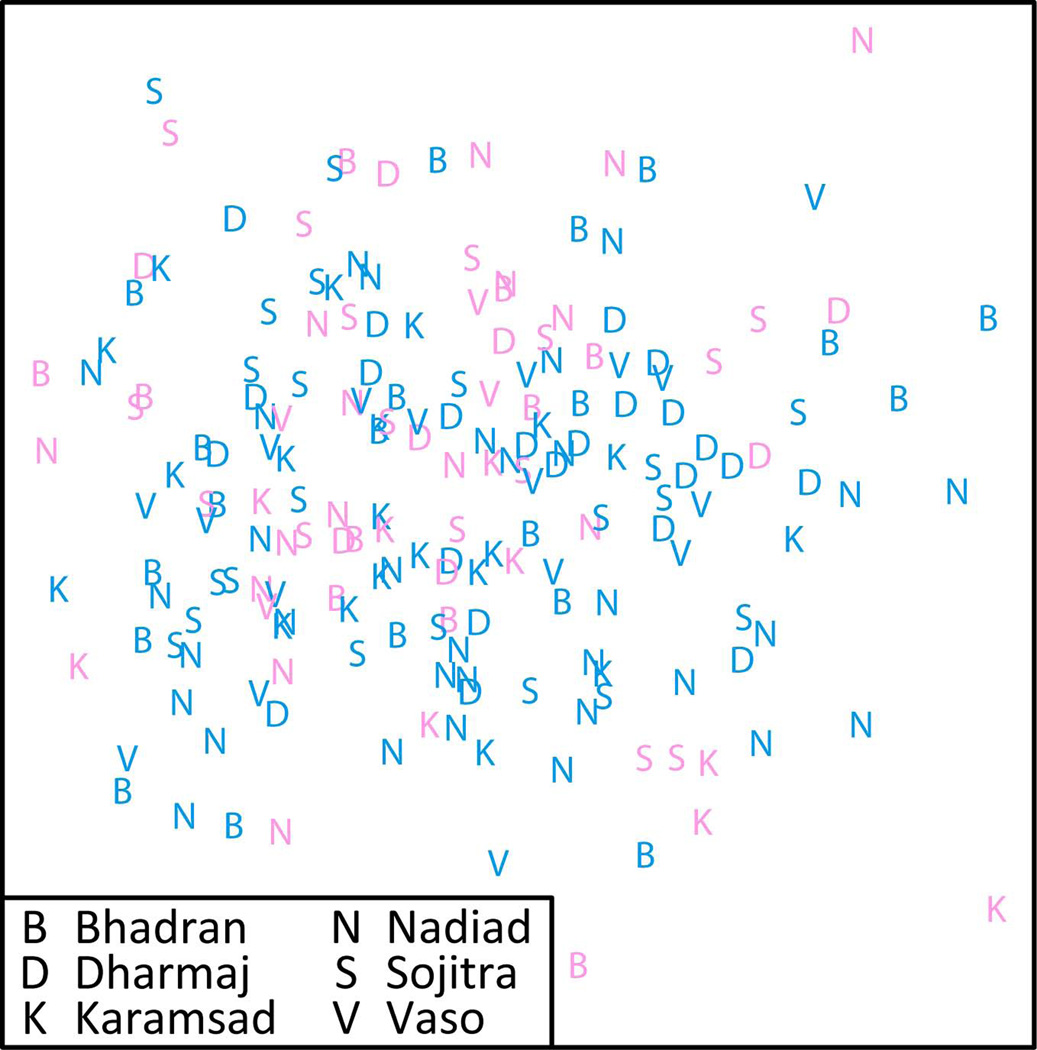
MDS analysis of individuals from the CGP produced no apparent clustering of individuals by village affiliation (Fig. 5). To evaluate the potential separation of individuals by village in the MDS plot, we examined the average pairwise linkage distance (L0) for all pairs of villages. In one pairwise comparison, between Bhadran and Vaso, there was a significant separation of individuals by village affiliation (P=0.004). All other pairwise comparisons had P>0.1, and a Fisher's combined probability test did not reveal a significant overall separation of individuals by village affiliation across the different village pairs (P=0.451).
Figure 5.
MDS analysis of pairwise autosomal allele-sharing distances between all 194 CGP individuals. Individuals are colored by their sex: males, blue; females, pink.
The autosomal MDS results are compatible with the practice of gotra exogamy within the CGP, and identify some evidence of gotra hypergamy among the six villages.
Signatures of patrilocality and patrilineality in the CGP
Patrilocality, in which male descendants remain in the village of their father while females move to the village of their husband, and patrilineality, in which property and surnames are inherited through the paternal line, have traditionally been practiced in the CGP. These practices are expected to introduce genetic dissimilarities between villages on the Y chromosome, but to maintain more uniform patterns across villages for mitochondrial DNA.
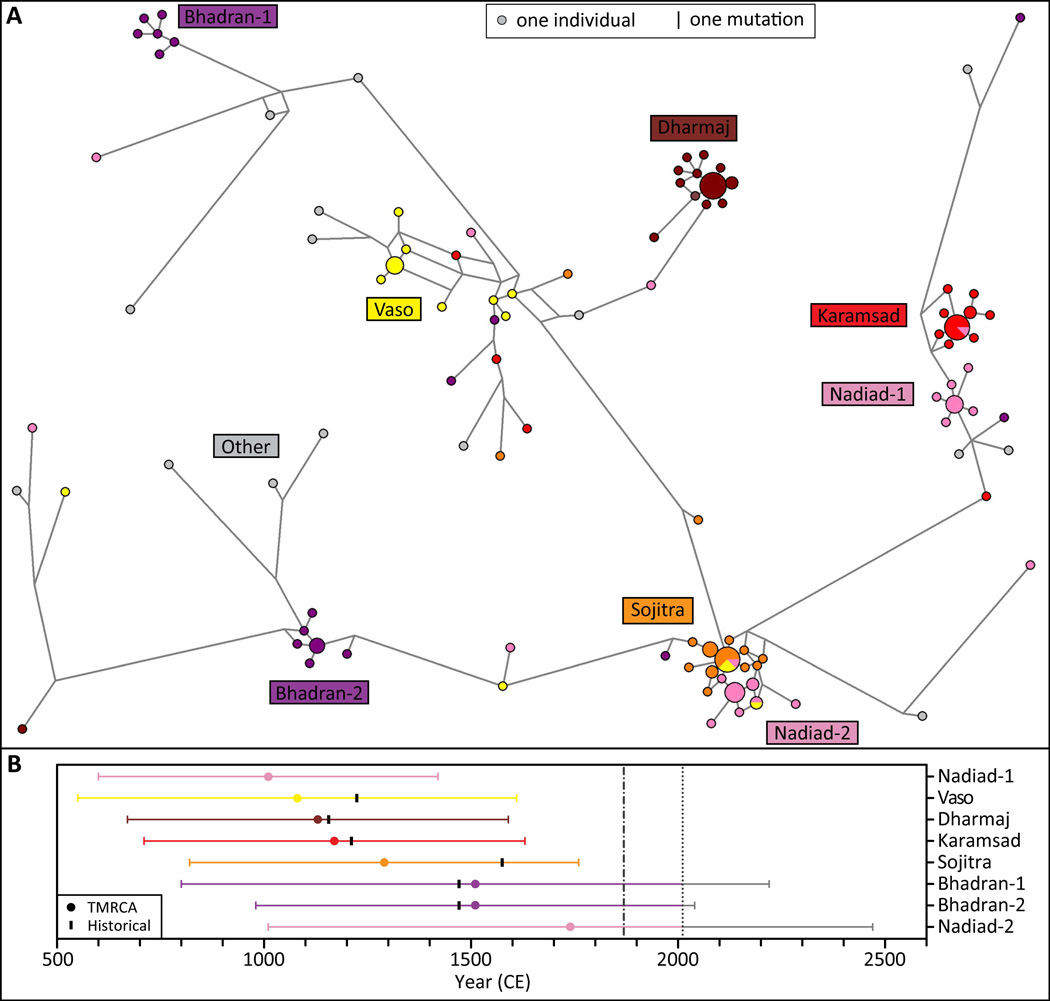
Analysis of Y-chromosomal haplotypes provides evidence for distinctive patterns in each of the six CGP villages (Fig. 6A). In each village, one (Dharmaj, Karamsad, Sojitra, Vaso) or two (Bhadran, Nadiad) clusters of haplotypes predominated. Across villages, these clusters together accounted for 103 of 124 males (83%). Clusters were highly specific to particular villages, and only five individuals within the major clusters originated from a village other than the predominant one. Estimated times to the most recent common ancestor (TMRCAs) of the clusters ranged from 1,010 ± 410 to 1,740 ± 730 years (Table 4).
Figure 6.
Relationships between Y-chromosomal haplotypes. (A) Median-joining network of 140 Y chromosomes from the six CGP villages and other Gujaratis. Twenty-four Y-chromosomal microsatellites were typed in 124 males from the six CGP villages and 16 Gujarati males from outside the gol. The relationships between the haplotypes were determined using the program NETWORK 4.516. Circles represent the microsatellite haplotypes and have areas proportional to the haplotype frequencies. Villages of origin are indicated by color; clusters of haplotypes from the same village are labeled. Line lengths represent the number of mutational differences between the haplotypes; the shortest line is one mutation. A line that abuts one or more haplotype circles may appear shorter than the number of mutations it represents, as part of the line may be covered by circles. (B) TMRCA estimates for each cluster in the Y-chromosome median-joining network. Clusters are ordered from top to bottom by increasing TMRCA estimates. Dots indicate TMRCA estimates, and colored lines represent their standard deviations. Black lines indicate the time of settlement of each village by Patels based on historical accounts, where available.
Table 4.
Estimates of the time to the most recent common ancestor (TMRCA) of Y-chromosomal haplotype clusters.
| Cluster | TMRCA (years) | Standard deviation (years) |
|---|---|---|
| Bhadran-1 | 1510 | 710 |
| Bhadran-2 | 1510 | 530 |
| Dharmaj | 1130 | 460 |
| Karamsad | 1170 | 460 |
| Nadiad-1 | 1010 | 410 |
| Nadiad-2 | 1740 | 730 |
| Sojitra | 1290 | 470 |
| Vaso | 1080 | 530 |
Haplotype clusters are as labeled in Figure 6.
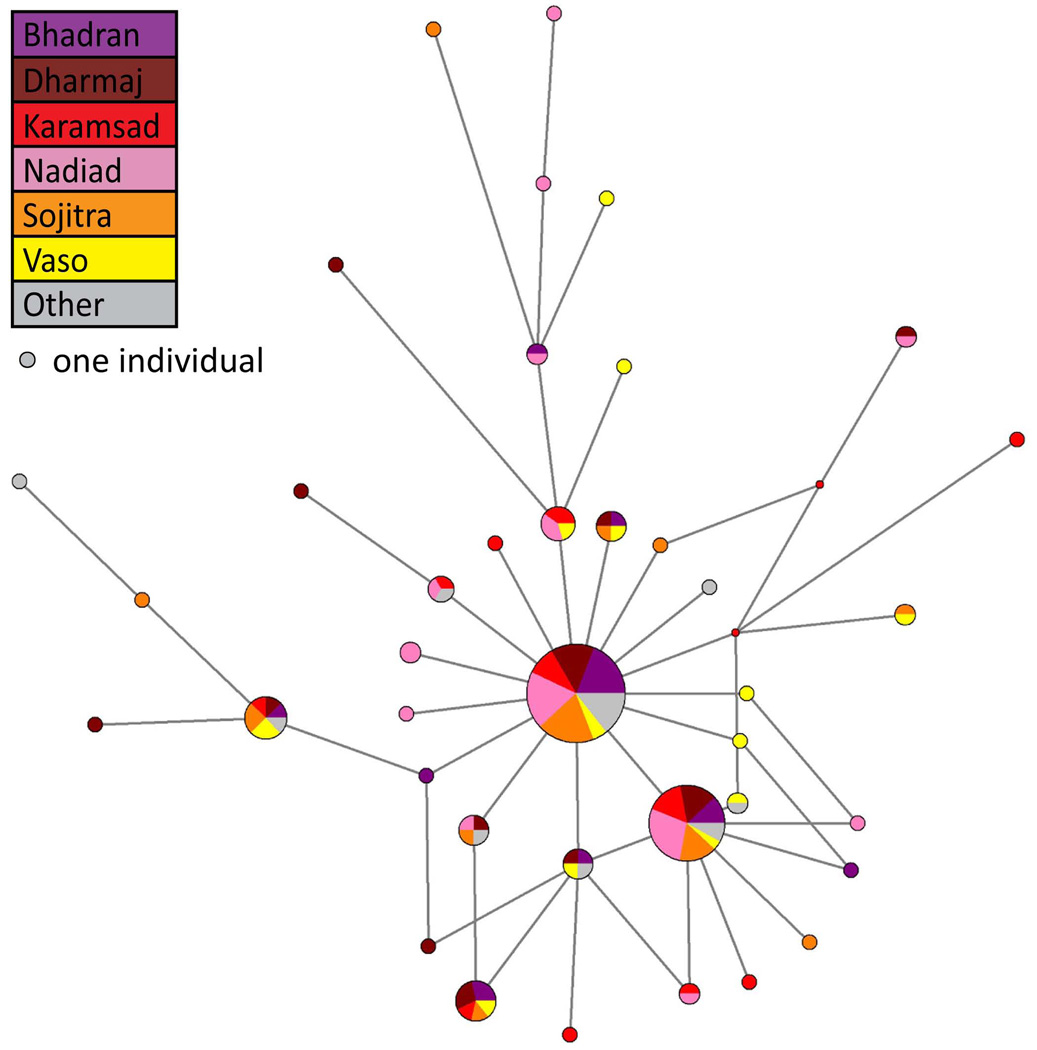
Conversely, analysis of mitochondrial haplotypes does not show major distinctions between villages, with all six villages represented in both of the most common haplotypes (Fig. 7). Additionally, we observe appreciable diversity in mitochondrial haplotypes, with numerous haplotypes present in only one or two individuals. This observation is compatible with the migration of females from gols of lower status into the CGP villages through the practice of gol hypergamy, as this practice would be expected to bring a diverse collection of mitochondrial haplotypes into the CGP from multiple sources.
Figure 7.
Median-joining network of 138 mitochondrial DNA HVS1 sequences from the six CGP villages and other Gujaratis. The HVS1 region of the mitochondrial genome was sequenced in 122 males from the six CGP villages and 16 Gujarati males from outside the gol. The relationships between the haplotypes were determined using the program NETWORK 4.600. Circles represent the haplotypes. Each has an area proportional to the haplotype frequency. Villages of origin are indicated by color. Each line represents one mutational difference.
Together, these observations support a scenario in which patrilocality and patrilineality are practiced within the CGP. They also provide evidence of the practice of gol hypergamy among Gujarati Patels.
This latter result is interesting, as gol hypergamy might also be expected to influence patterns of autosomal variation in the CGP; females entering the gol might originate from different sources, creating the expectation of a slightly greater level of genetic differentiation among the females of the different villages than among the males. The significant separation of females, but not males, from different villages in an autosomal MDS plot might therefore further indicate the presence of females from outside the gol who have migrated into the villages.
Examining, separately for males and females, the average pairwise linkage distance (L0) for all pairs of villages, we observed a significant separation of females by village affiliation in one of the comparisons: Nadiad and Vaso (P=0.042). All other pairwise comparisons had P>0.058 however, and there is no evidence of a significant overall separation of females by village affiliation across the different village pairs in the MDS plot (P=0.335, Fisher’s combined probability test). Interestingly, a significant separation of males by village affiliation was also observed in only one comparison: Karamsad and Sojitra (P=0.038). All other pairwise comparisons had P>0.087, and a Fisher's combined probability test did not reveal a significant overall separation of males by village affiliation across the different village pairs (P=0.245).
DISCUSSION
We have investigated how the practice of exogamic endogamy within a group of six villages in the state of Gujarat in western India has influenced genetic variation among its members. Consistent with the practice in which individuals from the six villages must marry within the six villages, our analysis of 1,200 autosomal polymorphisms has found the members of the CGP to be genetically distinguishable from other Indian individuals, including those from elsewhere in Gujarat. Furthermore, patterns of genetic variation and genetic diversity in the CGP villages are consistent with the practice in which individuals from the six villages must marry outside their own gotra.
Analysis of Y-chromosomal and mitochondrial haplotypes is consistent with strong patrilocality and patrilineality within the six CGP villages. Using a strict definition of haplotype clusters, 98 of 124 males (79%) lie within a haplotype cluster characteristic of their village. If the criteria for identifying a cluster are relaxed slightly (see Methods), 103 males (83%) fall within their characteristic village clusters. However, we find that the villages may not be strictly patrilocal; 21 males with 21 different haplotypes, including representatives from all six villages, lie far from any cluster and may represent rare village lineages or possible male gene flow from outside the gol. Five or six males (according to whether a strict or more relaxed cluster definition is used) carry haplotypes characteristic of a different village within the gol, accounting for 4–5% of the sampled gol haplotypes. While this result could provide evidence of a low level of male gene flow among the six villages, it might instead reflect the presence of multiple founding Y-STR haplotypes in these villages and a subsequent lack of male gene flow among them.
In some analyses, the results for one of the CGP villages (Vaso) were noticeably different from those for the other five villages. These differences accord with CGP societal norms, where factors such as social stigma may have led to males from Vaso being less sought after among CGP individuals. First, average heterozygosity was higher in Vaso than in the other five villages (Fig. 3), suggesting that individuals from Vaso might have more non-CGP admixture than individuals from the other villages. This would be consistent with Vaso males being more likely to marry females from outside the CGP than are males in the other five villages, purportedly through the practice of gol hypergamy. Second, Y-chromosomal haplotypes from Vaso did not cluster as tightly as did those from the other five villages (Fig. 6A); this result is compatible with a higher level of male gene flow into Vaso than into the other villages, possibly as a consequence of matrilocal marriage being more likely when the bride was from Vaso than when the bride was from one of the other villages. The apparent male gene flow into Vaso could also arise due to the fact that a husband from another CGP village may have moved to live with his wife's family in Vaso and become a “gharjamai,” a term describing a son-in-law who resides with his in-laws, because his in-laws either had no male heir or could not afford a dowry; hence, the husband would become heir-presumptive to the wife’s family’s inheritance (Pocock, 1972).
We observed that for autosomal loci, males were no more distinguishable across villages than were females. At the same time, mitochondrial haplotypes were more similar across villages than were Y-chromosomal haplotypes. These results are potentially incompatible, as patrilocality might be expected to generate a greater autosomal difference among males compared to females in addition to a greater difference in Y chromosomes compared to mitochondria, while gol hypergamy might be expected to generate the opposite patterns to patrilocality. It is possible that these apparently discrepant results reflect long-term patrilocality and gotra exogamy together with gol hypergamy in the most recent generations, with females entering the gol deriving primarily from the same broad pool of mitochondrial haplotypes. This female gene flow into the CGP might be at least partially attributable to the practice of female infanticide by Gujarati Patels during the nineteenth century (Clark, 1983; Nath, 1973). Female infanticide led to an imbalance in the sex ratio in many villages, particularly in villages of high status, such as those of the CGP. For example, in a census taken in 1849 CE, the sex ratio in Nadiad was reported as 70 females for every 100 males (Clark, 1983), while sex ratios in neighboring villages were in some cases even lower. The migration of females into the six villages might have occurred as a consequence of the dearth of eligible brides within the six villages, owing to their historical practice of female infanticide.
It is also possible that incompatibilities among our autosomal, mitochondrial, and Y-chromosomal analyses might have arisen from differences in levels of resolution; the 1200 autosomal markers potentially provide more information compared with the data available for the Y chromosome (24 microsatellites) and mtDNA (sequence of the HVS1 region). For example, although we have no reason to expect a bias toward specific groupings of populations, by limiting our mitochondrial analysis to the HVS1 region, we might have missed distinctions in mitochondrial haplotype variation across villages that would have been apparent had we analyzed complete mtDNA sequences (Gunnarsdottir et al., 2011).
An additional issue is that a variety of phenomena beyond marriage patterns might have affected the patterns we investigated. For example, a reduction in population size as a result of famine and plague in Gujarat at the end of the 19th century (Pocock, 1972), may have caused a decrease in genetic variation in the CGP villages. The lower average heterozygosities we observed in the CGP villages compared to the other language groups might partially reflect this event. However, mean heterozygosities for the six CGP villages are lower than for the other Gujaratis in our study (Table 1), most of whom emigrated to the United States from the same region of Gujarat where the CGP villages are located; consequently, our genetic evidence for gol endogamy within the CGP on the basis of heterozygosity patterns is not likely to be explained by a historic population size reduction.
While the TMRCA estimates for Y-chromosomal haplotype clusters have large uncertainties (Table 4), they predate or are otherwise consistent with historical accounts of the settlement of the six villages by Patels between 1155 CE and 1575 CE (Chh Gam Patidar Samaj Committee, 2010; Pocock, 1972) (Fig. 6B). Furthermore, the separate clustering of Y-chromosomal haplotypes from different villages is consistent with the view that at least four of the six villages (Bhadran, Dharmaj, Karamsad, and Vaso) were settled by Patels from different villages located in the neighboring state of Rajasthan (Anklav in 1471 CE, Jargal in 1156 CE (Pocock, 1972), Hilod near Adala in 1211 CE, and Undhhel in 1224 CE, respectively), and that one of the remaining two villages (Sojitra) was settled by Patels from a different village within Gujarat (Chapaner in 1575 CE). The historical record has not provided a hypothesis for the location of the origin of the Patels that first settled in the sixth village (Nadiad). However, the existence of two Y-chromosomal haplotype clusters associated with Nadiad could be compatible with this village having been settled by two distinct groups of migrants.
Information on the village of origin of the subjects in this study was self-reported, and the subjects are all first-generation immigrants to the United States. For the non-Gujaratis, village of origin was not known for most individuals, and as noted by Rosenberg et al. (2006), each language sample is not a random sample of individuals who speak that specific language. Among the CGP samples, the individuals are members of families who were originally from the CGP, but whose ancestors had at some point in the past migrated out of the six villages to urban areas elsewhere in India. As the CGP were among the earliest groups to migrate to urban areas of India and overseas, initially to East Africa and then to the United Kingdom and the United States beginning in the 1930s (Patel and Rutten, 1999), it is possible that our sampling in the United States contains some individuals who have provided imperfect information about their village of origin. However, there was and still is a strong motivation to maintain the strict endogamic, exogamic, and patrilineal marital practices of the CGP among its diaspora. Thus, even though the subjects in this study self-reported their village of origin and are first-generation immigrants primarily from urban India, the genetic data support patrilineality among the CGP diaspora, and likely reflect a high degree of compliance with the endogamic and exogamic rules established when the gol was founded.
While we have not explicitly investigated inter-gol differences, an early blood-group investigation focusing on between-gol differences among six Gujarati gols, including the CGP studied here, found significant genetic differences among gols (Vyas et al., 1958). Our results on the distinguishability of the CGP from other Gujaratis, together with the findings of Vyas et al., raise the possibility that the practice of exogamic endogamy has created isolated groups across India that are genetically distinguishable from other individuals in the Indian population, including those in their local vicinity.
A previous study of overlapping data found a low level of genetic differentiation among populations within India (Rosenberg et al., 2006). This previous study included a subset of the Gujarati individuals that we examined here, together with the same set of 382 individuals from 14 other linguistically-defined subgroups from across India. However, with the large sample of Gujaratis considered here, we were able to detect genetic differences among subgroups at a finer level of resolution.
CONCLUSIONS
Our findings highlight the important role that culturally-driven marital practices can have in shaping patterns of genetic variation in contemporary human populations. They provide further evidence on how caste-based social traditions have influenced genetic structure in India. In light of previous findings (Indian Genome Variation Consortium, 2008; Reich et al., 2009; Rosenberg et al., 2006; Wooding et al., 2004), a picture is emerging of different scales of genetic structure in the population of India, structure that has been introduced and reinforced by systems of endogamy over thousands of years. These scales of structure range from the large endogamous first-order divisions whose effects are observable at the national level, down to the small endogamous gols whose effects are observable locally. The findings of our study highlight the need to study genetic variation in first-, second-, and third-order divisions from across India to clarify the ways in which the rich variety of marital practices in India have influenced patterns of genetic structure.
Supplementary Material
ACKNOWLEDGEMENTS
This investigation was supported by a pilot grant award from the Center of Excellence in Genomic Science, University of Southern California (T.J.P.). This investigation was partially conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 (RR10600-01, CA62528-01, RR14514-01) from the National Center for Research Resources, National Institutes of Health. T.J.P. was partially supported by a postdoctoral fellowship from the University of Michigan Center for Genetics in Health and Medicine. N.A.R was partially supported by National Institute of Health grant GM081441, and by a grant from the Burroughs Wellcome Fund. C.T.S was supported by The Wellcome Trust. We thank Steven Se Fum Wong and Gustavo Mendoza-Fandino for assistance.
Grant sponsors: University of Southern California Center of Excellence in Genomic Science, University of Michigan Center for Genetics in Health and Medicine, National Institutes of Health, Burroughs Wellcome Fund, and The Wellcome Trust.
LITERATURE CITED
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Aoki K. Sexual selection as a cause of human skin colour variation: Darwin's hypothesis revisited. Ann Hum Biol. 2002;29:589–608. doi: 10.1080/0301446021000019144. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, et al. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH. Endogamy, consanguinity and community genetics. J Genet. 2002;81:91–98. doi: 10.1007/BF02715905. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ. Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. A classification of consensus methods for phylogenetics. In: Janowitz MF, Lapointe F-J, McMorris FR, Mirkin B, Roberts FS, editors. BioConsensus. Providence, RI: American Mathematical Society; 2003. pp. 163–183. [Google Scholar]
- Cailliez F. The analytical solution of the additive constant problem. Psychometrika. 1983;48:305–308. [Google Scholar]
- Census of India. 2001 http://www.censusindia.gov.in/.
- Chh Gam Patidar Samaj Committee. 2010 http://www.chhgamsamaj.com/.
- Clark A. Limitations on female life chances in rural central Gujarat. Indian Econ Soc Hist Rev. 1983;20:1–25. [Google Scholar]
- Cordaux R, Aunger R, Bentley G, Nasidze I, Sirajuddin SM, Stoneking M. Independent origins of Indian caste and tribal paternal lineages. Curr Biol. 2004;14:231–235. doi: 10.1016/j.cub.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Das V. Marriage and kinship in India: two recent studies. Contrib Indian Soc. 1973;7:135–139. [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, Laval LG, Schneide S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.68. Seattle: Department of Genome Sciences, University of Washington; 2008. [Google Scholar]
- Fisher RA. Statistical methods for research workers. Edinburgh, Scotland: Oliver and Boyd; 1925. [Google Scholar]
- Flood GD. An introduction to Hinduism. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- Frost P. European hair and eye color: a case of frequency-dependent sexual selection? Evol Hum Behav. 2006;27:85–103. [Google Scholar]
- Gadgil M, Joshi NV, Prasad UVS, Manoharan S, Patil S. Peopling of India. In: Balasubramanian D, Rao NA, editors. The Indian human heritage. Hyderabad, India: Universities Press; 1998. pp. 100–129. [Google Scholar]
- Ghebranious N, Vaske D, Yu A, Zhao C, Marth G, Weber JL. STRP screening sets for the human genome at 5 cM density. BMC Genomics. 2003;4:6. doi: 10.1186/1471-2164-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghurye GS. Caste and race in India. London: Routledge and Keegan Paul; 1969. [Google Scholar]
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- Gunnarsdottir ED, Nandineni MR, Li M, Myles S, Gil D, Pakendorf B, Stoneking M. Larger mitochondrial DNA than Y-chromosome differences between matrilocal and patrilocal groups from Sumatra. Nat Commun. 2011;2:228. doi: 10.1038/ncomms1235. [DOI] [PubMed] [Google Scholar]
- Hall A, Ballantyne J. The development of an 18-locus Y-STR system for forensic casework. Analyt Bioanalyt Chem. 2003;376:1234–1246. doi: 10.1007/s00216-003-2039-2. [DOI] [PubMed] [Google Scholar]
- Hanson EK, Berdos PN, Ballantyne J. Testing and evaluation of 43 "noncore" Y chromosome markers for forensic casework applications. J Forensic Sci. 2006;51:1298–1314. doi: 10.1111/j.1556-4029.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- Kashyap VK, Guha S, Sitalaximi T, Bindu GH, Hasnain SE, Trivedi R. Genetic structure of Indian populations based on fifteen autosomal microsatellite loci. BMC Genet. 2006;7:28. doi: 10.1186/1471-2156-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Cordaux R, Casto A, Lao O, Zhivotovsky LA, Moyse-Faurie C, Rutledge RB, Schiefenhoevel W, Gil D, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, Rosenberg NA. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 2009;10:80. doi: 10.1186/1471-2156-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laland KN, Odling-Smee J, Myles S. How culture shaped the human genome: bringing genetics and the human sciences together. Nat Rev Genet. 2010;11:137–148. doi: 10.1038/nrg2734. [DOI] [PubMed] [Google Scholar]
- Le Roux B, Rouanet H. Geometric data analysis: from correspondence analysis to structured data analysis. Dordrecht, Holland: Kluwer Academic Publishers; 2004. [Google Scholar]
- Minch E, Ruiz Linares A, Goldstein DB, Feldman MW, Cavalli-Sforza LL. MICROSAT (version 2.alpha): a program for calculating statistics on microsatellite data. Stanford, CA: Department of Genetics, Stanford University; 1998. [Google Scholar]
- Nasidze I, Quinque D, Rahmani M, Alemohamad SA, Stoneking M. Concomitant replacement of language and mtDNA in South Caspian populations of Iran. Curr Biol. 2006;16:668–673. doi: 10.1016/j.cub.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Nath V. Female infanticide and the Lewa Kanbis of Gujarat in the nineteenth century. Indian Econ Soc Hist Rev. 1973;10:386–404. [Google Scholar]
- Nei M. Molecular evoluionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Oota H, Settheetham-Ishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosome variation is correlated with matrilocal versus patrilocal residence. Nat Genet. 2001;29:20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- Patel PJ, Rutten M. Patels of central Gujarat in greater London. Econ Polit Weekly. 1999;34:952–954. [Google Scholar]
- Pocock DF. Factions in Indian and oversees Indian societies, Part 2: The bases of faction in Gujarat. Brit J Sociol. 1957;8:295–306. [Google Scholar]
- Pocock DF. Kanbi and Patidar: a study of the Patidar community of Gujarat. Oxford, UK: Clarendon Press; 1972. [Google Scholar]
- Pocock DF. The hypergamy of the Patidars. In: Uberoi P, editor. Family, kinship, and marriage in India. Delhi, India: Oxford University Press; 1993. pp. 331–340. [Google Scholar]
- R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2008. R: A language and environment for statistical computing. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Gonzales-Quevedo C, Nino-Rosales L, Ninis V, Das P, Hedge M, Molinari L, Zapata G, Weber JL, et al. Low levels of genetic divergence across populations of India. PLoS Genet. 2006;2:e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet. 2001;109:339–350. doi: 10.1007/s004390100577. [DOI] [PubMed] [Google Scholar]
- Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, Trivedi R, Endicott P, Kivisild T, Metspalu M, et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci USA. 2006;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schroder MS, Culhane AC, Quackenbush J, Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, et al. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AM. Division and hierarchy: an overview of caste in Gujarat. Contrib Indian Soc. 1982;16:1–33. [Google Scholar]
- Shah AM. The family in India: critical essays. New Delhi, India: Orient Longman Limited; 1998. [Google Scholar]
- Thakkar R. Gujaratis. In: Magocsi PR, editor. Encyclopedia of Canada's peoples. Toronto: University of Toronto Press; 1999. p. 635. [Google Scholar]
- Thangaraj K, Chaubey G, Kivisild T, Selvi Rani D, Singh VK, Ismail T, Carvalho-Silva D, Metspalu M, Bhaskar LV, Reddy AG, et al. Maternal footprints of Southeast Asians in North India. Hum Hered. 2008;66:1–9. doi: 10.1159/000114160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, Thangaraj K, Chaubey G, Singh VK, Bhaskar LV, Reddy BM, Reddy AG, Singh L. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 2006;7:42. doi: 10.1186/1471-2156-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm NH. Applied multivariate analysis. New York: Springer-Verlag; 2002. [Google Scholar]
- Tripathi M, Tripathi P, Chauhan UK, Herrera RJ, Agrawal S. Alu polymorphic insertions reveal genetic structure of north Indian populations. Hum Biol. 2008;80:483–499. doi: 10.3378/1534-6617-80.5.483. [DOI] [PubMed] [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci USA. 1989;86:9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas GN, Bhatia HM, Banker DD, Purandare NM. Study of blood groups and other genetical characters in six Gujarati endogamous groups in Western India. Ann Hum Genet. 1958;22:185–199. doi: 10.1111/j.1469-1809.1958.tb01413.x. [DOI] [PubMed] [Google Scholar]
- Watkins WS, Thara R, Mowry BJ, Zhang Y, Witherspoon DJ, Tolpinrud W, Bamshad MJ, Tirupati S, Padmavati R, Smith H, et al. Genetic variation in South Indian castes: evidence from Y-chromosome, mitochondrial, and autosomal polymorphisms. BMC Genet. 2008;9:86. doi: 10.1186/1471-2156-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, David D, Heil J, Fan Y, Zhao C, Marth G. Human diallelic insertion/deletion polymorphisms. Am J Hum Genet. 2002;71:854–862. doi: 10.1086/342727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Cockerham C. Estimating F-statitsics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wooding S, Ostler C, Prasad BV, Watkins WS, Sung S, Bamshad M, Jorde LB. Directional migration in the Hindu castes: inferences from mitochondrial, autosomal and Y-chromosomal data. Hum Genet. 2004;115:221–229. doi: 10.1007/s00439-004-1130-x. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.