Abstract
Background
A leptin-like immunoreactive substance has been found in chicken eggs and has been implicated in serving as a maternal signal to program offspring growth and metabolism. In the present study, we investigated the effects of in ovo leptin administration on hatch weight, serum and hepatic concentrations of metabolites and hormones, as well as on the expression of genes involved in hepatic lipid metabolism and the predicted microRNAs (miRNAs) targeting the affected genes. To this end we injected fertile eggs with either 0.5 μg of recombinant murine leptin or vehicle (PBS) before incubation.
Results
Prenatally leptin-exposed chicks showed lower hatch weight, but higher liver weight relative to the body weight, compared to the control group. In ovo leptin treatment increased the hepatic content and serum concentration of leptin in newly hatched chickens. The hepatic contents of triglycerides (TG) and total cholesterol (Tch) were decreased, whereas the serum levels of TG, Tch and apolipoprotein B (ApoB) were increased. The hepatic mRNA expression of sterol regulator element binding protein 1 (SREBP-1c), SREBP-2, hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) and cholesterol 7α-hydroxylase 1 (CYP7A1) was significantly up-regulated, as was the protein content of both SREBP-1c and SREBP-2 in hepatic nuclear extracts of leptin-treated chickens. Moreover, out of 12 miRNAs targeting SREBP-1c and/or HMGCR, five were significantly up-regulated in liver of leptin-treated chicks, including gga-miR-200b and gga-miR-429, which target both SREBP-1c and HMGCR.
Conclusions
These results suggest that leptin in ovo decreases hatch weight, and modifies hepatic leptin secretion and lipid metabolism in newly hatched broiler chickens, possibly via microRNA-mediated gene regulation.
Keywords: Broiler chickens, In ovo manipulation, Leptin, Lipid metabolism, Liver, MicroRNA, SREBPs
Background
Leptin is involved in the regulation of food intake and energy balance in mammals [1,2]. Despite the current controversy over the existence of a leptin gene in the chicken genome [3], the existence of a leptin-like immunoreactive substance [4,5] and a functional leptin receptor (LEPR) has been confirmed [6]. Furthermore, exogenous murine or human leptin exerts similar effects on poultry as it does on mammals [7,8].
Maternal leptin has been shown to program offspring obesity in mammals [9]. Fetal or neonatal abnormal nutritional environment induces leptin synthesis and secretion from adipocytes, and affects adipocyte morphology and metabolism, thus linking embryonic nutrition to adult obesity [10]. Manipulating either maternal plasma leptin or transplacental leptin transfer could impact the postnatal regulation of leptin synthesis and secretion in the offspring [11-13]. Previously, we have demonstrated the presence of a leptin-like immunoreactive substance in the yolk and albumen of chicken eggs [4]. Moreover, feeding hens with low-protein [14] or cysteamine-supplemented diets [4] affects leptin deposition in eggs, as well as early posthatch growth and metabolism of chickens. In ovo administration of leptin improved embryonic development and resulted in higher body weight at hatching of Japanese quails [15]. These findings imply possible roles of maternal leptin deposited in the egg in programming offspring growth and metabolism in poultry.
Leptin is mainly secreted by mammalian adipose tissue, and exerts direct effects on lipolysis and lipogenesis of adipose tissue [2]. In birds, liver is the major source of leptin [16] and the primary organ for lipogenesis [17]. Therefore, it is of interest whether in ovo administration of leptin affects hepatic leptin synthesis and secretion, as well as hepatic lipid homeostasis in newly hatched chickens.
Hepatic lipid homeostasis is regulated by a family of membrane-bound transcription factors, designated sterol regulatory element binding proteins (SREBPs) [18]. SREBP-1c, the most abundant isoform of SREBP-1 in the liver, preferentially enhances transcription of enzymes required for fatty acid synthesis, including acetyl CoA carboxylase (ACC), carnitine palmitoyltransferase I (CPT-I) and fatty acid synthase (FAS), whereas SREBP-2 is more selective for activating genes involved in cholesterol homeostasis, including HMG-CoA reductase (HMGCR) and cholesterol 7α-hydroxylase 1 (CYP7A1) [18]. Leptin has been shown to negatively regulate the expression of SREBPs [2], HMGCR [19], CYP7A1 [19], ACC and FAS [2], thus inhibiting lipogenesis in mammalian species.
MicroRNAs (miRNAs) are small noncoding RNAs with an average size of 22 nt. They bind to complementary sequences of target messenger RNA transcripts (mRNAs), usually resulting in translational repression and gene silencing [20]. A number of mammalian miRNAs that execute post-transcriptional regulation of genes involved in lipid homeostasis have been identified [21,22]. Recently, miR-33, which is located in the intron of SREBPs, was reported to regulate cholesterol metabolism [23,24]. Moreover, miRNAs have been shown to mediate the effects of leptin on adipocyte differentiation [25] and adipogenesis [26] in mammals. However, it is unknown whether the programming effects of maternal leptin involve miRNA-mediated post-transcriptional regulation. Relatively little is known about the function of miRNAs in chickens. More than 50 miRNAs have been identified in chicken liver [27-29], but only miR-33 has been experimentally verified to target the FAS gene in chicken fibroblast cells [30]. A correlation study linking the expression of miRNAs and their target genes, in association with hepatic lipid homeostasis may shed light on the potential functions of miRNAs in the chicken liver.
Therefore, the present study aimed to investigate the effects of in ovo leptin administration on hepatic leptin synthesis and secretion, as well as on hepatic lipid homeostasis in newly hatched chickens. The expression of genes involved in hepatic lipid metabolism, such as SREBPs and HMGCR, and of the predicted miRNAs targeting the relevant genes was also determined to reveal the possible mechanisms.
Methods
Animals and experimental design
Fertile breeder eggs purchased from Sanhuang broiler breeding farm (Wen’s group, Guangdong, China) were randomly divided into two groups and injected with 0 μg (Control, Con) or 0.5 μg (Leptin, Lep) of recombinant murine leptin (498-OB-01 M, R&D, Minneapolis, MN, USA) in 100 μL of phosphate buffered saline (PBS) before incubation. After injection, all eggs were incubated in a forced draught incubator with automatic turning every two hours at 37.5 ± 0.3°C and 50% to 60% humidity. At hatching (D0), chicks from each group were weighed and 12 chicks (6 males and 6 females) from each group were euthanized to take blood and liver samples. All tissue samples were snap frozen in liquid nitrogen and stored at -80°C. The experiments were undertaken following the guidelines of the Animal Ethics Committee of Nanjing Agricultural University, China.
Measurement of lipid parameters
Total lipid content in homogenized liver samples was extracted using a mixture of chloroform and methanol (2:1 v/v) according to a previously reported method [31]. The hepatic contents and serum concentrations of triglycerides (TG) and total cholesterol (TC) were determined using commercial kits (GPO-PAP and CHOD-PAP) purchased from Nanjing Jiancheng Biotechnology Institute (NJBI, Nanjing, Jiangsu, China) following the manufacturer’s protocols.
Tissue extraction and measurement of leptin and apolipoprotein B
Frozen liver samples (approximately 200 mg) were homogenized in 2 mL of ice-cold lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 0.2% Triton X-100 and protease inhibitor mixture) using a tissue grinder (Polytron PT1200E, Brinkman Instruments, Littau, Switzerland). The homogenate was then centrifuged at 5,000 × g for 20 min at 4°C to remove all insoluble material. The supernatant was collected, and the protein concentration was determined with a Bradford assay kit purchased from NJBI.
Leptin in serum and liver extracts was measured with a commercial multi-species RIA kit purchased from Beijing North Institute of Biotechnology (Beijing, China). The detection limit for leptin was 0.45 ng/mL. The intra- and inter-assay coefficients of variation were 5% and 10%, respectively. The commercial RIA kit was previously validated for measuring chicken samples [32]. Serum apolipoprotein B (ApoB) was determined using a commercial kit purchased from Biosino Biotechnology Company Ltd. (Beijing, China).
Western blot analysis
Nuclear and cytoplasmic protein was extracted from liver samples using Nuclear Protein Extraction Kit (PR116, Yuanpinghao Biotechnology Ltd., Beijing, China). Nuclear protein extracts were used for detecting SREBP-1c and SREBP-2, while cytoplasmic protein extracts were used for determining HMGCR and CYP7A1 levels. Whole cell lysates were prepared for quantitating leptin receptor (LEPR) by western blot analysis.
After electrophoresis, proteins were transferred to nitrocellulose membranes, which were then blocked with 5% fat-free milk or 3% BSA in Tween-Tris-buffer saline (TBST) for 2 h at room temperature. After repeated washing with TBST, the membranes were incubated with monoclonal antibodies against LEPR (diluted 1:1,000; a generous gift from Dr. Ohkubo, Faculty of Agriculture, Kagawa University, Japan), SREBP-1c (MA1-38651, Thermo, Waltham, MA, USA; diluted 1:200), SREBP-2 (ab30682, Abcam, UK; diluted 1:300), HMGCR (diluted 1:1,000; sc-33827, Santa Cruz, California, CA, USA) or CYP7A1 (diluted 1:200; ab79847, Cambridge, Abcam), followed by goat polyclonal horseradish peroxidase conjugated secondary antibody to rabbit IgG (diluted 1:5,000; ab6721, Abcam) or mouse IgG (diluted 1:4,000; GGHL-90P, Immunology Consultants Laboratory, Newberg, OR, USA), as previously described [6]. Finally, the membranes were washed and the specific signals were detected by chemiluminescence using the LumiGlo substrate (Super Signal West Pico Trial Kit, Pierce, Rockford, IL, USA). Enhanced chemiluminescence (ECL) signals recorded on x-ray film were scanned and analyzed with Kodak 1D Electrophoresis Documentation and Analysis System 120 (Kodak Photo Film Co. Ltd., Rochester, NY, USA). The membranes were stripped and reprobed with an antibody against β-actin (diluted 1:4,000; ab8227, Abcam) or LAMINA/C (diluted 1:500; BS1446, Bioworld Technology, Minneapolis, MN, USA), followed by horseradish peroxidase conjugated secondary antibodies (diluted 1:5,000; ab6721, Abcam). β-actin was used to normalize the band density of LEPR, HMGCR and CYP7A1, and LAMINA/C was used to normalize the band density of SREBP-1c and SREBP-2. The hepatic contents of LEPR, SREBP-1c, SREBP-2, HMGCR and CYP7A1 were presented as the fold change relative to the average values of the control group.
RNA extraction and mRNA quantification
Total RNA was extracted using TRIzol total RNA Kit (Tiangen Biotech Co., Ltd., Beijing, China), and 2 μg of total RNA were reverse-transcribed in a final volume of 25 μL with M-MLV reverse transcriptase (M1701, Promega, Madison, WI, USA) in a Bio-Rad DNA Engine Peltier Thermal Cycler PTC0200 (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed to quantitate LEPR, SREBP-1c/2, HMGCR and CYP7A1 mRNA with Mx3000P (Stratagene, Santa Clara, CA, USA). The primers were designed and synthesized by TaKaRa Biotechnology Co., Ltd. (Dalian, Shandong, China; Table 1).
Table 1.
Primer sequences for the target genes
| Target genes | GenBank accession numbers | PCR products (bp) | Primer sequences |
|---|---|---|---|
|
β-actin |
NM 205518 |
300 |
F: 5′- tgcgtgacatcaaggagaag -3′ |
| R: 5′- tgccagggtacattgtggta -3′ |
|||
|
LEPR |
NM 204323 |
87 |
F: 5′- gcatctctgcatctcaggaaaga -3′ |
| R: 5′- gcaggctacaaactaacagatcca -3′ |
|||
|
SREBP-1c |
NM 204126 |
104 |
F: 5′- gcagaagagcaagtccctcaa -3′ |
| R: 5′- tcggcatctccatcacctc -3′ |
|||
|
SREBP-2 |
XM 416222 |
108 |
F: 5′- cccagaacagcaagcaagg -3′ |
| R: 5′- gcgaggacaggaaagagagtg -3′ |
|||
|
HMGCR |
AB 109635 |
137 |
F: 5’-ttggatagagggaagagggaag -3’ |
| R: 5’- ccatagcagaacccaccaga-3’ |
|||
|
CYP7A1 |
AB 109636 |
106 |
F:5’- cattctgttgccaggtgatgtt -3’ |
| R:5’- gctctctctgtttcccgcttt -3’ |
Real-time PCR
Different controls were set to monitor the possible contamination of genomic and environmental DNA both at the stage of reverse transcription and PCR. A pooled sample made by mixing an equal quantity of total RNA from all samples was used for optimizing the PCR conditions and tailoring the standard curves. Two microliters of 8-64-fold dilutions of each reverse transcription product were used for PCR in a final volume of 25 μL. The PCR end products were verified with the melting curves that showed a single peak specific for the target gene.
miRNA quantification
Total RNA isolated from liver was treated with RNase-free DNase I (TaKaRa, Tokyo, Japan), and then polyadenylated (2 μg) by poly(A) polymerase (Ambion, Austin, TX, USA) at 37°C for 1 h in a 20 μL reaction mixture. After phenol-chloroform extraction and isopropyl ethanol precipitation, the RNA was dissolved and reverse-transcribed using a poly(T) adapter [33].
qPCR was performed using SYBR Green Real-time PCR Master Mix (TaKaRa) with a miRNA-specific forward primer and a universal reverse primer complementary to part of the poly(T) adapter sequence. Because no validated reference gene was available for chicken miRNA, random DNA oligonucleotides were added to the RNase-free Dnase I-treated total RNA samples before polyadenylation, as an exogenous reference, to normalize the expression of miRNAs. The sequences for all the primers, poly(T) adapter and the exogenous reference gene used are listed in Table 2.
Table 2.
Primer sequences for miRNAs used in the study
| Names | Primer sequences |
|---|---|
| gga-miR-15a |
F: 5′- TAGCAGCACATAATGGTTTGT -3′ |
| gga-miR-15b |
F: 5’- TAGCAGCACATCATGGTTTGCA -3’ |
| gga-miR-16 |
F:5’- TAGCAGCACGTAAATATTGGTG -3’ |
| gga-miR-27b |
F:5’- TTCACAGTGGCTAAGTTCTGC -3’ |
| gga-miR-99a |
F:5’- AACCCGTAGATCCGATCTTGTG -3’ |
| gga-miR-100 |
F: 5′- AACCCGTAGATCCGAACTTGTG -3′ |
| gga-miR-181a |
F: 5′- AACATTCAACGCTGTCGGTGAGT -3′ |
| gga-miR-183 |
F: 5′- AACATTCATTGCTGTCGGTGGG -3′ |
| gga-miR-200a |
F: 5′- TAACACTGTCTGGTAACGATGT -3′ |
| gga-miR-200b |
F: 5′- TAATACTGCCTGGTAATGATGAT -3′ |
| gga-miR-223 |
F: 5′- TGTCAGTTTGTCAAATACCCC -3′ |
| gga-miR-429 |
F: 5′- TAATACTGTCTGGTAATGCCGT -3′ |
| exogenous reference |
F: 5′- GTGACCCACGATGTGTATTCGC -3′ |
| universal reverse primer |
F: 5′- TAGAGTGAGTGTAGCGAGCA -3′ |
| poly(T) adapter | F:5′- TAGAGTGAGTGTAGCGAGCACAGAATTAATACGACTCACTATAGG(T)16VN-3′ |
No 5′UTR sequence was available for the chicken CYP7A1 gene, therefore we just predicted the miRNAs targeting SREBP-1cSREBP-2 and HMGCR with Targetscan 5.1 (http://www.targetscan.org/) [34-36]. Five miRNAs were predicted to target SREBP-1c, while nine miRNAs were predicted to target HMGCR. Only one miRNA, gga-miR-138, was predicted to target SREBP-2. However, we were unable to amplify specific gga-miR-138 from the chicken hepatic samples; therefore we excluded gga-miR-138 from the present study.
Statistical analysis
The method of 2-ΔΔCt was used to analyze the real-time PCR data expressed as the fold change relative to the control group [37]. All data were presented as means ± SEM. Statistical analyses were carried out with SPSS11.0 for windows (SPSS Inc., Chicago, IL, USA). The differences were tested with t-test for Independent-Samples. A P value of less than 0.05 was considered significant.
Results
Body weight and liver weight
As shown in Table 3, chicks that hatched from leptin-treated eggs exhibited significantly lower hatch weight (P = 0.000). No alterations were observed in the absolute liver weights, but the liver weight relative to the body weight, or the liver index, was significantly higher in the leptin-treated group at hatching (P = 0.018).
Table 3.
Effects ofin ovoleptin administration on body weight, liver weight and liver index (liver weight relative to body weight) in newly hatched chicks
| Parameters | Control | Leptin |
|---|---|---|
| Body weight (g) |
36.34 ± 0.32 |
33.30 ± 0.65** |
| Liver weight (mg) |
801.0 ± 22.0 |
801.7 ± 16.1 |
| Liver index (%) | 2.21 ± 0.06 | 2.42 ± 0.07* |
Values are presented as means ± SEM. *P < 0.05, **P < 0.01 compared with control, n = 12.
Serum and hepatic contents of leptin and hepatic LEPR expression
As shown in Table 4, in ovo administration of leptin significantly increased the serum concentration (P = 0.009) and liver content (P = 0.041) of leptin in newly hatched chicks.
Table 4.
Effects ofin ovoleptin administration on leptin concentration and lipid metabolic parameters in liver and serum of newly hatched chicks
| Parameter | Control | Leptin | |
|---|---|---|---|
| Liver |
leptin (ng/g T-protein) |
42.71 ± 4.06 |
58.06 ± 5.42* |
| TG (μmol/g liver) |
23.15 ± 1.38 |
20.12 ± 0.54 |
|
| Tch (μmol/g liver) |
16.88 ± 0.62 |
13.80 ± 0.13** |
|
| Serum |
leptin (ng/mL) |
1.10 ± 0.11 |
1.53 ± 0.10** |
| TG (mmol/L) |
0.67 ± 0.06 |
0.90 ± 0.07* |
|
| Tch (mmol/L) |
8.44 ± 0.22 |
9.93 ± 0.43* |
|
| ApoB (g/L) | 0.096 ± 0.009 | 0.138 ± 0.017* | |
Values are represented as means ± SEM, *P < 0.05, **P < 0.01 compared with control, n = 12.
Western blot analysis with a specific antibody against chicken LEPR detected a band of approximately 180 kDa in liver whole cell lysates (Figure 1A and B). In ovo leptin injection did not affect either LEPR mRNA abundance or LEPR protein content in the liver of newly hatched chicks (Figure 1).
Figure 1.
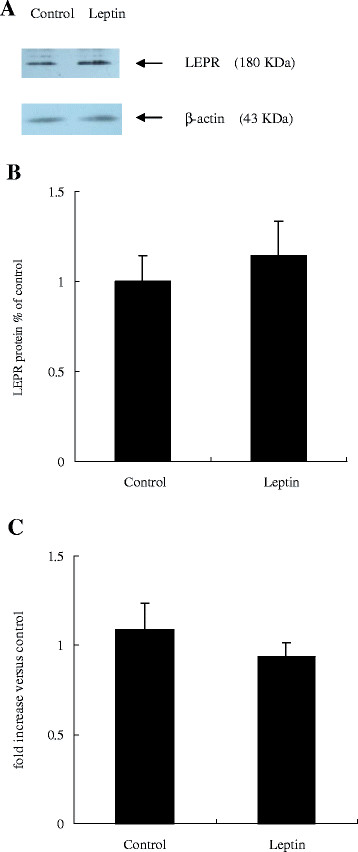
Effect ofin ovoleptin administration on hepatic mRNA and protein expression of LEPR in newly hatched chicks. A: Immunoreactive bands for LEPR and β-actin protein; B: LEPR protein content; C: LEPR mRNA. Values are presented as fold change relative to the control, expressed as means ± SEM, n = 12.
Lipid contents in serum and Liver
As shown in Table 4, the TG content in the liver was greatly decreased (P = 0.053), whereas the TG concentration in the serum significantly increased (P = 0.037) in leptin-treated chicks. The hepatic and serum contents of Tch showed the same pattern as TG, being greatly decreased (P = 0.001) in liver but increased in serum (P = 0.018) of leptin-treated chicks. Moreover, the serum content of ApoB significantly increased (P = 0.022) in newly hatched chicks from the leptin-treated group.
Hepatic expression of genes involved in the regulation of lipid metabolism
The hepatic expression of SREBP-1c (P = 0.027), SREBP-2 (P = 0.030), HMGCR (P =0.001) and CYP7A1 (P = 0.041) mRNA was significantly up-regulated in leptin-treated chicks at hatching (Figure 2A). In agreement with the mRNA abundances, the protein levels of both SREBP-1c (P = 0.032) and SREBP-2 (P = 0.001) in liver were significantly increased in leptin-treated chicks. However, the hepatic content of HMGCR and CYP7A1 protein was not affected by in ovo leptin treatment (Figure 2B).
Figure 2.
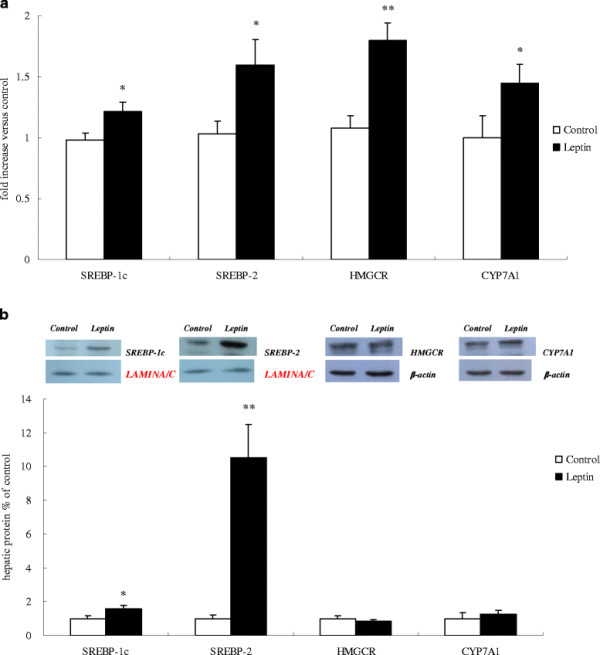
Effects ofin ovoleptin administration on hepatic expression of genes involved in lipid metabolism in newly hatched chicks. A: mRNA; B: protein. Values are presented as fold change relative to the control, expressed as means ± SEM. *P < 0.05, **P < 0.01, n = 12.
miRNA Quantification
Among the five miRNAs predicted to target SREBP-1c, gga-miR-99a, gga-miR-100, gga-miR-200b and gga-miR-429 were found to be significantly (P < 0.05) up-regulated in the liver of leptin-treated chickens. Among the nine miRNAs predicted to target HMGCR, the expression of gga-miR-200a, gga-miR-200b and gga-miR-429 was significantly increased (P < 0.05). It is worth noting that gga-miR-200b and gga-miR-429 were predicted to target both SREBP-1c and HMGCR (Table 5).
Table 5.
Effects ofin ovoleptin administration on hepatic expression of miRNAs predicted to targetSREBP-1candHMGCRin newly hatched chicks
| Target Genes | miRNA | Control | Leptin | P value |
|---|---|---|---|---|
|
SREBP-1c |
gga-miR-99a |
0.98 ± 0.11 |
2.11 ± 0.42 |
0.024 |
| gga-miR-100 |
1.00 ± 0.11 |
2.03 ± 0.38 |
0.026 |
|
| gga-miR-183 |
1.05 ± 0.23 |
1.70 ± 0.23 |
0.070 |
|
| gga-miR-200b |
0.97 ± 0.11 |
1.99 ± 0.40 |
0.033 |
|
| gga-miR-429 |
1.05 ± 0.08 |
2.04 ± 0.40 |
0.038 |
|
|
HMGCR |
gga-miR-15a |
1.02 ± 0.12 |
1.84 ± 0.41 |
0.086 |
| gga-miR-15b |
1.00 ± 0.10 |
1.24 ± 0.27 |
0.427 |
|
| gga-miR-16 |
1.03 ± 0.11 |
1.62 ± 0.36 |
0.147 |
|
| gga-miR-27b |
1.02 ± 0.08 |
1.22 ± 0.23 |
0.416 |
|
| gga-miR-181a |
0.96 ± 0.08 |
1.00 ± 0.18 |
0.663 |
|
| gga-miR-200a |
1.02 ± 0.11 |
2.52 ± 0.64 |
0.045 |
|
| gga-miR-200b |
0.97 ± 0.11 |
1.99 ± 0.40 |
0.033 |
|
| gga-miR-223 |
1.04 ± 0.07 |
1.39 ± 0.28 |
0.249 |
|
| gga-miR-429 | 1.05 ± 0.08 | 2.04 ± 0.40 | 0.038 |
Values are presented as fold change relative to the control, expressed as means ± SEM (n = 12).
Discussion
Increasing evidence suggests that maternal leptin programs fetal growth and development in mammalian species. In rats, dams receiving three injections of human recombinant leptin (3.5 mg/kg, Intraperitoneal Injection, i.p.) on days 8, 10 and 12 of gestation produced pups with reduced birth weight [13]. Accordingly, in the present study in ovo leptin administration reduced the hatch weight of broiler chickens. This observation, however, contradicts a previous publication in which in ovo injection of recombinant mouse leptin on day 5 of incubation advanced hatching by 5 to 24 h and improved the hatch weight of Japanese quails [15]. Perhaps the effect of leptin on Japanese quails is different than its effect on chickens. Furthermore, different doses of a hormone, like leptin, usually have diverse physical effects. These factors may contribute to the discrepancy in the results.
In ovo administration of leptin increased the proportional liver weight in the leptin-treated chickens at hatching. The increased liver index could be the consequence of decreased body weight, yet it has been suggested that alterations in proportional liver weight are usually associated with the change in hepatic lipid metabolism in the chicken [38-40]. Our results support this notion as the increased liver index correlated with alterations in hepatic and serum TG and Tch concentrations in newly hatched broiler chickens. Moreover, we observed, for the first time, a positive correlation between liver weight and hepatic leptin production and secretion in prenatally leptin-exposed chickens, which is in agreement with a previous report showing that the liver weight positively correlates with the plasma leptin concentrations in broiler breeder hens [41].
To date, the mechanism by which maternal leptin influences offspring leptin secretion has not been clarified. A transplacental transfer of maternal leptin to the fetus, which increases during late pregnancy in parallel with an up-regulation of expression of the shorter isoforms of the leptin receptor in the placenta, has been reported in rodents [42,43]. It is unknown how exogenous leptin injected into the albumen of the eggs is metabolized and/or transported to the developing embryos. We detected a significant down-regulation of LEPR mRNA expression in the yolk sac of leptin-treated embryos at 12 days of incubation (data not shown). However, the physiological significance of this down-regulation remains elusive. It is unlikely that the injected murine leptin remains active over 21 days of incubation, thus the increased hepatic and serum leptin contents in newly hatched chickens are most likely the consequences of an altered developmental program that may occur primarily at an early embryonic stage.
Leptin has been shown to exert direct effects on lipolysis and lipogenesis of adipose tissue in mammalian species [2]. Instead of the mammalian adipose tissue, liver is the primary organ for lipid metabolism in the chicken [17]. In the present study, the TG and Tch contents were greatly decreased in the liver, but were significantly increased in the serum of leptin-treated chickens, which was accompanied by a trend of increase in serum ApoB levels. ApoB is a major protein component of the plasma very low-density and low-density lipoproteins (VLDL and LDL, respectively) and plays an important role in transporting cholesterol and triglycerides from the liver [44]. The associated changes in serum ApoB and TG/Tch may indicate enhanced TG and Tch transport from the liver of leptin-treated chickens at hatching.
SREBPs directly activate the expression of more than 30 genes dedicated to cholesterol and fatty acid synthesis in the liver [18]. In the present study, the mRNA expression and protein contents of SREBP-1c and SREBP-2 were both enhanced in the liver of leptin-treated chicks at hatching. HMGCR and CYP7A1, which are dedicated respectively to the synthesis and uptake of cholesterol, were both significantly up-regulated in the liver of leptin-treated chickens, indicating a positive transcriptional regulation of SREBP-2 on HMGCR and CYP7A1 genes. SREBP-1c and SREBP-2 share a mechanism of feedback regulation, mediated by sterol response elements (SREs) present in the enhancer/promoters of each gene. Accumulation of hepatic TG and Tch has been reported to reduce SREBPs processing [18]. Therefore, the up-regulation of hepatic SREBP-1c and SREBP-2 expression may be the result of the reduced hepatic contents of TG and Tch in leptin-treated chickens.
Leptin could interact with leptin receptor to implement its biological functions [45,46]. Although the serum leptin concentration in the leptin-treated neonatal chicks was significantly increased, no significant alterations were detected in LEPR protein and mRNA expression in liver. Chicken leptin has been reported to desensitize its own response by decreasing the expression of its receptor mRNA [46]. The lack of change in hepatic LEPR expression in leptin-treated chickens implies a possible desensitizing effect of the elevated serum leptin on its receptor. In general, leptin plays an important role in the negative regulation of lipogenesis in mammals [2]. In contrast to its inhibitory effects on the expression of SREBPs [2], HMGCR and CYP7A1 [19] in mammals, in ovo leptin administration enhanced mRNA expression of SREBP-1c/2HMGCR and CYP7A1 in the present study, in parallel with elevated serum leptin concentration. It is possible that the effect of leptin on the hepatic expression of lipogenesis genes is species specific, being stimulatory in poultry yet inhibitory in mammals. Taking into consideration the irresponsiveness of hepatic LEPR, another possibility is that the enhanced hepatic expression of SREBPs, HMGCR and CYP7A1 may not be the direct effect of leptin. Other hormones or metabolites, altered by in ovo leptin treatment, may participate in the regulation of hepatic lipid metabolism and lipogenic gene expression, as well as hepatic leptin production and secretion.
Recently, considerable interest has been shown towards the role of miRNA in the regulation of lipid metabolism [23,24]. We detected concurrent changes in the expression of miRNAs and their target genes, which seems to contradict the common notion that the expression of miRNAs and their target genes are normally negatively correlated. However, the functions of miRNAs are much more complex and new mechanisms are emerging. For instance, although miRNAs are known to predominantly act in translational repression, miR-10 has been recently found to bind a group of transcripts containing a terminal oligo-pyrimidine (TOP) motif and to induce their translation [47]. Moreover, a time lag may exist between the expression of miRNAs and the endpoint of their actions in destruction of mRNAs or translational repression of target genes. We observed the uncoupling of HMGCR mRNA (up-regulated) and protein (unchanged) in the liver of leptin-treated chicks, which coincided with significantly increased expression of gga-miR-200agga-miR-200b and gga-miR-429 that were predicted to target the chicken HMGCR gene. However, further studies are required to clarify whether these miRNAs act as translational repressors to stabilize HMGCR protein content despite increased mRNA abundance.
In the present study, five out of 12 miRNAs predicted to target SREBP-1c and/or HMGCR, were significantly up-regulated in the liver of leptin-treated chicks. It is noteworthy that gga-miR-99a and gga-miR-100 belong to the miRNA gene family of miR-99, while the remaining three miRNAs, gga-miR-200a, gga-miR-200b and gga-miR-429, belong to the miRNA gene family of miR-8, located in the same miRNA cluster. In agreement with our results, it is presumed that miRNAs from the same family share similar biological functions and miRNAs from the same miRNA cluster normally have the same expression pattern. However, further experiments are necessary to validate the function of these five miRNAs on the expression of the target genes and hepatic lipid metabolism in the chicken.
Conclusions
We have shown for the first time that in ovo administration of leptin decreases the hatch weight, and modifies hepatic leptin synthesis and secretion, as well as hepatic lipid metabolism in newly hatched broiler chickens. A miRNA-mediated regulation of cholesterogenic and lipogenic genes, such as SREBPs and HMGCR, may be involved in these effects.
Abbreviations
miRNAs = MicroRNAs; LEPR = Leptin receptor; TG = Triglyceride; Tch = Total cholesterol; ApoB = Apolipoprotein B; SREBP-1c = Sterol regulator element binding protein 1c; SREBP-2 = Sterol regulator element binding protein 2; HMGCR = Hydroxy-3-methylglutaryl coenzyme A reductase; CYP7A1 = Cholesterol 7α-hydroxylase 1; SREBPs = Sterol regulatory element binding proteins; ACC = Acetyl CoA carboxylase; CPT-I = Carnitine palmitoyltransferase I; FAS = Fatty acid synthase; PBS = Phosphate buffered saline; SREs = Sterol response elements.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YH carried out the experiments, participated in the data collection, data analysis and interpretation, and drafted the manuscript. RZ performed the analysis of the hepatic microRNAs’ expression, helped in data interpretation and paper drafting. YZ and JL performed the protein quantification by western blot analysis. RG provided valuable advice for this study and helped in editing the manuscript. RZ contributed in conception, experimental design, data interpretation and finalized the manuscript. All authors approved the final version of the manuscript for publication.
Contributor Information
Yan Hu, Email: huyan0128@126.com.
Rui Zhang, Email: zhangrui666111@163.com.
Yanhong Zhang, Email: yuhui0906@126.com.
Jing Li, Email: lijing200522@126.com.
Roland Grossmann, Email: grossman@tzv.fal.de.
Ruqian Zhao, Email: zhao.ruqian@gmail.com.
Acknowledgements
This work was supported by the NSFC-Guangdong Joint Fund (U0931004), the Sino-German Cooperation in Agriculture (Project No. 28/04-05CHN7, 2010-2011), the Special Fund for Agro-scientific Research in the Public Interest (201003011), the National “948” project (2011-S11), the Natural Science Foundation of Jiangsu Province (BK2010296), the Doctor Foundation of the Institute of Poultry Science of Jiangsu Province (JQ201001) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Harvey J, Ashford ML. Leptin in the CNS: much more than a satiety signal. Neuropharmacology. 2003;44:845–854. doi: 10.1016/S0028-3908(03)00076-5. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Lynis DG. Peripheral metabolic actions of leptin. Best Pract Res Clin Endocrinol Metab. 2002;16:653–666. doi: 10.1053/beem.2002.0223. [DOI] [PubMed] [Google Scholar]
- Friedman-Einat M, Boswell T, Horev G, Girishvarma G, Dunn IC, Talbot RT. et al. The chicken leptin gene: has it been cloned? Gen Comp Endocrinol. 1999;115:354–363. doi: 10.1006/gcen.1999.7322. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ni Y, Ren L, Dai J, Zhao R. Leptin is involved in the effects of cysteamine on egg laying of hens, characteristics of eggs, and posthatch growth of broiler offspring. Poult Sci. 2008;87:1810–1817. doi: 10.3382/ps.2008-00040. [DOI] [PubMed] [Google Scholar]
- Neglia S, Arcamone N, Gargiulo G, de Girolamo P. Immunocytochemical detection of leptin-like immunoreactivity in the chicken gastroenteric tract. Gen Comp Endocrinol. 2008;155:432–437. doi: 10.1016/j.ygcen.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ohkubo T, Nishio M, Tsurudome M, Ito M, Ito Y. Existence of leptin receptor protein in chicken tissues: Isolation of a monoclonal antibody against chicken leptin receptor. Gen Comp Endocrinol. 2007;151:269–273. doi: 10.1016/j.ygcen.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Denbow DM, Meade S, Robertson A, McMurtry JP, Richards M, Ashwell C. Leptin-induced decrease in food intake in chickens. Physiol Behav. 2000;69(3):359–362. doi: 10.1016/S0031-9384(99)00258-9. [DOI] [PubMed] [Google Scholar]
- Macajova M, Lamosova D, Zeman M. Role of Leptin in Japanese Quail Development. Acta Vet. 2002;71:473–479. doi: 10.2754/avb200271040473. [DOI] [Google Scholar]
- Vickers MH. Developmental programming and adult obesity: the role of leptin. Curr Opin Endocrinol Diabetes Obes. 2007;14:17–22. doi: 10.1097/MED.0b013e328013da48. [DOI] [PubMed] [Google Scholar]
- McMillen IC, Edwards LJ, Duffield J, Muhlhausler BS. Regulation of leptin synthesis and secretion before birth: implications for the early programming of adult obesity. Reproduction. 2006;131:415–427. doi: 10.1530/rep.1.00303. [DOI] [PubMed] [Google Scholar]
- Stocker C, O'Dowd J, Morton NM, Wargent E, Sennitt MV, Hislop D. et al. Modulation of susceptibility to weight gain and insulin resistance in low birthweight rats by treatment of their mothers with leptin during pregnancy and lactation. Int J Obes Relat Metab Disord. 2004;28:129–136. doi: 10.1038/sj.ijo.0802476. [DOI] [PubMed] [Google Scholar]
- Yura S, Itoh H, Sagawa N, Yamamoto H, Masuzaki H, Nakao K. et al. Role of premature leptin surge in obesity resulting from intrauterine undernutrition. Cell Metab. 2005;1:371–378. doi: 10.1016/j.cmet.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Swolin-Eide D, Ohlsson C, Eriksson E, Ho HP, Bjorntorp P. et al. Reductions in adipose tissue and skeletal growth in rat adult offspring after prenatal leptin exposure. J Endocrinol. 2003;176:13–21. doi: 10.1677/joe.0.1760013. [DOI] [PubMed] [Google Scholar]
- Rao K, Xie J, Yang X, Chen L, Grossmann R, Zhao R. Maternal low-protein diet programmes offspring growth in association with alterations in yolk leptin deposition and gene expression in yolk-sac membrane, hypothalamus and muscle of developing Langshan chicken embryos. Br J Nutr. 2009;102:848–857. doi: 10.1017/S0007114509276434. [DOI] [PubMed] [Google Scholar]
- Lamosova D, Macajova M, Zeman M, Mozes S, Jezova D. Effect of in ovo leptin administration on the development of Japanese quail. Physiol Res. 2003;52:201–209. [PubMed] [Google Scholar]
- Ashwell CM, Richards MP, McMurtry JP. The ontogeny of leptin mRNA expression in growing broilers and its relationship to metabolic body weight. Domest Anim Endocrinol. 2001;21:161–168. doi: 10.1016/S0739-7240(01)00113-8. [DOI] [PubMed] [Google Scholar]
- Taouis M, Dridi S, Cassy S, Benomar Y, Raver N, Rideau N. et al. Chicken leptin: properties and actions. Domest Anim Endocrinol. 2001;21:319–327. doi: 10.1016/S0739-7240(01)00122-9. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpatten S, Karkanias GB, Rossetti L, Cohen DE. Intracerebroventricular leptin regulates hepatic cholesterol metabolism. Biochem J. 2004;379:229–233. doi: 10.1042/BJ20040134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernando C, Suarez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86–92. doi: 10.1097/MOL.0b013e3283428d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N. et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H. et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376:728–732. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao P, Zhou H, Xiao ZD, He JH, Huang MB, Chen YQ. et al. Identification of novel chicken microRNAs and analysis of their genomic organization. Gene. 2008;418:34–40. doi: 10.1016/j.gene.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang X, Du Z, Li N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006;580:3610–3616. doi: 10.1016/j.febslet.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Hicks JA, Tembhurne P, Liu HC. MicroRNA expression in chicken embryos. Poult Sci. 2008;87:2335–2343. doi: 10.3382/ps.2008-00114. [DOI] [PubMed] [Google Scholar]
- Hicks JA, Trakooljul N, Liu HC. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol Genomics. 2010;41:185–193. doi: 10.1152/physiolgenomics.00156.2009. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Li Y, Yuan L, Yang X, Ni Y, Xia D, Barth S. et al. Effect of early feed restriction on myofibre types and expression of growth-related genes in the gastrocnemius muscle of crossbred broiler chickens. Br J Nutr. 2007;98:310–319. doi: 10.1017/S0007114507699383. [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Chen J, Tang X, Zhang Y, Ma H, Zou S. Effects of maternal treatment of dehydroepiandrosterone (DHEA) on serum lipid profile and hepatic lipid metabolism-related gene expression in embryonic chickens. Comp Biochem Physiol B Biochem Mol Biol. 2010;155:380–386. doi: 10.1016/j.cbpb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Maurice DV, Bodine AB, Rehrer NJ. Metabolic effects of low aflatoxin B1 levels on broiler chicks. Appl Environ Microbiol. 1983;45:980–984. doi: 10.1128/aem.45.3.980-984.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KL, Chi WT, Chu C, Chen RS, Chiou PW. Effect of caponization and testosterone implantation on hepatic lipids and lipogenic enzymes in male chickens. Poult Sci. 2007;86:1754–1759. doi: 10.1093/ps/86.8.1754. [DOI] [PubMed] [Google Scholar]
- Bruggeman V, Onagbesan OM, Decuypere E. Body weight, fat content, liver weight and plasma leptin concentrations in broiler breeder females reared under ad libitum feeding, restricted feeding or combinations of both until age of first egg. Br Poultry Sci. 2000;41:57–59. doi: 10.1080/00071660050148912. [DOI] [Google Scholar]
- Smith JT, Waddell BJ. Leptin receptor expression in the rat placenta: changes in ob-ra, ob-rb, and ob-re with gestational age and suppression by glucocorticoids. Biol Reprod. 2002;67:1204–1210. doi: 10.1095/biolreprod67.4.1204. [DOI] [PubMed] [Google Scholar]
- Smith JT, Waddell BJ. Leptin distribution and metabolism in the pregnant rat: transplacental leptin passage increases in late gestation but is reduced by excess glucocorticoids. Endocrinology. 2003;144:3024–3030. doi: 10.1210/en.2003-0145. [DOI] [PubMed] [Google Scholar]
- Kirchgessner TG, Heinzmann C, Svenson KL, Gordon DA, Nicosia M, Lebherz HG. et al. Regulation of chicken apolipoprotein B: cloning, tissue distribution, and estrogen induction of mRNA. Gene. 1987;59:241–251. doi: 10.1016/0378-1119(87)90332-5. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA. The leptin receptor. J Biol Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- Cassy S, Derouet M, Crochet S, Dridi S, Taouis M. Leptin and insulin downregulate leptin receptor gene expression in chicken-derived leghorn male hepatoma cells. Poult Sci. 2003;82:1573–1579. doi: 10.1093/ps/82.10.1573. [DOI] [PubMed] [Google Scholar]
- Lund AH. miR-10 in development and cancer. Cell Death Differ. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]


