Abstract
Background
Daptomycin non-susceptible enterococci (DNSE) are emerging as an important cause of healthcare-associated infection, however little is known about the epidemiology of DNSE. At the University of Iowa Hospitals and Clinics (UIHC) an increase in the frequency of patients infected and/or colonized with DNSE has occurred. The goals of this study were to evaluate potential factors associated with the development of DNSE colonization and/or infection and to compare the characteristics of patients with prior daptomycin exposure to those without prior daptomycin exposure.
Methods
The study is a retrospective case-series involving all patients with DNSE infection and/or colonization at UIHC, a 734-bed academic referral center, from June 1, 2005 to June 1, 2011.
Results
The majority of patients with DNSE colonization and/or infection had prior daptomycin exposure (15 of 25; 60%), a concomitant gastrointestinal process (19 of 25; 76%), or were immunosuppressed (21 of 25; 84%). DNSE infection was confirmed in 17 of 25 (68%) patients, including 9 patients with bacteremia. Twelve of 17 (71%) patients with DNSE infection had prior daptomycin exposure, including 7 of 9 (78%) patients with bacteremia. Compared to patients without prior daptomycin exposure, patients with prior daptomycin exposure were less likely to harbor E. faecalis (0% vs. 33%; p = 0.019). A high proportion of patients (10 of 25; 40%) died during their hospitalizations. Most enterococcal isolates were E. faecium (86%), and were vancomycin-resistant (72%). Molecular typing revealed a diverse population of DNSE.
Conclusions
Prior daptomycin exposure, immunosuppression, and/or a concomitant gastrointestinal process, may be associated with the development of DNSE. PFGE revealed a diverse population of DNSE, which along with both increasing numbers of DNSE detected yearly and increasing annual rates of daptomycin usage, suggests the emergence of DNSE under antimicrobial pressure.
Keywords: Enterococcus, Daptomycin, Resistance, Non-Susceptible, DNSE
Background
Enterococci are Gram-positive, facultative anaerobes that reside primarily in the gastrointestinal tract. They are generally considered to be of low virulence, but are associated with serious bloodstream, joint, wound, cardiac, urinary, and gastrointestinal infections [1]. Enterococci are often multi-drug resistant, and are responsible for the transmission of various genetic resistance elements to other bacteria, including vancomycin resistance to other enterococci as well as Staphylococcus aureus[2,3].
Daptomycin is a lipopeptide antibiotic currently approved by the Food and Drug Administration (FDA) for the treatment of complicated infection of skin and/or subcutaneous tissue as well as bacteremia and right-sided endocarditis due to S. aureus[3]. In addition, it is often used in the treatment of infections due to vancomycin-resistant enterococci (VRE), although it is not approved for these conditions [3,4]. Shortly after its FDA approval in 2003, reports of infections due to daptomycin non-susceptible enterococci (DNSE) emerged and cases have been described both with and without prior daptomycin exposure [4-9]. Despite this, little is known about the potential causes or risk factors for the development of DNSE colonization or infection. Here, we report the findings at our institution, The University of Iowa Hospitals and Clinics (UIHC), over a 6-year period (June 1, 2005 through June 1, 2011).
Methods
The UIHC is a 734-bed, academic, and major referral center for the state of Iowa and surrounding region, with over 28,000 inpatient admissions and 900,000 outpatient visits each year. The UIHC Institutional Review Board granted approval for this study.
By searching our clinical microbiology laboratory database, we identified all DNSE isolates from the time the first clinical isolate was identified (June 1, 2005) to June 1, 2011. An extensive review of the medical record was performed for all patients with DNSE isolated from any source. Infection due to DNSE was defined as isolation from a sterile source, or isolation from a non-sterile source if accompanied by documented symptoms or signs of infection and/or treating clinician explicit diagnosis. Daptomycin usage during the study period was determined yearly by dividing the number of inpatients receiving daptomycin (based on discharge billing data) by the total number of patient discharges for a given year. P-values were calculated with the use of the chi-square.
At UIHC, daptomycin susceptibility testing is performed on all enterococcal isolates (both VSE and VRE) according to standard Clinical and Laboratory Standards Institute (CLSI) method [10], with daptomycin non-susceptibility defined as a daptomycin minimum inhibitory concentration (MIC) > 4 micrograms/mL. All enterococcal isolates not susceptible to daptomycin upon initial testing were re-tested using the CLSI broth microdilution and Etest methods. All available DNSE isolates underwent pulsed-field gel electrophoresis (PFGE) to assess for genetic relatedness, using previously described methods [11]. PFGE patterns were analyzed using Bionumerics software (Applied Maths, Kortrijk, Belgium). The unweighted pair group method with arithmetic averages and DICE coefficient (0.5% optimization, 1.0% position tolerance) were used for dendrogram construction. A similarity coefficient of 0.8 was used to define PFGE types, and subtypes were defined as isolates sharing indistinguishable banding patterns.
Results
The median age of patients with DNSE was 52.2 years, with 14 of 25 (56%) being female (Table 1). DNSE was identified in 25 patients and found in 32 clinical samples from 4 specimen sources including; blood (9 patients; 12 isolates), urine (12 patients; 14 isolates), peritoneal fluid or intra-abdominal abscess (3 patients; 4 isolates), and wounds (2 patients; 2 isolates). One patient had DNSE identified from both urine and blood (Table 1). Multiple isolates from the same patient were always of the same species, PFGE type or subtype, and had an identical antimicrobial susceptibility pattern.
Table 1.
Clinical characteristics of patients with DNSE colonization or infection
| Year, Age, Sex | Past Medical History | Total Days of Daptomycin Exposure | Species | Source | Reason for Admission and/or Complications |
|---|---|---|---|---|---|
| 2011 |
|
|
|
|
|
| 55F |
MDS, SCT |
5 |
E. faecium |
Blood, urine |
Neutropenia, GVHD of gut |
| 55F |
ALL, BMT, DM, PVD |
No |
E. faecium |
Blood |
Neutropenia, CDI |
| 58F |
Gynecological malignancy |
9 |
E. faecium |
Blood |
GI inflammation due to metastatic disease |
| 59F |
AML |
19 |
E. faecium |
Blood |
Neutropenia, appendicitis |
| 51F |
AML, SCT |
29 |
E. faecium |
Urinea |
Neutropenia, colitis |
| 2010 |
|
|
|
|
|
| 74F |
MDS, SBO, CHF, Pulmonary Htn |
10 |
E. faecium |
Urinea |
Small bowel resection, sepsis |
| 80F |
CAD, COPD, PVD |
No |
E. faecalis |
Urinea |
Bowel ischemia after surgery |
| 42 M |
DM, bipolar disorder |
12 |
E. faecium |
Wound, lumbar |
Chronic lumbar abscess |
| 56F |
AML, SCT, DM |
67 |
E. faecium |
Blood |
Neutropenia, sigmoid colon perforation |
| 2009 |
|
|
|
|
|
| 50F |
AML, SCT |
20 |
E. faecium |
Urinea |
Neutropenia, GVHD of gut, colitis |
| 50 M |
DM, Hip SSTI/osteomyelitis |
19 |
E. faecium |
Wound, hip |
Hip SSTI / osteomyelitis |
| 49 M |
CAD, CHF, COPD, ESRD/HD, PVD |
52 |
Not able to identify |
Peritoneal fluid |
GI perforation, peritonitis |
| 37 M |
Kidney-pancreas transplant, DM |
11 |
E. faecium |
Blood |
Recurrent cholangitis |
| 49 M |
Kidney-pancreas transplant, DM |
12 |
E. faecium |
Blood |
Intraabdominal abscess after surgery |
| 2008 |
|
|
|
|
|
| 39 M |
Hydrocephalus, VPS |
25 |
E. faecium |
Peritoneal fluid |
GI perforation following MVA |
| 61F |
DM, endometrial malignancy |
No |
E. faecium |
Urine |
Pelvic exenteration, kidney abscess |
| 78 M |
CAD, CKD, CVA, DM |
No |
E. faecium |
Urinea |
Septic arthritis due viridans streptococci |
| 57 M |
Urinary bladder malignancy |
No |
E. faecium |
Peritoneal fluid |
Pelvic abscess after surgery |
| 2007 |
|
|
|
|
|
| 59 M |
MDS, BMT |
8 |
E. faecium |
Blood |
Neutropenic fever, GVHD of gut |
| 59F |
DM, ESRD/HD, VHD |
16 |
E. faecium |
Urinea |
Cryptococcal meningitis |
| 2006 |
|
|
|
|
|
| 6F |
Recurrent UTI |
No |
Not Available |
Urine |
UTI |
| 46F |
Cholangitis, DM, ESRD/HD |
No |
E. faecalis |
Blood |
Recurrent polymicrobial cholangitis |
| 62 M |
CAD, COPD |
No |
Not available |
Urinea |
Lumbar pain and sciatica |
| 2005 |
|
|
|
|
|
| 38 M |
DM, metastatic malignancy of colon |
No |
Not available |
Urine |
Chemotherapy, neutropenia, pyelonephritis |
| 35F | DM, gastroparesis | No | Not available | Urinea | Recurrent gastroparesis |
Patient characteristics, past medical history, daptomycin exposure history (total days in prior year), isolate species, isolate source, and abbreviated reason for admission and/or complications arising during index admission for the 25 patients identified to be colonized or infected with DNSE during the study period.
NOTE:
a – Represents DNSE colonization.
Abbreviations: ALL = acute lymphoblastic leukemia; AML = acute myelogenous leukemia; BMT = bone marrow transplant; CAD = coronary artery disease; CDI = Clostridium difficile infection; CHF = congestive heart failure; CKD = chronic kidney disease; COPD = chronic obstructive pulmonary disease; CVA = cerebro-vascular accident; DM = diabetes mellitus; ESRD/HD = end stage renal disease requiring hemodialysis; GI = gastrointestinal; GVHD = graft versus host disease; MDS = myelodysplastic syndrome; MVA = motor vehicle accident; PVD = peripheral vascular disease; SBO = small bowl obstruction; SCT = stem cell transplant; SSTI = skin and soft tissue infection; UTI = urinary tract infection; VHD = valvular heart disease; VPS = ventriculoperitoneal shunt; Pulmonary Htn = pulmonary hypertension.
Infection due to DNSE was confirmed by both laboratory and chart review in 17 of 25 (68%) patients. In the remaining 8 patients, DNSE was isolated from urine alone and represented colonization based on the absence of documented clinical symptoms and/or negative urinalysis. A bloodstream infection was identified in 9 patients, a genitourinary infection such as UTI, pyelonephritis, or kidney abscess in 3 patients, bacterial peritonitis in 3 patients, and a skin and soft tissue infection and/or osteomyelitis in 2 patients.
A concomitant gastrointestinal or intra-abdominal process was identified in 19 of 25 (76%) patients, including Clostridium difficile infection, graft-versus-host-disease of the gut, neutropenic enterocolitis/perforation, traumatic bowel perforation, bowel ischemia, bacterial peritonitis, ascending cholangitis, gastroparesis, pyelonephritis and/or kidney abscess, or other gastrointestinal surgery with complications. Twenty-one patients (84%) were immunosuppressed, including: 12 (48%) with underlying cancer and/or ongoing chemotherapy; 12 (48%) with diabetes mellitus (including 4 with an associated malignancy, 2 requiring hemodialysis for end-stage renal disease, 2 requiring immunosuppressive therapy for prior kidney-pancreas transplants, and 1 with hip osteomyelitis); and 1 patient requiring dialysis for end-stage renal disease due to hypertension. In-hospital mortality of patients with DNSE infection or colonization was high, occurring in 10 of 25 patients (40%).
Daptomycin exposure was confirmed in 15 of 25 (60%) patients prior to the isolation of DNSE. Of these, 10 of 15 (67%) had DNSE isolated during treatment with daptomycin after receiving an average of 13.9 days of therapy (range 3–40 days). The remaining 5 patients had recently received daptomycin with an average drug-free interval of 7.8 days (range 3–14 days) prior to DNSE isolation. In patients with prior daptomycin exposure, the total days of daptomycin therapy in the year prior to isolation of DNSE varied between 5–67 days, with a mean of 20.9 days and median of 16 days. Ten patients had no documented daptomycin exposure at our institution, and no evidence in their medical records that they received daptomycin prior to admission at UIHC. However, detailed records of care prior to UIHC admission were not always available and therefore daptomycin exposure could not be completely excluded in these cases.
Of patients with prior daptomycin exposure, 8 of 15 (53%) were female, compared to 6 of 10 (60%) of patients without prior daptomycin exposure (Table 2). Compared to patients without prior daptomycin exposure, patients with prior daptomycin exposure were less likely to harbor E. faecalis (0% vs. 33%; p = 0.019). In patients with DNSE and prior daptomycin exposure, there was a non-significant trend toward having a bloodstream isolate (47% vs. 20%; p = 0.174), a history of immunosuppression from any cause (93% vs. 70%; p = 0.119), and death (53% vs. 20%; p = 0.096).
Table 2.
Characteristics of patients with respect to prior daptomycin exposure
| Prior Daptomycin Exposure | No Daptomycin Exposure | P value* | |
|---|---|---|---|
| Number of Isolates |
15 (60%) |
10 (40%) |
-- |
| Age |
52.5 |
51.8 |
-- |
| Sex – no (%) |
|
|
|
| -Female |
8/15 (53%) |
6/10 (60%) |
0.742 |
| Isolate |
|
|
|
| -E. faecium |
14 (93%) |
4 (67%) |
0.115 |
| -E. faecalis |
0 (0%) |
2 (33%) |
0.019 |
| -Species Not Identified |
1 (7%) |
0 (0%) |
-- |
| -Isolate Not Availablea |
0 |
4 |
-- |
| Source |
|
|
|
| -Bloodb |
7 (47%) |
2 (20%) |
0.174 |
| -Urineb |
5 (33%) |
7 (70%) |
0.072 |
| -Peritoneal Fluid |
2 (13%) |
1 (10%) |
0.802 |
| -Wound |
2 (13%) |
0 (0%) |
0.229 |
| -Colonizationc |
4 (27%) |
4 (40%) |
0.484 |
| Past Medical History |
|
|
|
| -Diabetes |
6 (40%) |
6 (60%) |
0.327 |
| -ESRD/Dialysis |
2 (13%) |
1 (10%) |
0.802 |
| -Cancer/Chemotherapyd |
8 (53%) |
4 (40%) |
0.513 |
| -History of Transplant |
2 (13%) |
0 (0%) |
0.229 |
| -CVDe |
3 (20%) |
4 (40%) |
0.275 |
| -Lung Diseasef |
2 (13%) |
2 (20%) |
0.656 |
| GI or Intra-abdominal Processg |
12 (80%) |
7 (70%) |
0.566 |
| Immunosuppressionh |
14 (93%) |
7 (70%) |
0.119 |
| Deathi | 8 (53%) | 2 (20%) | 0.096 |
Note:
* - All P values were calculated with the use of the chi-square test.
a – Four isolates were not available for species identification.
b – One patient with DNSE and prior daptomycin exposure had DNSE isolated from both blood and urine.
c – All colonizing isolates were obtained from urine of patients without symptoms and/or negative urinalysis.
d – includes all patients with acute lymphoblastic leukemia, acute myelogenous leukemia, myelodysplastic syndrome, bone marrow transplant, stem cell transplant, solid organ tumor, and those undergoing chemotherapy.
e – includes all patients with coronary artery disease, congestive heart failure, cerebro-vascular accident, peripheral vascular disease, and valvular heart disease.
f – includes all patients with chronic obstructive pulmonary disease and pulmonary hypertension.
g – includes all patients with Clostridium difficile infection, graft-versus-host-disease of the gut, neutropenic enterocolitis/perforation, traumatic bowel perforation, bowel ischemia, bacterial peritonitis, ascending cholangitis, gastroparesis, pyelonephritis and/or kidney abscess, or other gastrointestinal surgery with complications.
h – includes all patients with any cancer or undergoing chemotherapy, diabetes mellitus, ESRD/HD, organ transplant.
i – Hospital and/or thirty-day mortality from any cause.
The number of patients with DNSE colonization or infection at UIHC increased from 2.33 cases per year for the time period 2005–2007 to 4.33 cases per year for the time period 2008–2010 (Figure 1). An additional five cases were identified in the first 6 months of 2011 alone – as many as had been noted over any 12-month period previously. Rates of daptomycin usage at UIHC increased during the study period as well, from approximately 0.2% of inpatients in 2005 receiving daptomycin to 0.9% in 2011 (Figure 1).
Figure 1.
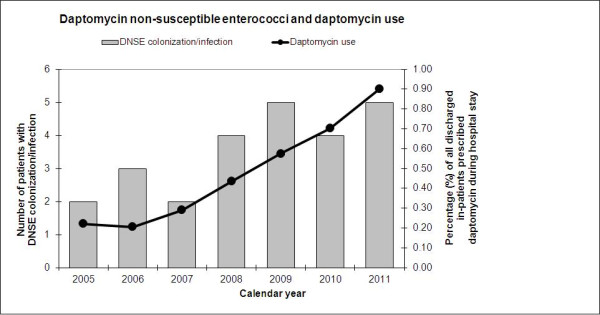
Number of patients with colonization/infection due to daptomycin non-susceptible enterococci (solid bars; left vertical axis), and percentage of all discharged in-patients prescribed daptomycin during their hospital stay (solid line; right vertical axis) for each study year. Information for 2011 based on information for the first 6 months of calendar year.
Twenty-one patients had enterococcal isolates available for species identification. Of these, eighteen patients (86%) had E. faecium, and two (10%) had E. faecalis (Table 3). Vancomycin resistance was common, occurring in DNSE isolates from 18 of 25 patients (72%), including 16 of 18 (89%) E. faecium isolates. Ampicillin resistance was detected in DNSE isolates from 18 of 25 patients (72%), and in 17 of 18 (94%) E. faecium isolates. Linezolid resistance was uncommon, occurring in only 1 DNSE isolate from 21 patients. Linezolid susceptibility was not performed for 4 patient’s isolates, as they were not available for further susceptibility testing.
Table 3.
Antimicrobial resistance profile of enterococcal isolates to selected antimicrobial agents by species
| Enterococcus species |
Proportion of isolates resistant to selected antimicrobial agents |
|||
|---|---|---|---|---|
| |
Daptomycin |
Vancomycin |
Ampicillin |
Linezolid |
| (%) | (%) | (%) | (%) | |
|
E. faecium (n = 18) |
100 |
89 |
94 |
0 |
|
E. faecalis (n = 2) |
100 |
50 |
0 |
0 |
| Other (n = 5)a | 100 | 20 | 20 | 5b |
NOTE:
a – One Enterococcus isolate was identified as non-faecium / non-faecalis, and four were not available for species identification or linezolid susceptibility testing.
b – Of the 21 patient isolates available for susceptibility testing; only the non-faecium / non-faecalis Enterococcus isolate was resistant to linezolid.
PFGE revealed significant genetic heterogeneity, with 16 PFGE types and 24 subtypes represented among the 29 patient isolates available for typing (results not shown). Multiple isolates from the same patient were always of the same type or subtype. There were only four instances in which more than one patient shared the same PFGE type with the most common PFGE type shared among 4 patients. In only one instance did two patients share the same PFGE subtype.
Discussion
In the past 20 years, there have been increases in both the frequency of enterococcal infections and in rates of enterococcal drug resistance [12]. VRE is now the third most common cause of nosocomial bloodstream infection [13], and compared to vancomycin-susceptible enterococci (VSE), is associated with increased healthcare costs, morbidity, and mortality [14]. Enterococci are of particular concern due to their ability to spread drug resistance to other bacteria via mobile genetic elements, resulting in beta-lactam, aminoglycoside, and glycopeptide resistance.
Vancomycin resistance in both Enterococcus species and S. aureus is well described, mediated by van-type plasmids from VRE [15]. In S. aureus, daptomycin resistance is possibly mediated by several mechanisms, including cell wall thickening and charge alterations [16,17]. The exact mechanism of daptomycin resistance in enterococci is not entirely known, but may be related to alterations in cell membrane charge, thickness, and permeability via mutations to cell membrane cardiolipin synthetase and/or other proteins involved in regulating phospholipid metabolism or the stress response to antimicrobial agents [18,19]. Daptomycin exposure is likely to be essential to the development of resistance in enterococci as suggested by the findings in the aforementioned studies and our finding of a high rate (60%) of confirmed prior daptomycin exposure in patients harboring a DNSE isolate as well as the genetic heterogeneity noted on PFGE.
Little is known regarding risk factors or causes for the development of DNSE. Prior daptomycin exposure has been described in case reports of DNSE bacteremia; however cases describing resistance developing de novo have been reported [5-9]. In the largest case series to date of DNSE bacteremia, performed at Memorial Sloan-Kettering Cancer Center during a 3 year period (2007–2009), the authors found prior daptomycin exposure to be an uncommon event with only 2 of 18 (11%) patients having had documented daptomycin exposure prior to the development of DNSE bacteremia [7]. These findings are in contrast to our observation at UIHC, where during a similar time period (2005–2011), the majority of patients with DNSE colonization or infection had confirmed daptomycin exposure prior to the isolation of a non-susceptible isolate (any clinical sample; 15 of 25 or 60%, blood stream 7 of 9 or 78%). This may be an under-estimate however, as the rate of confirmed daptomycin exposure in the era of electronic order entry (February 2009 to present) at UIHC was substantially higher than before its institution (86% vs. 27%). A recent case series and case control study comparing DNSE in patients either with or without prior daptomycin exposure showed results similar to ours, with prior daptomycin exposure in 59%, immunosuppression in 78%, an average age of 58.9 years, E. faecium in 78%, and death in 44% [8,9].
Since daptomycin was introduced for clinical use in 2003, significant increases in rates of resistance or MIC creep have not been noted in the United States with an overall prevalence of DNSE estimated at < 1% [4,20,21]. However in other areas, in particular Asia and parts of Europe, resistance rates may be significantly higher than in the US [4]. Over the past 6 years at our institution, we have noticed an increase in the absolute number of patients colonized or infected with DNSE, which has taken place in the setting of increasing rates of daptomycin usage during the same period (Figure 1).
In the US, approximately 90% of DNSE isolates are also resistant to vancomycin [20,21]. And like VRE, daptomycin non-susceptible isolates are more often E. faecium than E. faecalis (88% vs. 9%) [4]. At our institution a large majority (99%) of clinical enterococcal isolates remain daptomycin susceptible, and the observed species identification and susceptibility of the daptomycin non-susceptible isolates observed in our study align with that reported nationally (Table 3) [4,20].
In addition to prior daptomycin exposure, several findings in patients with DNSE colonization or infection at our institution are worth noting and may provide clues to potential factors associated with the development of DNSE. The majority of patients were immunosuppressed (21 of 25; 84%) or had a concomitant gastrointestinal inflammatory process (19 of 25; 76%). Also, a genitourinary process was noted in 3 of 25 (12%) patients. These results are not surprising given that enterococci commonly colonize the gastrointestinal and genitourinary tracts, and that the majority of these patients had risk factors for the development of gastrointestinal or genitourinary complications or infections.
Molecular typing revealed that daptomycin non-susceptibility emerged among diverse strains of enterococci, with patient-to-patient transmission of DNSE occurring less often. This suggests that cases of DNSE were likely the result of mutations in patient’s own flora under antimicrobial pressure, as opposed to patient-to-patient transfer of an outbreak or common strain. Finally, in-hospital mortality of patients with DNSE infection or colonization was high, at 40%. We are not aware of outcome studies related to the presence of daptomycin non-susceptibility versus daptomycin susceptibility in enterococcal colonization or infection.
Conclusion
To our knowledge, this is the largest study to date to evaluate potential factors associated with the development of DNSE colonization and/or infection and to compare the characteristics of patients with prior daptomycin exposure to those without prior daptomycin exposure. We found that prior daptomycin exposure, immunosuppression, or a concomitant gastrointestinal or intra-abdominal process, preceded the development of DNSE. During the study period, we noticed an increase in the number of cases of DNSE yearly that corresponded to increasing annual rates of daptomycin usage at our institution. Molecular typing revealed a diverse population of isolates suggesting the development of resistance under antimicrobial pressure. Also, a high percentage of patients with DNSE colonization or infection died during their hospitalization. Limitations of this study include its retrospective nature and small number of patients. Future case–control or prospective studies comparing patients with and without DNSE and/or prior daptomycin exposure would be helpful in better identifying factors associated with DNSE infection and/or colonization.
Competing interest
Dr. Diekema has received research funding from Merck, Pfizer, Cerexa, Innovative Biosensors, and bioMérieux. All other authors have no disclosures or conflict of interest.
Authors’ contribution
All authors have read and approve the submission of the manuscript. The manuscript has not been published elsewhere and that it is not currently under consideration for publication by another journal.
Contributor Information
Jeremy C Storm, Email: jcstorm79@gmail.com.
Daniel J Diekema, Email: daniel-diekema@uiowa.edu.
Jennifer S Kroeger, Email: jennifer-kroeger@uiowa.edu.
Sarah J Johnson, Email: sarah-j-johnson@uiowa.edu.
Birgir Johannsson, Email: birgir-johannsson@uiowa.edu.
Acknowledgements
The authors thank Kristopher Heilmann for his assistance with laboratory testing of these isolates.
Financial support
None.
References
- Gold HS. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis. 2001;33(2):210–219. doi: 10.1086/321815. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Staphylococcus aureus resistant to vancomycin—United States, 2002. MMWR Morb Mortal Wkly Rep. 2002;51(26):565–567. [PubMed] [Google Scholar]
- Linden PK. Optimizing therapy for Vancomycin Resistant Enterococci (VRE) Sem Resp Crit Care Med. 2007;28(6):632–645. doi: 10.1055/s-2007-996410. [DOI] [PubMed] [Google Scholar]
- Kelesidis T, Humphries R, Uslan DZ, Pegues DA. Daptomycin nonsusceptible enterococci: an emerging challenge for clinicians. Clin Infect Dis. 2011;52(2):228–234. doi: 10.1093/cid/ciq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesho EP, Wortmann GW, Craft D, Moran KA. De Novo daptomycin nonsusceptibility in a clinical isolate. J Clin Microbiol. 2006;44(2):673. doi: 10.1128/JCM.44.2.673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher MH, Corcoran GD, Creagh S, Feeney E. Daptomycin non-susceptible enterococci faecium in a patient with no prior exposure to daptomycin. J Hosp Infect. 2007;65(4):376–378. doi: 10.1016/j.jhin.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Kamboj M, Cohen N, Gilhuley K, Babady NE, Seo SK, Sepkowitz KA. Emergence of daptomycin-resistant VRE: experience of a single institution. Infect Control Hosp Epidemiol. 2011;32(4):391–394. doi: 10.1086/659152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelesidis T, Humphries R, Uslan DZ, Pegues D. De-novo daptomycin nonsusceptible Enterococcal infections. Emerg Infect Dis. 2012;18(4):674–676. doi: 10.3201/eid1804.110932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelesidis T, Chow ALP, Humphries R, Uslan DZ, Pegues D. Case–control study comparing de novo and daptomycin-exposed daptomycin-nonsusceptible Enterococcus infections. Antimicrob Agents Chemother. 2012;56(4):2150–2152. doi: 10.1128/AAC.05918-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility testing – Twentieth Information Supplement. CLSI, Wayne, PA; 2010. CLSI document M100-S20. [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–485. doi: 10.1016/j.ajic.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seiferd H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Carmeli Y, Eliopoulos G, Mozaffari E, Samore M. Health and economic outcomes of vancomycin-resistant enterococci. Arch Intern Med. 2002;162:2223–2238. doi: 10.1001/archinte.162.19.2223. [DOI] [PubMed] [Google Scholar]
- Arthur M, Reynolds PE, Depardieu F, Evers S, Dutka-Malen S, Quintiliani R, Courvalin P. Mechanisms of glycopeptide resistance in enterococci. J Infect. 1996;32:11–16. doi: 10.1016/S0163-4453(96)80003-X. [DOI] [PubMed] [Google Scholar]
- Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(6):2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother. 2008;52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother. 2011;55(7):3345–3356. doi: 10.1128/AAC.00207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365(10):892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader HS, Jones RN. Antimicrobial susceptibility of gram-positive bacteria isolated from US medical centers: results of the Daptomycin Surveillance Program (2007–2008) Diagn Microbiol Infect Dis. 2009;65:158–162. doi: 10.1016/j.diagmicrobio.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Sader HS, Moet GJ, Farrell DJ, Jones RN. Antimicrobial susceptibility of daptomycin and comparator agents tested against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: trend analysis of a 6-year period in US medical centers (2005–2010) Diagn Microbiol Infect Dis. 2011;70:412–416. doi: 10.1016/j.diagmicrobio.2011.02.008. [DOI] [PubMed] [Google Scholar]


