Abstract
Motivation: Recent advances in flow cytometry enable simultaneous single-cell measurement of 30+ surface and intracellular proteins. CytoSPADE is a high-performance implementation of an interface for the Spanning-tree Progression Analysis of Density-normalized Events algorithm for tree-based analysis and visualization of this high-dimensional cytometry data.
Availability: Source code and binaries are freely available at http://cytospade.org and via Bioconductor version 2.10 onwards for Linux, OSX and Windows. CytoSPADE is implemented in R, C++ and Java.
Contact: michael.linderman@mssm.edu
Supplementary Information: Additional documentation available at http://cytospade.org.
1. INTRODUCTION
Recent advances in flow cytometry (Bendall et al., 2011) enable simultaneous single-cell measurement of 30+ surface and intracellular proteins. With such instruments, it is possible to measure, in a single experiment, enough markers to identify and compare functional immune activities across nearly all cell types in the hematopoietic lineage. However, practical approaches to analyze and visualize cytometry data at this scale are only now becoming available. Qiu et al. (2011) proposed a novel algorithm, termed Spanning-tree Progression Analysis of Density-normalized Events (SPADE), which organizes cells into hierarchies of related phenotypes. The resulting hierarchies, or trees, facilitate visualization of developmental lineages, identification of rare cell types and comparison of functional markers across stimuli.
The SPADE algorithm has four phases: density-dependent downsampling to increase representation of rare cell types, agglomerative clustering to identify related cells, minimum spanning-tree construction to link those clusters and upsampling to assign previously removed cells to clusters. SPADE has been successfully applied to fluorescent and mass cytometry data to automatically recover and display the architecture of the hematopoietic lineage and other complex continuums of phenotypes from surface protein expression levels. The resulting tree representation provides an intuitive structure on which to overlay measurements of surface and functional proteins to identify populations and behaviors of interest.
As cytometry datasets increase in size and dimensionality, the performance of the computational tools researchers apply are of increasing importance; long waits for results, particularly for exploratory tools such as SPADE, negatively impact researcher productivity. In this note, we present CytoSPADE, a robust, modular, cross-platform and high-performance implementation of the SPADE algorithm and an accompanying graphical user interface that improves performance by 12–19-fold relative to the SPADE prototype, enabling gigabyte-scale datasets to be analyzed and effectively visualized in hours or minutes, not days.
2. CYTOSPADE IMPLEMENTATION
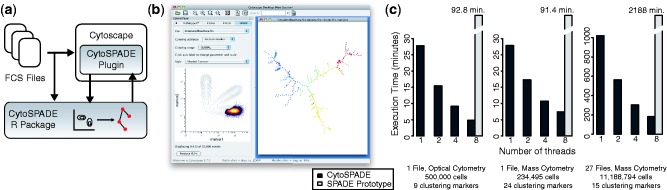
Figure 1 shows the structure, use and execution time of CytoSPADE. The SPADE workflow is orchestrated by our plugin for the Cytoscape network visualization platform (Cline et al., 2007). The plugin imports local FCS files, invokes our multicore-optimized SPADE R package and enables interactive visualization of the resulting SPADE trees in the context of the underlying cytometry data. The R package can be used independently of the Cytoscape plugin, and other interfaces, specifically an HTML5-based web client integrated with the Cytobank online flow cytometry platform (Kotecha et al., 2010), are under development.
Fig. 1.
Structure (a) of CytoSPADE, including the R-package and the user interface (b) implemented as a Cytoscape plugin. Using the Cytoscape plugin, users can simultaneously view the SPADE tree (right panel) and the underlying cytometry data (biaxial plot in left panel). The R package can be used independently of the Cytoscape plugin. Bar charts (c) show the performance of the multi-threaded R package on a dual-socket Intel Xeon 2.27 GHz server with 12 GB of RAM for a variety of cytometry datasets and clustering parameters, including high-dimensional mass cytometry datasets with millions of cells.
The common feature of these interfaces is the capability to simultaneously view the resulting SPADE trees and the underlying cytometry data and then interactively ‘gate’ the cytometry data by their cluster assignment. In Figure 1b, the user has selected the lower branch of the tree; the cells associated with those clusters or nodes are shown in the biaxial plot of the left-hand side of the interface. The size of a node reflects the relative number of cells assigned to that node, whereas the color reflects the median, fold-change or other statistic for a given parameter for that node. This multi-modal, multi-scale visualization enables users to interactively visualize the behavior of and relationships between many different cell types in the immune system in a single graphic, as opposed to hundreds, and to do so in the context of the underlying cytometry data.
Alongside interactively ‘gating’, researchers can use the Cytoscape plugin to manipulate the tree by moving nodes and changing the node color and size mappings; create ‘nested nodes’ that collapse uniform phenotypes into a single node; interactively view statistical tests of parameter significance for groups of nodes and apply other visual or quantitative operations to the SPADE tree. A researcher might use these various capabilities to (1) identify different cell types, e.g. T cells and B cells, and visually organize them in a familiar pattern (as performed in Bendall et al., 2011), then (2) overlay various surface and functional parameters to quickly visually identify differential cell populations or behavior that may be associated with a particular disease and (3) explore the underlying flow cytometry data for populations of interest to generate hypotheses for follow-up experiments or for analyses with other tools such as Gemstone or flowClust (Lo et al., 2009).
The CytoSPADE R package is a faithful implementation of the SPADE algorithm that also incorporates substantial performance improvements and alternative optional implementations for specific components, e.g. clustering. CytoSPADE builds on top of the widely used flowCore R package (Hahne et al., 2009) and thus incorporates the many different data transforms and visualization modalities it provides. The R package is modular in design so that users can run the standard SPADE workflow—density-dependent downsampling, clustering and tree construction, and upsampling—or individual phases as needed. Examples of alternative workflows currently in use include assigning newly collected data to previously generated clusters or incorporating additional parameters, such as time, in tree construction.
The computationally demanding components of the SPADE algorithm are implemented in C++ using OpenMP to enable multi-threaded execution. As shown in Figure 1c, the combination of a careful C++ implementation and multi-threaded execution results in a substantial speedup relative to the prototype implementation. Users can run CytoSPADE on most datasets in just minutes. If OpenMP is not available, as is often the case on the Windows platform, CytoSPADE will default to using a single core. However, even in single-threaded mode, the C++ implementation offers a significant performance improvement for SPADE users. Additional information about enabling OpenMP is available in the online documentation.
3. CONCLUSION
CytoSPADE is a high-performance, modular and cross-platform implementation of the SPADE algorithm and an accompanying graphical user interface. Source code and binaries for CytoSPADE are available under the GPL license from http://cytospade.org and as part of Bioconductor release 2.10 onwards. Documentation, including the package vignette, wiki and getting started tutorials are also available at http://cytospade.org.
Funding: This work is supported by the Rachford and Carlota A. Harris Endowed Professorship; National Institutes of Health U19AI057229, P01 CA034233, HHSN272200700038C, 1R01CA130826; CIRM DR1-01477 and RB2-01592; NCI RFA CA 09-011; NHLBI-HV-10-05(2); European Commission HEALTH.2010.1.2-1 and the Bill and Melinda Gates Foundation (GF12141-137101) and NLM Training Grant 5T15LM007033-27.
Conflict of Interest: none declared.
REFERENCES
- Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, et al. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10, Unit10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K, et al. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics. 2009;10:145. doi: 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]