Abstract
Summary: To address the impending need for exploring rapidly increased transcriptomics data generated for non-model organisms, we developed CBrowse, an AJAX-based web browser for visualizing and analyzing transcriptome assemblies and contigs. Designed in a standard three-tier architecture with a data pre-processing pipeline, CBrowse is essentially a Rich Internet Application that offers many seamlessly integrated web interfaces and allows users to navigate, sort, filter, search and visualize data smoothly. The pre-processing pipeline takes the contig sequence file in FASTA format and its relevant SAM/BAM file as the input; detects putative polymorphisms, simple sequence repeats and sequencing errors in contigs and generates image, JSON and database-compatible CSV text files that are directly utilized by different web interfaces. CBowse is a generic visualization and analysis tool that facilitates close examination of assembly quality, genetic polymorphisms, sequence repeats and/or sequencing errors in transcriptome sequencing projects.
Availability: CBrowse is distributed under the GNU General Public License, available at http://bioinfolab.muohio.edu/CBrowse/
Contact: liangc@muohio.edu or liangc.mu@gmail.com; glji@xmu.edu.cn
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Web-based genome browsers, such as GBrowse (Stein et al., 2002) and UCSC Genome Browser (Kent et al., 2002), are widely utilized for visualizing genomes and their sequence features to facilitate various data analyses that address interesting biological questions. For non-model organisms without sequenced genomes, transcriptome sequencing is definitely the most efficient way to explore the transcribed portions of genomes and determine their dynamics (Bräutigam et al., 2011; Feldmeyer et al., 2011; Parchman et al., 2010). Many bioinformatics programs have been developed/improved to address the challenges in transcriptome assembly, especially de novo assembly without a reference genome, using complementary DNA (cDNA)/messenger RNA (mRNA) data from next-generation sequencing and Sanger sequencing (Bräutigam et al., 2011; Feldmeyer et al., 2011; Martin and Wang, 2011; Zheng et al., 2011). So far, there is no open-source, web-based contig browser yet that allows users to navigate transcript assembly, visualize contigs and examine genetic polymorphisms, simple sequence repeats and sequencing errors embedded in the assembly. To address the impending need for exploring rapidly increased transcriptomics data for non-model organisms, we developed CBrowse (contig browser), an AJAX-based web browser to visualize and analyze transcriptome assemblies and their individual contigs.
2 IMPLEMENTATION
As shown in Supplementary Figure S1, CBrowse is designed to follow a standard three-tier software architecture composed of Data Layer, Business Logic Layer and Presentation layer, with a data pre-processing pipeline. The data pre-processing pipeline detects simple sequence repeats for contigs, makes inferences from read alignments about putative polymorphisms and sequencing errors and stores resultant data in a hard-drive file system (HDFS), which can be optionally imported into a SQL-based database (e.g. MySQL or PostgreSQL). Data layer enables data accessing through HDFS or a database, Business Logic Layer processes users' requests submitted from Presentation Layer and Presentation Layer displays the desired data in different web interfaces.
Since Sequence Alignment/Map (SAM) format and its sister format Binary Sequence Alignment/Map (BAM) are widely adopted in presenting sequence alignment information for both genome and transcriptome assembly (Barnett et al., 2011; Li et al., 2009), the input files for the pre-processing pipeline are as follows: (i) a SAM/BAM file that contains alignment information for all individual cDNA/mRNA reads mapped to the contigs, (ii) a sequence file in FASTA format that contains all contigs within a transcriptome assembly and (iii) a Extensible Markup Language (XML) configure file that provides necessary information (e.g. species name, assembly name and data location) for data processing (Supplementary Fig. S1). Implemented in C++ with Perl wraps, the pipeline can process input data; detect polymorphisms, simple sequence repeats and sequencing errors and generate image, JSON and database-compatible CSV text files that are utilized by different web viewers of CBrowse (Fig 1). Our C++ program relies on the application programming interface (API) of BamTools (Barnett et al., 2011) to access BAM files, uses tinyXML library (http://www.grinninglizard.com/tinyxml/) to generate and parse configuration files and map index files in XML format and utilizes GD library (http://www.libgd.org) to draw alignment graphics in PNG format. The pipeline not only extracts overall information for a transcriptome assembly (e.g. total number of contigs and associated reads, average reads per contig and contig length distribution) and calculates its N50 length but also retrieves summary information for each contig and computes its sequence coverage. For simple sequence repeats, our pipeline invokes Phobos (Mayer et al., 2010) to identify perfect/imperfect repeats and generates results in GFF format. The repeat unit size and the minimum repeat number are customizable using the configuration XML file. By default, the repeat unit size is between 1 and 12 nt, while the minimum repeat number is set to be 8 for mono-nucleotides, 5 for dimers, 4 for triplets and 3 for repeats with a unit size of 4–12 nt. For putative polymorphisms and sequencing errors, our C++ program examines base by base for any discrepancy between each contig and its component sequence reads. Along a given contig, the C++ program identifies all putative polymorphic positions, which must be covered by ≥10 individual sequence reads and the accumulated occurrence of any polymorphic type is ≥5. The valid polymorphism types include single-nucleotide polymorphisms (SNPs, single-base mismatch), single base indel and multiple-base mismatch and indels. The frequency of any valid polymorphism type needs to be at least 2 for any putative polymorphic position along a contig. Our pipeline also invokes SAMTools and BCFTools to call SNPs and short indels and generate results in VCF format, which can be explored through our Polymorphism Viewer (see below).
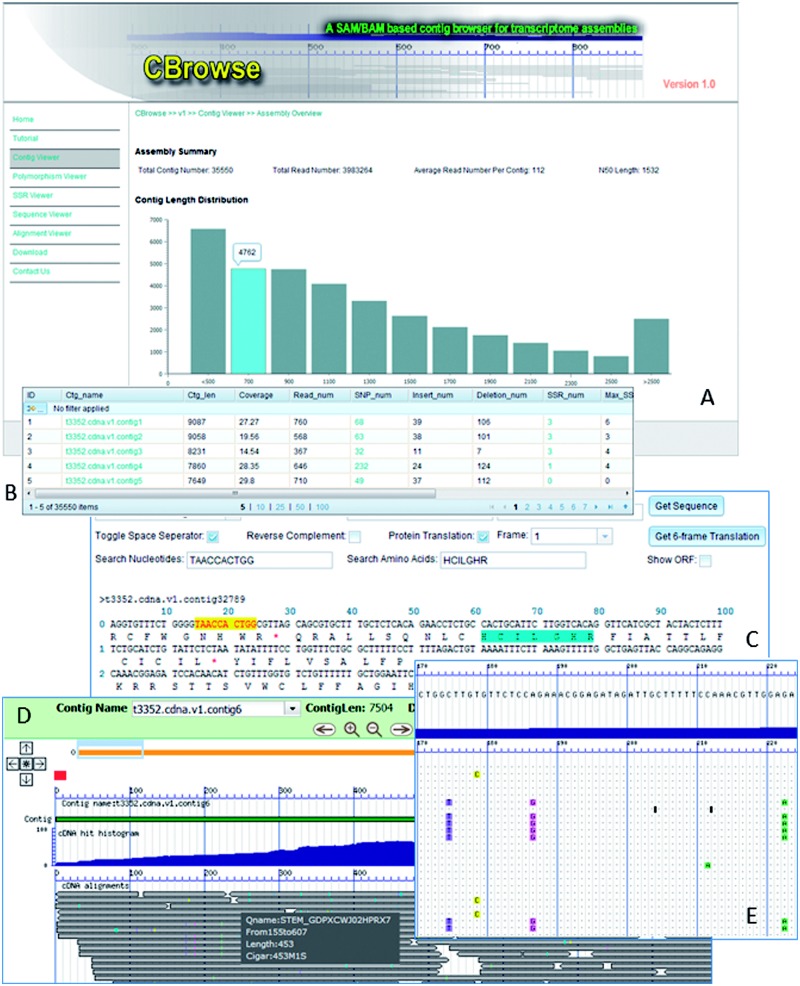
Implemented in PHP and JavaScript (i.e. Dojo and MapEasy libraries), CBrowse web interfaces are AJAX-based and compatible with Mozilla Firefox (8.0 or above), Google Chrome and Internet Explorer (9.0 or above). As shown in Figure 1, there are five major viewers in CBrowse: (i) Contig Viewer presents summary information about the assembly as well as individual contigs; (ii) Sequence Viewer allows users to manipulate, examine and scan individual nucleotides of each contig; (iii) Polymorphism Viewer provides the polymorphism types (SNPs, single indels and other multiple-base mismatches/indels), positions and frequencies for each contig; (iv) SSR Viewer displays the detected simple sequence repeats in each contig and (v) Alignment Viewer offers both bird's eye and nucleotide resolution of sequence alignments in Google-map style, with color-coded nucleotide differences between contigs and their component sequence reads. In particular, all these interfaces allow users to search and navigate data easily.
3 CONCLUSION
Similar to GBrowse 2.0 (http://gmod.org/wiki/GBrowse), X-MAP (Yates et al., 2008), Genome Projector (Arakawa et al., 2009) and JBrowse (Skinner et al., 2009), CBrowse essentially is an AJAX-based Rich Internet Application that decouples interactions with users from interactions with the server. Such decoupling empowers web applications with rich graphic user interface (GUI) characteristics such as desktop application, enables a asynchronous client–server communication and offers faster and smoother user experience by partial updates in web pages. Different from these genome browsers, CBrowse is designed for analyzing and visualizing transcriptome assembly and contigs, with unique functionality such as seamlessly integrated data grid viewer, alignment viewer and sequence viewer that allow users to navigate, sort, filter, search and visualize transcriptome data efficiently. As an open source project, our web-based CBrowse can be utilized by the research community to disseminate and release transcriptome data over the Internet.
Fig. 1.
The snapshots of CBrowse web interfaces Panel A: the major web portal shows contig length distribution for an assembly. Panel B: The data grid view shows contig summary information. Panel C: the sequence viewer shows its functionality with one contig. Panel D: the alignment view in bird's-eye resolution. Panel E: the alignment view in nucleotide resolution that displays and highlights the differences between contig sequence and individual sequence reads
Funding: This project was funded partially by the NIH-AREA (1R15GM94732-1 A1 to C.L.), the Key Project of Chinese National Programs for Fundamental Research and Development (973 Program: No. 2009CB118902), the National Natural Science Foundation of China (No. 61174161 to J.G.), the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20090121110022 to J.G.), and the Fundamental Research Funds for the Central Universities in China (Xiamen University: No. 2011121047, No. 201112G018 and No. CXB2011035 to J.G.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Arakawa K, et al. Genome Projector: zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31. doi: 10.1186/1471-2105-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett DW, et al. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, et al. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C(3) and C(4) species. J. Exp. Bot. 2011;62:3093–3102. doi: 10.1093/jxb/err029. [DOI] [PubMed] [Google Scholar]
- Feldmeyer B, et al. Short read Illumina data for the de novo assembly of a non-model snail species transcriptome (Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics. 2011;12:317. doi: 10.1186/1471-2164-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Wang Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Mayer C, et al. Genome-wide analysis of tandem repeats in Daphnia pulex–a comparative approach. BMC Genom. 2010;11:277. doi: 10.1186/1471-2164-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchman TL, et al. Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genom. 2010;11:180. doi: 10.1186/1471-2164-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, et al. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T, et al. X-Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–D786. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, et al. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics. 2011;12:453. doi: 10.1186/1471-2105-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.