Abstract
Dystroglycan is a major cell surface glycoprotein receptor for the extracellular matrix in skeletal muscle. Defects in dystroglycan glycosylation cause muscular dystrophy and alterations in dystroglycan glycosylation can impact extracellular matrix binding. Here we describe an immunoprecipitation technique that allows isolation of beta dystroglycan with members of the dystrophin-associated protein complex (DAPC) from detergent solubilized skeletal muscle.
Immunoprecipitation, coupled with shotgun proteomics, has allowed us to identify new dystroglycan-associated proteins and define changed associations that occur within the DAPC in dystrophic skeletal muscles. In addition, we describe changes that result from overexpression of Galgt2, a normally synaptic muscle glycosyltransferase that can modify alpha dystroglycan and inhibit the development of muscular dystrophy when it is overexpressed. These studies identify new dystroglycan-associated proteins that may participate in dystroglycan’s roles, both positive and negative, in muscular dystrophy.
INTRODUCTION
Muscular dystrophy refers to a group of diseases affecting skeletal muscles that are characterized by progressive muscle weakness leading to loss of motor function and often to premature death1-6. A subset of muscular dystrophies have been associated with mutations in genes coding for a group of proteins that form a large oligomeric complex, anchored by dystrophin, at the sarcolemmal membrane of muscle fibers - the dystrophin associated protein complex (DAPC)7, 8. The core DAPC comprises intracellular (dystrophin, dystrobrevins, syntrophins), transmembrane (beta dystroglycan, alpha, beta, gamma and delta sarcoglycans, sarcospan) and extracellular (alpha dystroglycan) proteins. The DAPC plays an important structural role in skeletal muscle, as it links the F-actin cytoskeleton to the extracellular matrix via dystrophin, beta dystroglycan and alpha dystroglycan. Mutations that destabilize this complex typically lead to loss of muscle membrane integrity, dysregulation of intracellular calcium homeostasis and ultimately myofiber necrosis1. As a result, intense research efforts have focused on understanding the biology and structure of the DAPC complex in order to develop approaches to stabilize it at the muscle membrane and to preserve muscle membrane integrity and function.
Within the DAPC, alpha dystroglycan is the primary protein responsible for attachment to the extracellular matrix8, 9. It is subject to extensive and complex post translational processing involving addition of unique carbohydrate moieties that affect ligand affinity10-14. We have previously shown that in skeletal muscle the beta1,4-N-acetylgalactosaminyltransferase Galgt2 is one such modifier of alpha dystroglycan, increasing ECM binding through addition of the CT carbohydrate antigen13, 15, 16. Galgt2 is normally concentrated at the neuromuscular junction, the region of the myofiber membrane innervated by the motor nerve terminal, as is the CT glycan Galgt2 helps to synthesize15, 17, 18. Since the synaptic DAPC is preserved in most muscular dystrophies, we generated transgenic mice that overexpress Galgt2 to determine whether glycosylation of alpha dystroglycan by Galgt2 could stabilize the DAPC throughout the muscle fiber membrane, offering a potential therapeutic avenue for muscular dystrophies15, 16. Indeed, transgenic expression of Galgt2 increased the extrasynaptic expression of normally synaptic DAPC proteins and inhibited the development of muscle pathology in dystrophin-deficient mdx mice16, 19, a model for Duchenne muscular dystrophy5, 20, as well as in several other mouse muscular dystrophy models21-24. In Galgt2 transgenic mdx mice, inhibition of muscular dystrophy correlates with reduced muscle damage as well as increased specific force of isolated skeletal muscles to levels exceeding wild type muscles25.
Our previous studies indicated that Galgt2 may act by modulating binding of muscle extracellular matrix proteins to alpha dystroglycan and favor intracellular interactions of beta dystroglycan with cytoskeletal proteins other than dystrophin, such as utrophin and plectin, that might stabilize the transmembrane DAPC complex 13, 21, 24. To gain further insights into the mechanisms by which alpha dystroglycan glycosylation by Galgt2 may reinforce the muscle membrane and increase muscle strength, we sought an approach that would allow us to isolate and compare the composition of the DAPC in wild type, dystrophin-deficient mdx and Galgt2 transgenic mice. Mass spectrometry (MS) offers some very unique advantages for protein identification such as specificity, sensitivity, and resolving power, and has been applied successfully to the study of skeletal muscle26, 27. Most comparative analyses of dystrophic muscle tissues have relied on 2D gel electrophoresis followed by matrix assisted laser desorption/ionization (MALDI/MS) or micro liquid chromatography/electrospray ionization (uLC/ESI-MS) to identify proteins whose expression is affected by disease 28-32. These approaches, however, typically fail to detect dystrophin and fail to detect other DAPC members as differentially expressed in control versus dystrophin-deficient muscle. This is in part due to the fact that integral membrane proteins, low abundance proteins, and proteins with extreme pH values are inherently difficult to detect by MS following protein separation by gel electrophoresis33, 34. To date, detection of dystrophin and DAPC members by MS has required a protocol involving large amounts of starting material, tissue sub fractionation to isolate the membrane fraction, and lectin-pull downs to enrich for glycosylated membrane proteins35. Here we describe an optimized, rapid, approach that combines the specificity of antibody immunoprecipitation to purify proteins complexed to dystroglycan with on line nanoflow reverse phase LC-MS/MS for the reliable identification of DAPC members from small amounts of muscle tissue. This approach has allowed us, for the first time, to compare by immunoprecipitation the composition of the DAPC complex between muscles that are altered by deletion or overexpression of single genes relevant to muscular dystrophy.
EXPERIMENTAL PROCEDURES
Products
Sequencing grade–modified trypsin was obtained from Promega. C18 (Zorbax SB, 5 μm) column resins were purchased from Agilent. Dynabead resin conjugated with Protein G and Protein G sepharose were purchased from Invitrogen. Anti-beta dystroglycan (MANDAG236) and species-matched IgG control (MW837) antibodies used for immunoprecipitations were produced in-house from hybridoma cell lines (Developmental Studies Hybridoma Bank, DSHB) and concentrated using the Amicon ultra-filtration cell (Millipore). Antibodies used for immunoblotting were: anti-beta dystroglycan (43DAG or MANDAG2), anti-dystrophin (Dys1 or MANEX1011b) from Novo Castra and DSHB, anti-utrophin from DSHB, anti alpha dystroglycan (clone IIH6C4) from Upstate Biotechnology and anti alpha dystrobrevin (610766) from BD Bioscience. Antibodies specific to alpha- and beta-syntrophins were a kind gift of Dr. Stanley Froehner (University of Washington) and have been previously characterized by his group 38-40. Antibodies to Interleukin 15 receptor alpha were from Bioss or R & D systems, antibodies to Spire2 were from Abgent or Proteintech, and antibody to alpha B crystallin was from Enzo Life Sciences. Antibody to synaptopodin 2 was from Prosci, antibody to rhomboid related protein 1 was from Abcam. Secondary HRP-conjugated anti-mouse or anti-rabbit IgG or IgM were purchased from Jackson Immunoresearch.
Animals
Wild type mice were maintained as a C57Bl/6 strain. Mice deficient in dystrophin (mdx, C57Bl/10ScSn-Dmdmdx/J, stock 001801) were obtained from Jackson laboratory and maintained as a C57Bl/10 strain. Galgt2 transgenic (wild type and mdx) mice were made and described previously15, 16. While originally made as C57Bl/6 × Balb/c hybrid mice, Galgt2-WT mice were bred for more than 10 generations on a C57Bl/6 background, while Galgt2-mdx were bred for more than 10 generations on a C57Bl/10 background. Mice with hindlimb muscle deleted for dystroglycan were made by crossing P3Pro-Cre transgenic mice, which show a caudal to rostral gradient of Cre transgene expression in skeletal muscles, to Dag1loxP/loxP mice41. Mice were bred and cared for in a clean barrier facility and all animal care and experiments were done under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Nationwide Children’s Hospital. Only littermates from identical crosses were compared for each experiment. Mice were given free access to water and food, maintained on a 12h:12h light dark cycle, and were housed in cages with 2-4 animals per cage.
Sample Preparation and Immunoprecipitation
After mice were sacrificed, skeletal muscles (100 mg of quadriceps and gastrocnemius muscles) were cleanly dissected and minced until no large pieces remained. Dissected tissue was then homogenized in protein extraction buffer (Tris-Cl (pH 7.4, 50 mM), NaCl (150 mM), NP-40 (0.05%), protease inhibitors (1:25 cOmplete UTLRA, Roche), phosphatase Inhibitors (1:10, PhosSTOP, Roche) and digitonin (1%) at 4°C for 1hr. Soluble proteins were separated from undissolved tissue by centrifugation at 80,000g for 30 minutes at 4°C. Supernatant was then precleared of mouse antibody by incubation with Protein G sepharose for 30 minutes at 4°C, with centrifugation at 16,000g to remove resin. Protein levels were measured using a BCA protein assay kit (Pierce) and protein amounts normalized to 2 mg/mL solutions for each sample by dilution in protein homogenization buffer.
Magnetized Beads (DynaBeads, Invitrogen) were conjugated with antibody 1 day prior to immunoprecipitation. DynaBeads (300 μL with Protein G) were resuspended in sodium phosphate buffer (50mM, pH 5.0, 1 mL) and washed for 1 min. The beads were then placed on a magnet, supernatant decanted and beads were washed three times with sodium phosphate buffer (50mM, pH 5.0, 0.01% Tween 20). Dynabead slurry (Protein G, binding capacity 0.64mg/mL) was then incubated with MANDAG2 (384 μg for DG IP) and MW8 (for control IP) antibody in sodium phosphate buffer (50mM, pH 5.0) overnight. This is double the estimated binding capacity of the Dynabeads to allow for saturation.
To cross-link MANDAG2 or MW8 to the resin, Dynabead-Protein G-MANDAG2 or –MW8 complexes were washed twice in triethanolamine (pH 8.2, 0.2 M, 1mL) containing Tween 20 (0.01%) and then incubated with DMP (dimethyl pimelimidate × 2HCl, 20 mM) in triethanolamine (pH 8.2, 0.2 M) with gentle mixing for 30 min at 20°C. After the supernatant was discarded, ethanolamine blocking buffer (0.1M, pH 8.2, 0.01% Triton) was added. Beads were washed once and incubated with blocking buffer at RT for 1 hr with agitation. Beads were then washed three times with PBS containing NP-40 (0.01 %). Protein lysate (700 μL of 2 mg/mL) was added to the cross-linked beads and incubated at 4°C on rocker for 3 hr. The solution was then placed on a magnet. After discarding supernatant, beads were washed three times with Tris-Cl (pH 7.4, 50 mM) containing NaCl (150 mM), NP-40 (0.05%).
After the last wash, on bead trypsin digestion was performed. Proteins were first reduced in dithiothreitol (DTT, 10mM in 100 mM ammonium bicarbonate) at 37 °C for 30 minutes. To block reduced cysteine residues, iodoacetimide solution (55 mM in 100 mM NH4HCO3) was added to the samples at room temperature for 15 minutes in the dark. The tryptic digestion was performed by addition of sequencing grade trypsin in NH4HCO3 (50 mM) at 1:20 ratios overnight at 37 °C, after which trypsinization was quenched by formic acid. Beads were removed by centrifugation at 1000 g for 5 min and the supernatant used for LC-MS/MS analysis.
For reverse IL15 receptor alpha or Spire 2 Ips, muscle proteins were solubilized as above and polyclonal antibody to IL15Rα or Spire 2 were conjugated to protein G agarose, as above, instead of MANDAG2. Antibodies used were anti IL15Rα (bs-2605R, Bioss) and anti Spire 2 (17757-1-AP, Proteintech). Precipitations were otherwise carried out as for MANDAG2.
Nano-LC-ESI-MS/MS and data analysis
All MS/MS experiments for peptide identification were performed using an LTQ-MS (Thermo Finnigan) equipped with a nano-ESI source. Samples (10 μL) were loaded by the autosampler (Agilent 1100) onto reversed phase columns (100 μm i.d. × 12 cm). Solvent A consisted of H2O/CH3CN (95:5 v/v) + 0.1 % formic acid and solvent B of H2O/CH3CN (20:80 v/v) + 0.1 % formic acid. Samples were loaded on the column (split closed) at 2 μL/min (100% A) and eluted at ~200 nL/min (HPLC pump at 10 μL/min, splitopen). The RP LC gradient consisted of: 0 to 10% B (0-1 min), 10-45 % B (1-95 min), 45 to 60% B (95-110 min), 60 to 100 % B (110-115 min), 100 % B (115-120 min), 100 to 0 % B (120-121 min), and 100% A (121-150 min).
Data dependent MS acquisition conditions were as follows: 1 MS scan (3 microscans averaged) and 1 MS2 on the top 13 most intense peaks; dynamic exclusion was enabled at repeat count 2, repeat duration 45 s, exclusion list size 150, exclusion duration 45 s and exclusion mass width 1.5 m/z; collision-induced dissociation (CID) parameters were set at isolation width 3 m/z, normalized collision energy 35%, activation Q 0.25, activation time 30 ms. Both spectra were obtained at a heated capillary temperature of 200°C and an ESI voltage of 2.3 kV. The fragment mass spectra were searched against the SEQUEST IPI.MOUSE.v3.27 database (67528, http://www.ebi.ac.uk/IPI/). The database search parameters included: 1. Only fully tryptic fragments were considered for peptide matching, 2. The number of allowed missed cleavage sites was 2, 3. The peptide tolerance was 2 Da, 4. The fragment ion tolerance was 1 Da and 5. The capability to match one peptide sequence to multiple references within database was set at 20. The calculations of FDR from our IP results were not obtained by applying the databases (original database and decoy database) due to the insufficient data distribution. Instead, FDR for peptide matches in SEQUEST were estimated using the reverse sequence database strategy with transitional threshold at Xcorr = 1.9 for z = 1, Xcorr = 2.5 for z =2, Xcorr = 3.5 for z = 342-44. IPI accession numbers for identification proteins were entered from the UniProt database (www.pir.uniprot.org).
Western Blotting
Immunoblots were done as previously described13. Aliquots of immunoprecipitated samples were separated on a 4-12% SDS PAGE gel. For immunoblotting, the gel was transferred onto nitrocellulose membranes blotted with antibodies as previously described13.
Immunostaining and Confocal Microscopy
Skeletal muscles from wild type (WT), Galgt2 transgenic (CT), mdx, Galgt2 transgenic mdx (mdxCT), or dystroglycan deleted mice (from hindlimb muscles of P3ProCreDag1loxP/loxP mice) were dissected and snap frozen in liquid nitrogen cooled isopentane. Muscles were cut in cross section at 8-10 μm on a cryostat and mounted on coated glass slides. Sections were co-stained with antibody to β dystroglycan (MANDAG2) or dystrophin (Dys1) and with antibody to αB crystallin, IL15Rα or Spire2 with species specific Cy2 or Cy3 labeled secondary antibodies. For Spire2/MANDAG2 or Spire2/Dys1 co staining, sections were first fixed in 2% paraformaldehyde for 20 minutes and then denatured with 0.2% SDS for 30 minutes. All other antibodies were used to immunostain unfixed sections. All sections were blocked in 10% horse serum with 0.1% Triton X-100 for 60 minutes prior to addition of primary antibody. Sections were stained with primary antibody in blocking buffer overnight, washed 3 times for 10 minutes each with PBS, incubated with secondary antibodies for 60 minutes, washed and mounted in glycerol with paraphenylenediamene to inhibit fluorescence quenching. Co-localization was determined by visualizing staining using rhodamine- or fluorescein-specific optics on a Zeiss LSM710 or Zeiss LSM510 confocal microscope. Only single z plane co-stained images are shown and all comparative stains shown used time-matched exposures.
RESULTS
DAPC isolation from skeletal muscle by immunoprecipitation
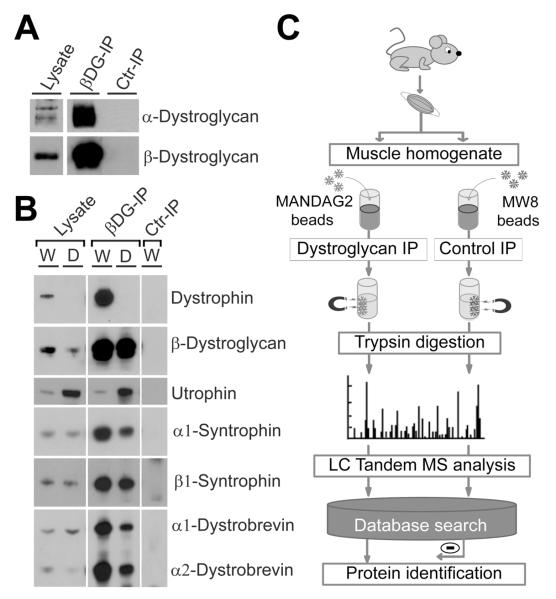
Prior studies have enriched for the DAPC by affinity purification with Wheat germ agglutinin (WGA), a lectin that binds to glycosylated proteins, including alpha dystroglycan7, 8. WGA, however, does not exclusively isolate the DAPC, as this lectin has generic affinity for sialic acids and N-acetylglucosamine. In addition, WGA does not bind well, if at all, to the CT carbohydrate antigen when it is present on alpha dystroglycan15, 16. This is likely due to steric hindrance by the additional beta1,4GalNAc made by Galgt2, which is known to block sialic acid binding of other sialic acid binding lectins (e.g. Selectins)45. To overcome these challenges, we optimized conditions for the isolation of the DAPC by antibody-based immunoprecipitation. We targeted dystroglycan because it is expressed, albeit at reduced levels, in dystrophin-deficient muscle16, allowing analysis of DAPC composition in dystrophic mdx mice. The monoclonal antibody MANDAG2, whose binding epitope is directed against a small peptide region within the intracellular domain of beta dystroglycan36, was able to immunoprecipitate the majority of beta dystroglycan and alpha dystroglycan from mdx muscles (Figure 1A). Using a very mild detergent mixture containing 1% digitonin and 0.05% NP-40, we were able to co-precipitate other DAPC members that bind directly, such as dystrophin and utrophin, or indirectly to beta dystroglycan, such as syntrophins and alpha dystrobrevins (Figure 1B). Dystrophin was present in beta dystroglycan immunoprecipitations (Ips) from wild type muscle (labeled W in Figure 1B), while utrophin was concentrated in beta dystroglycan Ips from mdx muscle (labeled D in Figure 1B), where dystrophin is absent. Control Ips using the monoclonal antibody MW8, which recognizes huntingtin, a brain protein not commonly found in skeletal muscle, did not result in isolation of dystroglycan or DAPC members (Ctr IP in Figures 1A and B).
Figure 1. Dystroglycan immunoprecipitation and scheme for shotgun proteomics by LC tandem MS analysis of dystroglycan-associated proteins.
(A) Both alpha- and beta-dystroglycan were immunoprecipitated from wild type skeletal muscle with the MANDAG2 monoclonal antibody to beta-dystroglycan (βDG-IP), but not with the MW8 monoclonal control antibody (Ctr-IP). (B) beta-dystroglycan and associated members of the DAPC were specifically immunoprecipitated by the MANDAG2 antibody (βDG-IP) from lysates of both wild type (W) and dystrophic (D) mdx mouse skeletal muscles. (C) Scheme for analysis of dystroglycan-associated proteins. Mouse skeletal muscles were dissected and homogenized in a non-ionic detergent buffer that preserves protein interactions. Lysates were incubated with magnetic Dynabeads cross linked to the MANDAG2 antibody against beta dystroglycan or to the control MW8 antibody. After magnetic separation, proteins bound to the Dynabeads were directly digested with trypsin. Resulting peptides were separated and analyzed by liquid chromatography (LC) coupled to tandem mass spectrometry (MS). Peptides were identified using SEQUEST and UNITPROT databases and non specific proteins identified in control MW8 Ips were subtracted to identify proteins specifically associated with dystroglycan. Molecular weights in A and B are 160 kDa (alpha-dystroglycan), 43 kDa (beta-dystroglycan), 400 430 kDa (dystrophin and utrophin), 54kDa (alpha1-syntrophin), 58kDa (beta1 syntrophin), 90kDa (alpha1-dystrobrevin) and 65kDa (alpha2-dystrobrevin).
Identification of DAPC members by shotgun proteomics using tandem MS
For DAPC identification by tandem MS, protocol steps were further optimized to increase protein yield from small amounts of starting tissue material and to enhance identification of relevant proteins. This was achieved primarily by saturating the protein G beads with antibody, cross linking the antibody to the beads using conditions that preserve antigen recognition, and by maximizing depletion of alpha and beta dystroglycan during immunoprecipitation. To maximize protein recovery, on bead digestion with trypsin was performed followed by peptide separation by reverse phase LC and shotgun proteomics using tandem mass spectrometry.
Figure 1C shows the general scheme for sample preparation and analysis that was used on dystroglycan and control Ips from skeletal muscle samples obtained from four experimental groups: wild type, dystrophin-deficient (mdx), Galgt2 transgenic wild type (Galgt2-WT) and Galgt2 transgenic mdx (Galgt2-mdx) mice. We used optimal conditions from our previous studies for a number of parameters related to the LC/MS experimental setup, including data acquisition, database searching, and data filtering systems46. The primary database was a mouse IPI database (release v3.27) with added common contaminant proteins such as trypsin. This database also contained reversed sequences for each entry so that the number of false positives could be assessed. All results we reported were for peptides with less than 5% FDR except for one (Galgt2-WT#1). The FDRs for each experimental group were as follows: wild type [#1 (4.2%), #2 (1.5%)], mdx, [#1 (2.5%), #2 (2.9%), #3 (3.7%)], Galgt2 WT [#1(6.4%), #2 (2.1%)] and for Galgt2-mdx [#1 (3.4%), #2 (2.8%)].
Using this approach we achieved reproducible and specific identification of known DAPC members in all dystroglycan IP replicates performed on skeletal muscles taken from all four genotypes of mice (Table 1 and Supplementary 1-5). These data are consistent with robust isolation of an intact DAPC. Of note, we obtained high peptide hits for DAPC proteins that are known to interact with beta dystroglycan either directly or indirectly. This was achieved without tissue sub-fractionation to enrich for membranes and required only 50 mg of starting material per MS sample analysis. Peptides identified in dystroglycan ranged from amino acid 516-821 in all samples (not shown). This is consistent with immunoprecipitation of both alpha and beta dystroglycan, as confirmed by Western blot analysis (Figure 1A), and with the known cleavage of the N-terminal third of alpha dystroglycan (which would therefore be absent) and with the abundant O-linked glycosylation present in the middle third of alpha dystroglycan (which would mask these peptides from detection)7, 8. Multiple splice variants of alpha dystrobrevin were detected by tandem MS in all dystroglycan Ips, a finding that was validated by Western blot analysis (Figure 1B). A few DAPC proteins were identified only in a subset of muscle genotypes. For example, dystrophin was not found in mdx or Galgt2-mdx muscles, as dystrophin protein is not normally expressed in mdx mice, while utrophin, which can be upregulated in mdx muscle, was only identified in these genotypes (Table 1). In addition, beta2 syntrophin was only identified in Galgt2-mdx muscles (Table 1).
Table 1. Immunoprecipitation of DAPC complex proteins with the MANDAG2 monoclonal antibody.
MANDAG2, a monoclonal antibody specific for beta dystroglycan, immunoprecipitated most members of the dystrophin-associated protein complex (DAPC) from skeletal muscles of wild type (WT), mdx, Galgt2 transgenic WT and Galgt2 transgenic mdx skeletal muscles. Examples of best scores taken from DG IP replicates reported in Supplemental Tables S1, S3, S4, and S5, are shown, with the genotype of the best sample shown in bold.
| NO | Protein Name |
Accession | MW | Score | Pep # (unique) |
% Cov. | WT | mdx | Galgt2- WT |
Galgt2- mdx |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Dystrophin | IPI00474450 | 425552.12 | 510.32 | 97(57) | 22.68 | + | + | ||
| 2 | Utrophin | IPI00353420 | 392460.31 | 90.30 | 12(9) | 3.91 | + | + | ||
| 3 | Dystroglycan | IPI00122273 | 96844.54 | 80.26 | 20(7) | 11.42 | + | + | + | + |
| 4 | α-sarcoglycan | IPI00322911 | 43259.41 | 70.27 | 19(7) | 24.41 | + | + | + | + |
| 5 | β-sarcoglycan | IPI00124836 | 34850.71 | 40.29 | 8(4) | 23.23 | + | + | + | + |
| 6 | α1-syntrophin | IPI00267848 | 53860.82 | 40.28 | 9(3) | 10.52 | + | + | + | + |
| 7 | γ-sarcoglycan | IPI00110503 | 31930.41 | 40.26 | 7(3) | 19.00 | + | + | + | + |
| 8 | δ-sarcoglycan | IPI00124831 | 32112.89 | 30.20 | 6(3) | 14.90 | + | + | + | + |
| 9 | Dystrobrevin α | IPI00135042 | 83996.19 | 20.31 | 6(2) | 6.50 | + | + | + | + |
| 10 | β2-syntrophin | IPI00118895 | 56346.30 | 20.30 | 3(2) | 8.30 | + | |||
| 11 | Dystrobrevin α isoform 2 | IPI00222696 | 41949.91 | 20.30 | 5(2) | 12.00 | + | + | + | + |
| 12 | Dystrobrevin α isoform 1 | IPI00222694 | 76899.52 | 20.30 | 6(2) | 6.51 | + | + | + | + |
| 13 | β1-syntrophin | IPI00116414 | 58045.06 | 10.20 | 2(1) | 3.50 | + |
No DAPC members were detected in any control MW8 Ips. These controls were performed in parallel on the same muscle extract used for the dystroglycan Ips (Supplementary 2). Immunoglobulins, trypsin and abundant muscle structural proteins (e.g. myosins) were the primary proteins identified in control Ips. To better visualize proteins specific to the dystroglycan Ips and facilitate comparisons among experimental groups, proteins identified in any of the control Ips were subtracted from all of the corresponding dystroglycan Ips. In addition, we performed immunoprecipitations using MANDAG2 anti beta dystroglycan antibody on identical amounts of muscle protein extract made from P3ProCreDag1loxP/loxP mouse hindlimb muscles (quadriceps) (Supplementary 6). These mice have dystroglycan deleted in hindlimb muscles due to a caudal to rostral gradient of Cre expression in the P3ProCre transgenic line41. These proteins were also removed from results for MANDAG2 in WT, Galgt2-WT, mdx, and Galgt2-mdx muscles. None of the known DAPC members were identified in such control Ips. A number of other muscle proteins, however, were bound to MANDAG2 in control Ips (Supplementary 6) despite the fact that MANDAG2 appears to exclusively recognize β dystroglycan by immunoblotting and by immunostaining.
The resulting protein lists were used for all further analyses described below. Comparison of replicate experiments within each experimental group revealed good agreement and a significant proportion of overlapping proteins (Table 2). mdx muscle Ips were performed an additional time compared to other conditions due to the fact that dystrophic muscles are more variable in their protein composition. Proteins were considered positive here if present in two of three Ips.
Table 2. Reproducibility of protein identification using beta-dystroglycan immunoprecipitation.
Number of proteins identified in dystroglycan (DG) Ips compared to control Ips for each experimental replicate. Unique proteins that overlapped between DG IP experiments were 25% of total proteins identified for wild type (WT), 33% for mdx, 46% for Galgt2 transgenic mdx (Galgt2 mdx), and to 74% for Galgt2 transgenic wild type (Galgt2-WT) muscles.
| Skeletal Muscle | Control IP | DG IP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | 1st Trial |
2nd Trial |
3rd Trial |
Over- lapped |
Total | 1st Trial |
2nd Trial |
3rd Trial |
Over- lapped |
Total |
| WT | 31 | 58 | - | 17 | 72 | 51 | 120 | - | 34 | 137 |
| Galgt2-WT | 36 | 24 | - | 12 | 48 | 56 | 57 | - | 48 | 65 |
| Galgt2-mdx | 50 | 45 | - | 31 | 64 | 73 | 70 | - | 45 | 98 |
| mdx | 43 | 35 | 58 | 14 | 87 | 50 | 35 | 85 | 34 | 102 |
Identification of dystroglycan-associated proteins in wild type muscle that are not part of the core DAPC
Cytoskeletal proteins known to associate directly or indirectly with the DAPC were also identified in dystroglycan Ips from wild type skeletal muscle (Table 3, Table 5 and Supplementary 1). These include nebulin, alpha-actinin 3, myozenin-1 and Troponin T (fast and slow muscle isoforms). Interestingly, none of these proteins were also found in mdx muscle, suggesting their association may be diminished or lost as a result of dystrophin deficiency (Tables 3-5). By contrast, synaptopodin 2 and Rhomboid-related protein 1 were identified in both wild type and mdx muscle DG Ips (Table 3). Neither of these proteins, however, showed high co-localization with beta dystroglycan at the sarcolemmal membrane by immunostaining (not shown). Of note, we were unable to identify utrophin in wild type dystroglycan Ips even though a small amount was detected by Western blot analysis (Figure 1B). Utrophin interacts with a small pool of muscle dystroglycan specifically at the neuromuscular junction and therefore does not represent an abundant protein in skeletal muscle, which may have made detection more difficult.47 Similarly, we did not identify rapsyn, another dystroglycan interacting protein localized at the neuromuscular junction48, 49, in wild type dystroglycan Ips (Supplementary 1).
Table 3. Protein in common to wild type (WT) and mdx dystroglycan (DG) immunoprecipitations (Ips).
| WT Replicate | mdx Replicate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| No | Protein Name/Gene ID | Accession | Protein Code |
MW | p-value | Score | Pep # |
p-value | Score | Pep # |
| 1 | Dystroglycan (Dagl) | IPI00122273 | Q62165 | 96844.5 | 8.22E-14 | 80.28 | 16 | 3.44E-14 | 40.29 | 7 |
| 2 | Alpha-sarcoglycan (Sgca) | IPI00322911 | P82350 | 43259.4 | 7.94E-12 | 70.27 | 19 | 7.40E-11 | 40.23 | 6 |
| 3 | Beta-sarcoglycan (Sgcb) | IPI00124836 | P82349 | 34850.7 | 4.04E-11 | 40.29 | 7 | 2.19E-10 | 20.22 | 3 |
| 4 | Alpha-1-syntrophin (Snta1) | IPI00267848 | Q61234 | 53860.8 | 1.27E-10 | 40.28 | 9 | 2.36E-11 | 20.60 | 3 |
| 5 | Gamma-sarcoglycan (Sgcg) | IPI00110503 | P82348 | 31930.4 | 2.76E-12 | 40.26 | 7 | 2.22E-15 | 30.27 | 5 |
| 6 | Delta-sarcoglycan (Sgcd) | IPI00124831 | P82347 | 32112.9 | 5.93E-07 | 30.20 | 5 | 4.28E-07 | 10.24 | 2 |
| 7 | Dystrobrevin alpha (Dtna) | IPI00135042 | Q9D2N4 | 83996.2 | 8.88E-15 | 20.30 | 6 | 8.01 E-05 | 10.16 | 4 |
| 8 | Synaptopodin-2 (Synpo2) | IPI00670490 | Q91YE8 | 80858.0 | 3.13E-06 | 10.13 | 2 | 1.01 E-05 | 10.15 | 2 |
| 9 | Rhomboid-related protein 1 (Rhbdl1) | IPI00121565 | Q8VC82 | 41758.6 | 4.44E-04 | 10.13 | 1 | 3.33E-04 | 10.14 | 1 |
Table 5. Protein unique to wild type DG IPs not found in mdx DG IPs.
| Replicate 1 | Replicate 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | ||||||||||
| No | Protein Name/Gene ID | Accession | Protein Code |
MW | p-value | Score | Pep # |
p-value | Score | Pep # |
| 1 | Dystrophin (Dmd) | IPI00474450 | P11531 | 425552.1 | 1.10E-13 | 300.31 | 46 | 1.67E-14 | 510.32 | 97 |
| 2 | Troponin T fast skeletal muscle isoform (Tnnt3) | IPI00153943 | Q8R563 | 36247.2 | 2.84E-12 | 58.27 | 6 | 8.32E-10 | 50.24 | 10 |
| 3 | Alpha-actinin-3 (Actn3) | IPI00136701 | O88990 | 102978.0 | 1.14E-09 | 29.50 | 3 | 8.85E-09 | 20.21 | 4 |
| 4 | Tripartite motif-containing protein 47 (Trim47) | IPI00480235 | Q8C0E3 | 69939.0 | 8.10E-04 | 10.13 | 2 | 1.38E-05 | 20.16 | 2 |
| 5 | Keratin, type I cytoskeletal 13 (Krt13) | IPI00136056 | P08730 | 47724.3 | 3.32E-05 | 20.16 | 3 | 1.33E-14 | 10.32 | 3 |
| 6 | Nebulin (Neb) | IPI00720238 | A2AQA9 | 800082.4 | 4.67E-09 | 16.23 | 4 | 2.36E-05 | 10.22 | 6 |
| 7 | Myozenin-1 (Myoz1) | IPI00459945 | Q9JK37 | 31437.6 | 2.22E-07 | 15.2 | 4 | 1.54E-07 | 10.18 | 2 |
| 8 | Troponin T, slow skeletal muscle (Tnnt1) | IPI00130005 | O88346 | 30526.6 | 9.91 E-06 | 10.26 | 2 | 1.31E-05 | 10.22 | 2 |
| 9 | Beta-1-syntrophin (Sntb1) | IPI00116414 | Q99L88 | 58045.1 | 2.29E-04 | 8.10 | 2 | 1.77E-06 | 20.20 | 2 |
| 10 | AT-hook DNA-binding motif-containing protein 1 (Ahdc1) |
IPI00420683 | Q6PAL7 | 168072.5 | 4.44E-04 | 10.13 | 1 | 9.52E-05 | 10.14 | 2 |
| 11 | GDP-L-fucose synthase (Tsta3) | IPI00133690 | P23591 | 35855.0 | 8.93E-04 | 10.09 | 1 | 3.67E-04 | 9.15 | 1 |
Alterations in dystroglycan-interacting proteins following loss of dystrophin expression in mdx skeletal muscle
We next compared the repertoire of proteins identified from muscles of mdx mice, a dystrophin-deficient animal model for Duchenne muscular dystrophy, to wild type5, 20. Loss of dystrophin in mdx muscle resulted in its replacement by utrophin (Table 4), a protein highly homologous to dystrophin50. This was evident in Ips analyzed by Western blot (Figure 1B) and by LC tandem MS (Table 4 and Supplementary Table 3). This result agrees with the reported up regulation of utrophin in mdx muscle, in part due to its expression in regenerating muscle fibers where it associates with dystroglycan51, 52. Among other known DAPC members, beta1 syntrophin was the only one that was not identified in mdx muscle beta dystroglycan Ips (Table 1 and 5).
Table 4. Proteins unique to mdx DG IPs not found in WT DG IPs.
| Replicate 1 | Replicate 2 | Replicate 3 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| No | Protein Name /Gene ID | Accession | Protein Code |
MW | p-value | Score | Pep # |
p-value | Score | Pep # |
p-value | Score | Pep # |
| 1 | Utrophin (Utrn) | IPI00353420 | E9Q6R7 | 392460.3 | 1.71E-05 | 10.17 | 2 | 4.97E-05 | 10.18 | 2 | 1.00E-30 | 90.29 | 12 |
| 2 | Ankyrin-1 (Ankl) | IPI00755993 | Q02357 | 209417.9 | 1.16E-04 | 10.11 | 1 | 6.04E-10 | 20.24 | 2 | |||
| 3 | Alpha-internexin (Ina) | IPI00135965 | P46660 | 55708.5 | 2.17E-04 | 9.03 | 1 | 1.28E-05 | 18.13 | 2 | |||
| 4 | Neurofilament heavy polypeptide (Nefh) | IPI00114241 | P19246 | 116923.8 | 3.33E-04 | 14.21 | 1 | 6.48E-05 | 10.14 | 2 | |||
| 5 | GTP-binding protein 6 (Gtpbp6) | IPI00469381 | Q3U6U5 | 56440.3 | 4.33E-06 | 10.21 | 2 | 5.78E-04 | 10.13 | 1 | |||
| 6 | Peripherin (Prph) | IPI00230444 | P15331 | 57798.4 | 1.01 E-05 | 10.14 | 2 | 9.12E-05 | 10.16 | 1 | |||
| 7 | Neurofilament light polypeptide (Nefl) | IPI00554928 | P08551 | 61340.1 | 9.43E-04 | 8.13 | 1 | 6.48E-05 | 10.14 | 2 | |||
| 8 | Retinal-specific ATP-binding cassette transporter (Abca4) |
IPI00132595 | O35600 | 260367.3 | 7.60E-04 | 10.11 | 2 | 2.00E-04 | 10.13 | 1 | |||
| 9 | Chromodomain helicase DNA binding protein 3 (Chd3) |
IPI00675483 | B1AR17 | 232598.1 | 4.15E-04 | 8.11 | 1 | 2.77E-04 | 6.11 | 2 | |||
Several other proteins were specifically identified in mdx muscle as compared to wild type (Table 4). beta-dystroglycan Ips from mdx muscles indicated enrichment for associations with cytoskeletal elements, in particular ankyrin-1 and neurofilaments (H and L). In addition, we identified alpha-internexin, peripherin, Retinal specific ATP binding cassette transporter and GTP binding protein 6.
Effects of Galgt2 transgene overexpression on dystroglycan-associated proteins in wild type and mdx muscles
We have previously shown that transgenic expression of Galgt2 increases glycosylation of alpha-dystroglycan with the CT carbohydrate and leads to increased extrasynaptic expression of normally synaptic dystroglycan-binding partners, including laminin alpha4, laminin alpha5, and utrophin 13, 15, 16, 25. This is accompanied by increased resistance of Galgt2 transgenic muscles to injury and inhibition of muscular dystrophy16, 19, 21, 24, 25. Here we sought to identify additional alterations in dystroglycan-associated proteins that are triggered by the addition of the CT carbohydrate on alpha-dystroglycan that may be important to stabilize the DAPC in the absence of dystrophin.
The profile of proteins identified by tandem MS in dystroglycan Ips from wild type (Supplementary 1) and Galgt2-WT (Supplementary 4) muscles were highly similar. Amongst known DAPC proteins, Galgt2 expression led to a loss of identification of beta1-syntrophin (Table 1). Additional proteins uniquely identified in Ips from Galgt2 transgenic muscles, both wild type and mdx, included protein spire 2 homologue and interleukin 15 receptor α chain isoform 2D (Tables 6 and 7 and Supplementary 1-8).
Table 6. Proteins unique to Galgt2 Tg DG IPs not found in WT DG IPs.
| No | Protein Name | Accession | Protein Code |
Gene Name |
p-value | Score | MW | Peptide Sequence |
|---|---|---|---|---|---|---|---|---|
| 1 | General transcription factor 3C polypeptide 1 | IPI00403414 | Q8K284 | Gtf3c1 | 8.22E-05 | 20.26 | 237324.5 | ERKDITSSIR GYYTVPGMVSTR SSALPSTSAPANAPAR |
| 2 | TATA element modulatory factor 1 | IPI00668529 | E9QPJ3 | Tmf1 | 7.23E-05 | 20.2 | 121728.6 | QVLDGK ERAATEELLANK |
| 3 | Interleukin 15 receptor alpha chain isoform 2D |
IPI00402905 | Q810T6 | Il15ra | 6.57E-04 | 10.14 | 11154.68 | MTKASQDLRVPCHR |
| 4 | Actin-binding LIM protein 3 | IPI00464316 | Q69ZX8 | Ablim3 | 7.66E-04 | 10.12 | 77580.55 | DLAALPKVKSIYEVQR EEMKAR |
| 5 | G1 to S phase transition 1 | IPI00230355 | Q8K2E1 | Gspt1 | 1.21 E-04 | 8.11 | 64383.49 | LPIVDKYK |
| 6 | Protein spire homolog 2 | IPI00173139 | Q8K1S6 | Spire2 | 4.23E-04 | 8.1 | 80158.68 | FLQDKELFSSLKR |
Table 7. Proteins unique to Galgt2-mdx DGIPs not found in mdx DG IPs.
| No | Protein Name | Accession | Protein Code |
Gene Name |
p-value | Score | MW | Peptide Sequence |
|---|---|---|---|---|---|---|---|---|
| 1 | Beta-2-syntrophin | IPI00118895 | Q61235 | Sntb2 | 7.19E-07 | 20.3 | 56346.26 | KPSLVSDLPWEGASPQSPSFSGSEDSGSPK SPSLGSDLTFATR |
| 2 | Four and a half LIM domains protein 1 | IPI00309997 | P97447 | Fhl1 | 2.24E-06 | 20.14 | 31866.83 | AIVAGDQNVEYK NPITGFGK |
| 3 | E3 ubiquitin-protein ligase HERC4 | IPI00453903 | Q6PAV2 | Herc4 | 8.46E-05 | 20.13 | 118350.3 | QFLLFLTGSDRIPILGMK CSDPSK |
| 4 | Early endosome antigen 1 | IPI00453776 | Q8BL66 | Eea1 | 7.56E-05 | 20.11 | 160816.5 | FQEKQEHCIQLESHLK GKAAVLDLEK |
| 5 | Ahnak | IPI00553798 | Q8VDN3 | Ahnak | 5.21 E-05 | 18.16 | 603870.6 | VDIDVPDVNIEGPDAK VDIDVPDVNIEGPEGK |
| 6 | Troponin T fast skeletal muscle isoform | IPI00153943 | Q8R563 | Tnnt3 | 2.03E-11 | 10.21 | 36247.24 | ELWDTLYQLETDKFEFGEK |
| 7 | Coiled-coil domain-containing protein 40 | IPI00648466 | Q8BI79 | Ccdc40 | 6.22E-04 | 10.21 | 136686.7 | STDSEIEVCKKSIMQEEEK |
| 8 | Prolactin regulatory element-binding protein |
IPI00124980 | Q9WUQ2 | Preb | 1.71E-06 | 10.18 | 45408.98 | AHEGEIGDLTLGPDGK FGQVPDQLGGLRLFTVQIPHK |
| 9 | Major vault protein | IPI00111258 | Q9EQK5 | Mvp | 1.35E-06 | 10.17 | 96698.72 | LAQDPFPLYPGELLEK SFFLQPGER |
| 10 | Pyrroline-5-carboxylate reductase 2 | IPI00123278 | Q922Q4 | Pycr2 | 2.55E-05 | 10.17 | 33637.63 | LGAQALLGAAK MLLDSEDHPGQLK |
| 11 | Heparan-sulfate 6-O-sulfotransferase 2 | IPI00471111 | Q80UW0 | Hs6st2 | 3.68E-04 | 10.15 | 61586.14 | HVLSAIFSKIFGPLASVGNMDEKSNK |
| 12 | Transmembrane protein 121 | IPI00330696 | Q80XA0 | Tmem121 | 4.07E-06 | 10.13 | 35660.16 | MMLYPVLSLATVNVVAVLARAANMALFR DSRVSAIFVGK |
| 13 | Interleukin 15 receptor alpha chain isoform 2D |
IPI00402905 | Q810T6 | Il15ra | 8.29E-04 | 10.1 | 11154.68 | MTKASQDLRVPCHR |
| 14 | Protein spire homolog 2 | IPI00173139 | Q8K1S6 | Spire2 | 1.61 E-04 | 8.11 | 80158.68 | FLQDKELFSSLKR |
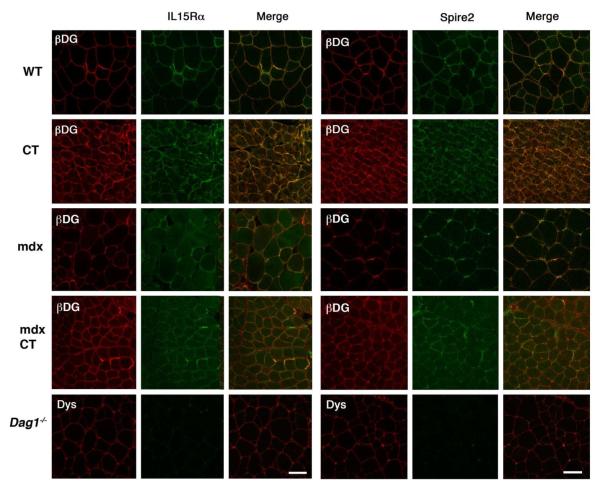
We performed co-immunostaining of beta dystroglycan with IL15R alpha and Spire2 to determine their subcellular localization in wild type, Galgt2-WT, mdx, Galgt2-mdx and Dag1−/− muscles (Figure 2). Interestingly, much like Galgt2 and the CT carbohydrate themselves (and also dystroglycan), IL15R alpha and Spire2 were concentrated at the neuromuscular junctions in the sarcolemmal membrane of wild type muscle, with additional extrasynaptic expression that co-localized with beta dystroglycan. In Galgt2-WT and Galgt2-mdx transgenic muscles, both IL15R alpha and Spire2 expression was increased along the sarcolemmal membrane relative to non transgenic muscles, and this expression was largely coincident with increased beta dystroglycan staining. In Dag1−/− hindlimb muscles (from P3ProCreDag1loxP/loxP mice), beta dystroglycan is absent from skeletal myofibers41, while dystrophin is reduced but still present. Therefore we co stained for IL15R alpha and Spire2 with dystrophin in the Dag1−/− muscle in order to mark the sarocolemmal membrane. We found that the expression of IL15R alpha and Spire2 were dramatically decreased or absent in Dag1−/− muscle (Fig. 2), which was consistent with reduced overall protein expression (Fig. 3A). By contrast, antibodies for a number of other proteins (alpha B crystallin, for example) showed no difference between sections taken from the same muscles of these three different genotypes of mice (not shown).
Figure 2. Co-localization of Spire2 and IL15R alpha with beta dystroglycan in Galgt2 transgenic wild type and mdx skeletal muscles.
(A) Confocal images of muscle cross-sections co-stained with antibody to beta dystroglycan (βDG, using MANDAG2) and with antibody to IL15 receptor alpha (IL15Rα) or Spire2. Muscles were taken from wild type (WT), Galgt2 transgenic (CT), mdx, Galgt2 transgenic mdx (mdxCT) or muscle dystroglycan deficient (P3ProCreDag1loxP/loxP, Dag1−/−) mice. For Dag1−/− muscles, where no muscle beta dystroglycan is present, muscle sections were co-stained for dystrophin (Dys) and IL15Rα or Spire 2. Bar is 50 μm.
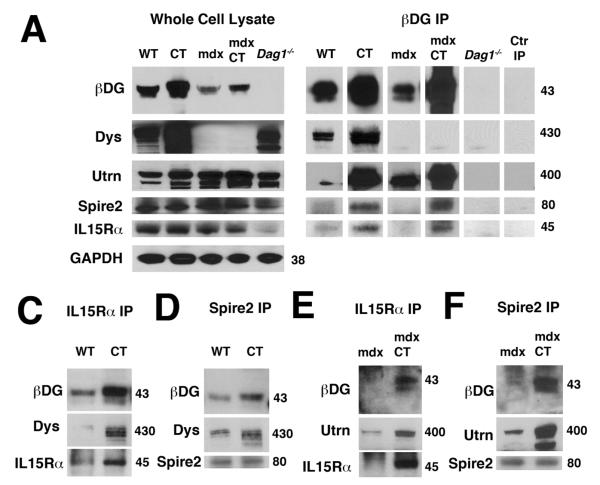
Figure 3. Co-precipitation of Spire2 and IL15R alpha with beta dystroglycan and utrophin or dystrophin in Galgt2 transgenic wild type and mdx skeletal muscles.
(A) Western blotting was used to identify proteins precipitated by beta dystroglycan (βDG) antibody MANDAG2, compared to immunoblots of whole cell lysate starting material, with GAPDH as a loading control. (B-E) Western blotting was used to identify proteins precipitated by IL15R alpha (B, D) or Spire2 (C, E) antibody in wild type (WT) and Galgt2 transgenic wild type (CT) (B, C) or mdx and Galgt2 transgenic mdx (mdxCT) (D, E) skeletal muscle. Approximate molecular weights of bands on Western blots are 43kDa for βDG, 430kDa for dystrophin, 400kDa for utrophin, 45kDa for IL15Rα and 80kDa for Spire2. Control IP antibody used in A was MW8.
Co-immunoprecipitation studies also confirmed the association of IL15Rα and Spire2 with beta dystroglycan in Galgt2-WT and Galgt2-mdx skeletal muscle (Figure 3); IL15R alpha and Spire2 co-precipitated with antibody to beta dystroglycan (Fig. 3A), while beta dystroglycan co-precipitated with antibodies to either IL15R alpha (Fig. 3C, E) or Spire2 (Fig. 3D, F). Such associations were more apparent in Galgt2-WT and Galgt2-mdx muscle than in WT or mdx muscle, respectively, and were absent from Dag1−/− muscle (Fig. 3A). Dystrophin or utrophin also co-precipitated with IL15R alpha and Spire2 in Galgt2-WT or Galgt2-mdx muscles, respectively, for both forward and reverse immunoprecipitations (Fig. 3). In addition, immunoprecipitation of beta dystroglycan and IL15R alpha appeared enriched in Galgt2 transgenic muscles relative to non-transgenic controls (Fig. 3A, B, D). For beta dystroglycan, this somewhat correlated with changed expression in whole cell detergent lysates (Fig. 3A).
Analysis of dystroglycan protein interaction profiles between mdx and Galgt2-mdx mice also revealed a significant degree of overlap (Supplementary 3 and 5). Some differences, however, were evident (Table 7). Among DAPC members, Galgt2 overexpression led to identification of beta2 syntrophin, which is normally only expressed at the neuromuscular junction39 (Table 1 and 7 and Supplementary 7). In addition, Galgt2-mdx muscles showed associations of dystroglycan with protein spire homologue 2 and interleukin 15 receptor alpha. These proteins were also differentially enriched between Galgt2-WT and WT samples for proteomics analysis (Tables 6 and 7). Other proteins specific to Galgt2-mdx muscles were Ahnak protein, major vault protein, heparin-sulfate 6-O-sulfotransferase, prolactin regulatory element-binding protein, transmembrane protein 121, pyrroline-5-carboxylate reductase 2 and four and half LIM domains 1. These proteins may also participate in unique associations with dystroglycan as a result of glycosylation by Galgt2 and therefore are candidates to convey the biological effects of Galgt2 overexpression on muscular dystrophy.
DISCUSSION
We have optimized a simple and rapid protocol for antibody-based purification of dystroglycan and interacting proteins directly from skeletal muscle tissues followed by direct protein identification by tandem MS. This method has allowed us to reproducibly and specifically isolate and identify dystroglycan with extremely high confidence as well as to purify it within the context of almost the entire known DAPC 29, 30, including dystrophin, alpha, beta, gamma and delta sarcoglycan, alpha1- and beta1-syntrophin, and alpha dystrobrevin (Table 1). This is a significant advance for the field of muscular dystrophy since identification of DAPC members by MS has been challenging and has required a laborious protocol involving differential centrifugation to isolate membrane fractions followed by lectin pull-downs 35. Even following such enrichment strategies only a few DAPC members were confidently detected and peptide hits for identified DAPC members were appreciably lower than reported here. Furthermore, these procedures require large amounts of starting material while our protocol only requires 100 mg of muscle tissue for a full MS analysis (dystroglycan IP + control IP on the same sample). Therefore, our approach represents a significant improvement over current methods and is expected to facilitate the biochemical characterization of the DAPC in skeletal muscle in order to further our understanding of its function in wild type muscle and its dysfunction in muscular dystrophies.
While this approach is efficient and reliable, not all known DAPC members were identified. These include neuronal nitric oxide synthase (nNOS) and sarcospan. These proteins do not interact directly with dystroglycan but are removed by at least 2 protein interactions from dystroglycan. These interactions have been detected in muscle Ips using an antibody directed against dystrophin (Johnson et al., submitted), indicating that our detergent extraction conditions preserve these interactions. Lack of detection in the dystroglycan Ips may be related to the use of the MANDAG2 antibody. This antibody recognizes a peptide sequence (SPPPYVP) in the cytoplasmic domain of beta dystroglycan that contains a known protein-binding motif (PXXP). It is therefore conceivable that this antibody may disrupt some protein interactions or may enrich for dystroglycan complexes where this intracellular domain is more accessible. Alternatively, nNOS and sarcospan may not be sufficiently abundant in the dystroglycan Ips to be detected using the LTQ. Detection of low abundance proteins in a sample with a large dynamic range is known to represent a challenge for identification by MS. Nevertheless, we were able to detect by tandem MS most direct and indirect protein associations of dystroglycan with a high level of confidence and high inter-sample reproducibility.
One curious aspect of our results is that MANDAG2 can precipitate dystrophin and utrophin, despite the fact that these proteins are thought to bind via the SPPPYVP motif recognized by this monoclonal antibody53. We believe that this is due to the fact that the DAPC complex likely contains multimers of dystroglycan, some of which may bind utrophin or dystrophin and some of which may be available for binding by MANDAG2. Alternatively, there may be aspects of these protein-protein interactions, or of protein antibody interactions, that are not sufficiently appreciated. For example, Winder has described tyrosine phosphorylation of the PPXY motif on beta dystroglycan and shown that this phosphorylation can inhibit utrophin and dystrophin binding54, 55. It may well be that Galgt2 glycosylation inhibits such tyrosine phosphorylation of beta dystroglycan, allowing enriched immunoprecipitation with MANDAG2 and enriched co-precipitation of dystrophin or utrophin and associated proteins.
This achievement has allowed us to identify a few additional candidate proteins as new potential DAPC members. For example associations with synaptopodin 2 and Rhomboid related protein 1 were detected in both wild type and mdx muscle. These proteins, however, are not localized in the muscle cell in such a way that they could significantly interact with dystroglycan at the membrane. A number of cytoskeletal proteins, however, were also identified, and these would be more likely to functionally interact. Some such proteins were identified in wild type muscles but not mdx, including Troponin T, alpha actinin 3, keratin 13, nebulin and myozenin-1, or in mdx but not wild type, including ankyrin-1. Further studies are needed to verify these new DAPC associated proteins by alternative methods. Interestingly, several membrane-associated proteins were highly identified by MANDAG2 in dystroglycan-deficient muscles. Some of these proteins may also contain the SPPPYVP motif recognized by this monoclonal antibody. Thus, while immunoblots and immunostains show MANDAG2 to be highly specific for beta dystroglycan, proteomics studies suggest that this antibody can indeed interact with other proteins under the appropriate detergent conditions. Therefore, making use of dystroglycan-deficient muscle controls was an essential step needed to clarify dystroglycan binding specificity.
The main focus of this study was the identification of changes in DAPC associations following Galgt2 transgene expression that could offer insights into the mechanisms by which Galgt2 inhibits development of muscular dystrophy and prevents muscle damage13, 16, 19, 21, 24, 25. We identified only two proteins enriched in dystroglycan Ips from both Galgt2-WT and Galgt2-mdx transgenic muscles compared to non-transgenic controls. These were protein spire homolog 2 (Spire2) and interleukin 15 receptor alpha chain (IL15rα). Loss of IL15rα in mice leads to increased resistance to fatigue and altered calcium homeostasis, suggesting that this receptor may impact muscle function56. The association of inactivating mutations of IL15rα and IL15 with obesity also suggests a metabolic role that may involve skeletal muscle57. Protein spire homolog 2 (Spire2), along with Spire1, is involved in nucleating actin filaments58. As such, association of dystroglycan with Spire2 may also affect muscle strength or resistance to injury by organizing the F-actin cytoskeleton. We have shown here that both of these proteins are expressed at the sarcolemmal membrane in a pattern overlapping with beta dystroglycan, both in wild type and in Galgt2 transgenic muscles, and co-precipitation experiments suggested an association as well. Thus, their association with dystroglycan may have functional consequences, for example in Galgt2-mediated resistance to muscle injury25. Several additional proteins were uniquely found in Galgt2-mdx vs. mdx comparisons, but not in Galgt2-WT versus wild type Ips. These include Ahnak protein, beta2-syntrophin, major vault protein, heparin-sulfate 6-O-sulfotransferase 2, prolactin regulatory element binding protein, transmembrane protein 121, pyrroline-5-carboxylate reductase 2 and four and half LIM domain proteins 1. Several of these proteins may also be of significance to Galgt2’s biological effects in mdx skeletal muscle16, 25.
The results presented here describe a new method to identify novel dystroglycan-associated proteins and to identify changes in the DAPC that may occur in pathological muscle states and following therapeutic interventions. Future work will be required to determine how the associations and changes found relate to dystroglycan function in skeletal muscle biology and disease.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH grant AR049722 to PTM, by internal support from TRINCH to FM and LTM, and by an American Heart Association fellowship to EKJ. We express our thanks to Drs. Kun Cho and Gun Wook Park at the mass spectrometry center of the Korea Basic Science Institute (South Korea) for helpful discussion of the manuscript and advice for the SEQUEST software analysis. We thank Jeffery Minor (Washington University) for P3ProCreDag1loxP/loxP mice and Kevin Campbell (U. Iowa) and Jonathan Epstein (U. Penn) for creating the original mouse lines.
REFERENCES
- 1.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin related proteins in muscle. Physiol Rev. 2002;82(2):291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 2.Guglieri M, Straub V, Bushby K, Lochmuller H. Limb girdle muscular dystrophies. Curr Opin Neurol. 2008;21(5):576–84. doi: 10.1097/WCO.0b013e32830efdc2. [DOI] [PubMed] [Google Scholar]
- 3.Mendell JR, Boue DR, Martin PT. The congenital muscular dystrophies: recent advances and molecular insights. Pediatr Dev Pathol. 2006;9(6):427–43. doi: 10.2350/06-07-0127.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manzur AY, Muntoni F. Diagnosis and new treatments in muscular dystrophies. J Neurol Neurosurg Psychiatry. 2009;80(7):706–14. doi: 10.1136/jnnp.2008.158329. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman EP, Brown RH, Jr., Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–28. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 6.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–17. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 7.Ervasti JM, Campbell KP. Membrane organization of the dystrophin glycoprotein complex. Cell. 1991;66(6):1121–31. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 8.Ervasti JM, Campbell KP. A role for the dystrophin glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122(4):809–23. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13(8):55R–66R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
- 10.Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H, Straub V, Mathews KD, Moore SA, Campbell KP. Post translational disruption of dystroglycan ligand interactions in congenital muscular dystrophies. Nature. 2002;418(6896):417–22. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. O mannosyl phosphorylation of alpha dystroglycan is required for laminin binding. Science. 2010;327(5961):88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba A, Matsumura K, Yamada H, Inazu T, Shimizu T, Kusunoki S, Kanazawa I, Kobata A, Endo T. Structures of sialylated O linked oligosaccharides of bovine peripheral nerve alpha dystroglycan. The role of a novel O mannosyl type oligosaccharide in the binding of alpha dystroglycan with laminin. J Biol Chem. 1997;272(4):2156–62. doi: 10.1074/jbc.272.4.2156. [DOI] [PubMed] [Google Scholar]
- 13.Yoon JH, Chandrasekharan K, Xu R, Glass M, Singhal N, Martin PT. The synaptic CT carbohydrate modulates binding and expression of extracellular matrix proteins in skeletal muscle: Partial dependence on utrophin. Mol Cell Neurosci. 2009;41(4):448–63. doi: 10.1016/j.mcn.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrasekharan K, Yoon JH, Xu Y, deVries S, Camboni M, Janssen PM, Varki A, Martin PT. A human specific deletion in mouse Cmah increases disease severity in the mdx model of Duchenne muscular dystrophy. Sci Transl Med. 2010;2(42):42–54. doi: 10.1126/scitranslmed.3000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia B, Hoyte K, Kammesheidt A, Deerinck T, Ellisman M, Martin PT. Overexpression of the CT GalNAc transferase in skeletal muscle alters myofiber growth, neuromuscular structure, and laminin expression. Dev Biol. 2002;242(1):58–73. doi: 10.1006/dbio.2001.0530. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen HH, Jayasinha V, Xia B, Hoyte K, Martin PT. Overexpression of the cytotoxic T cell GalNAc transferase in skeletal muscle inhibits muscular dystrophy in mdx mice. Proc Natl Acad Sci U S A. 2002;99(8):5616–21. doi: 10.1073/pnas.082613599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin PT, Scott LJ, Porter BE, Sanes JR. Distinct structures and functions of related pre and postsynaptic carbohydrates at the mammalian neuromuscular junction. Mol Cell Neurosci. 1999;13(2):105–18. doi: 10.1006/mcne.1999.0737. [DOI] [PubMed] [Google Scholar]
- 18.Hoyte K, Kang C, Martin PT. Definition of pre and postsynaptic forms of the CT carbohydrate antigen at the neuromuscular junction: ubiquitous expression of the CT antigens and the CT GalNAc transferase in mouse tissues. Brain Res Mol Brain Res. 2002;109(1 2):146–60. doi: 10.1016/s0169-328x(02)00551-x. [DOI] [PubMed] [Google Scholar]
- 19.Xu R, Camboni M, Martin PT. Postnatal overexpression of the CT GalNAc transferase inhibits muscular dystrophy in mdx mice without altering muscle growth or neuromuscular development: evidence for a utrophin independent mechanism. Neuromuscul Disord. 2007;17(3):209–20. doi: 10.1016/j.nmd.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sicinski P, Geng Y, Ryder Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244(4912):1578–80. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 21.Xu R, Chandrasekharan K, Yoon JH, Camboni M, Martin PT. Overexpression of the cytotoxic T cell (CT) carbohydrate inhibits muscular dystrophy in the dyW mouse model of congenital muscular dystrophy 1A. Am J Pathol. 2007;171(1):181–99. doi: 10.2353/ajpath.2007.060927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Christmas P, Wu XR, Wewer UM, Engvall E. Defective muscle basement membrane and lack of M laminin in the dystrophic dy/dy mouse. Proc Natl Acad Sci U S A. 1994;91(12):5572–6. doi: 10.1073/pnas.91.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Wu XR, Wewer UM, Engvall E. Murine muscular dystrophy caused by a mutation in the laminin alpha 2 (Lama2) gene. Nat Genet. 1994;8(3):297–302. doi: 10.1038/ng1194-297. [DOI] [PubMed] [Google Scholar]
- 24.Xu R, DeVries S, Camboni M, Martin PT. Overexpression of Galgt2 reduces dystrophic pathology in the skeletal muscles of alpha sarcoglycan deficient mice. Am J Pathol. 2009;175(1):235–47. doi: 10.2353/ajpath.2009.080967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin PT, Xu R, Rodino Klapac LR, Oglesbay E, Camboni M, Montgomery CL, Shontz K, Chicoine LG, Clark KR, Sahenk Z, Mendell JR, Janssen PM. Overexpression of Galgt2 in skeletal muscle prevents injury resulting from eccentric contractions in both mdx and wild type mice. Am J Physiol Cell Physiol. 2009;296(3):C476–88. doi: 10.1152/ajpcell.00456.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon JH, Yea K, Kim J, Choi YS, Park S, Lee H, Lee CS, Suh PG, Ryu SH. Comparative proteomic analysis of the insulin induced L6 myotube secretome. Proteomics. 2009;9(1):51–60. doi: 10.1002/pmic.200800187. [DOI] [PubMed] [Google Scholar]
- 27.Isfort RJ. Proteomic analysis of striated muscle. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771(1 2):155–65. doi: 10.1016/s1570-0232(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 28.Doran P, Dowling P, Donoghue P, Buffini M, Ohlendieck K. Reduced expression of regucalcin in young and aged mdx diaphragm indicates abnormal cytosolic calcium handling in dystrophin deficient muscle. Biochim Biophys Acta. 2006;1764(4):773–85. doi: 10.1016/j.bbapap.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Doran P, Dowling P, Lohan J, McDonnell K, Poetsch S, Ohlendieck K. Subproteomics analysis of Ca+ binding proteins demonstrates decreased calsequestrin expression in dystrophic mouse skeletal muscle. Eur J Biochem. 2004;271(19):3943–52. doi: 10.1111/j.1432-1033.2004.04332.x. [DOI] [PubMed] [Google Scholar]
- 30.Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K. Proteome analysis of the dystrophin deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics. 2006;6(16):4610–21. doi: 10.1002/pmic.200600082. [DOI] [PubMed] [Google Scholar]
- 31.Lewis C, Jockusch H, Ohlendieck K. Proteomic Profiling of the Dystrophin Deficient MDX Heart Reveals Drastically Altered Levels of Key Metabolic and Contractile Proteins. J Biomed Biotechnol. 2010;2010:648501. doi: 10.1155/2010/648501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis C, Ohlendieck K. Proteomic profiling of naturally protected extraocular muscles from the dystrophin deficient mdx mouse. Biochem Biophys Res Commun. 2010;396(4):1024–9. doi: 10.1016/j.bbrc.2010.05.052. [DOI] [PubMed] [Google Scholar]
- 33.Lu B, McClatchy DB, Kim JY, Yates JR., 3rd Strategies for shotgun identification of integral membrane proteins by tandem mass spectrometry. Proteomics. 2008;8(19):3947–55. doi: 10.1002/pmic.200800120. [DOI] [PubMed] [Google Scholar]
- 34.Rabilloud T, Vaezzadeh AR, Potier N, Lelong C, Leize Wagner E, Chevallet M. Power and limitations of electrophoretic separations in proteomics strategies. Mass Spectrom Rev. 2009;28(5):816–43. doi: 10.1002/mas.20204. [DOI] [PubMed] [Google Scholar]
- 35.Lewis C, Ohlendieck K. Mass spectrometric identification of dystrophin isoform Dp427 by on membrane digestion of sarcolemma from skeletal muscle. Anal Biochem. 2010;404(2):197–203. doi: 10.1016/j.ab.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Pereboev AV, Ahmed N, thi Man N, Morris GE. Epitopes in the interacting regions of beta dystroglycan (PPxY motif) and dystrophin (WW domain) Biochimica et biophysica acta. 2001;1527(1 2):54–60. doi: 10.1016/s0304-4165(01)00147-7. [DOI] [PubMed] [Google Scholar]
- 37.Ko J, Ou S, Patterson PH. New anti huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain research bulletin. 2001;56(3 4):319–29. doi: 10.1016/s0361-9230(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 38.Peters MF, Kramarcy NR, Sealock R, Froehner SC. beta 2 Syntrophin: localization at the neuromuscular junction in skeletal muscle. Neuroreport. 1994;5(13):1577–80. [PubMed] [Google Scholar]
- 39.Peters MF, Adams ME, Froehner SC. Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol. 1997;138(1):81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters MF, Sadoulet Puccio HM, Grady MR, Kramarcy NR, Kunkel LM, Sanes JR, Sealock R, Froehner SC. Differential membrane localization and intermolecular associations of alpha dystrobrevin isoforms in skeletal muscle. J Cell Biol. 1998;142(5):1269–78. doi: 10.1083/jcb.142.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarad G, Miner JH. The Pax3 Cre transgene exhibits a rostrocaudal gradient of expression in the skeletal muscle lineage. Genesis. 2009;47(1):1–6. doi: 10.1002/dvg.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elias JE, Gygi SP. Target decoy search strategy for increased confidence in large scale protein identifications by mass spectrometry. Nature methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 43.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large scale proteomics investigations. Nature methods. 2005;2(9):667–75. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 44.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC MS/MS) for large scale protein analysis: the yeast proteome. Journal of proteome research. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura YI, Kawashima R, Fukunaga R, Hirai K, Toyama Sorimachi N, Tokuhara M, Shimizu T, Dohi T. Introduction of Sd(a) carbohydrate antigen in gastrointestinal cancer cells eliminates selectin ligands and inhibits metastasis. Cancer Res. 2005;65(14):6220–7. doi: 10.1158/0008-5472.CAN-05-0639. [DOI] [PubMed] [Google Scholar]
- 46.Sarvaiya HA, Yoon JH, Lazar IM. Proteome profile of the MCF7 cancer cell line: a mass spectrometric evaluation. Rapid communications in mass spectrometry: RCM. 2006;20(20):3039–55. doi: 10.1002/rcm.2677. [DOI] [PubMed] [Google Scholar]
- 47.Ohlendieck K, Ervasti JM, Matsumura K, Kahl SD, Leveille CJ, Campbell KP. Dystrophin related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron. 1991;7(3):499–508. doi: 10.1016/0896-6273(91)90301-f. [DOI] [PubMed] [Google Scholar]
- 48.Apel ED, Roberds SL, Campbell KP, Merlie JP. Rapsyn may function as a link between the acetylcholine receptor and the agrin binding dystrophin associated glycoprotein complex. Neuron. 1995;15(1):115–26. doi: 10.1016/0896-6273(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 49.Cartaud A, Coutant S, Petrucci TC, Cartaud J. Evidence for in situ and in vitro association between beta dystroglycan and the subsynaptic 43K rapsyn protein. Consequence for acetylcholine receptor clustering at the synapse. J Biol Chem. 1998;273(18):11321–6. doi: 10.1074/jbc.273.18.11321. [DOI] [PubMed] [Google Scholar]
- 50.Love DR, Hill DF, Dickson G, Spurr NK, Byth BC, Marsden RF, Walsh FS, Edwards YH, Davies KE. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339(6219):55–8. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 51.Hirst RC, McCullagh KJ, Davies KE. Utrophin upregulation in Duchenne muscular dystrophy. Acta Myol. 2005;24(3):209–16. [PubMed] [Google Scholar]
- 52.Rybakova IN, Humston JL, Sonnemann KJ, Ervasti JM. Dystrophin and utrophin bind actin through distinct modes of contact. J Biol Chem. 2006;281(15):9996–10001. doi: 10.1074/jbc.M513121200. [DOI] [PubMed] [Google Scholar]
- 53.Chung W, Campanelli JT. WW and EF hand domains of dystrophin family proteins mediate dystroglycan binding. Mol Cell Biol Res Commun. 1999;2(3):162–71. doi: 10.1006/mcbr.1999.0168. [DOI] [PubMed] [Google Scholar]
- 54.Ilsley JL, Sudol M, Winder SJ. The interaction of dystrophin with beta dystroglycan is regulated by tyrosine phosphorylation. Cell Signal. 2001;13(9):625–32. doi: 10.1016/s0898-6568(01)00188-7. [DOI] [PubMed] [Google Scholar]
- 55.James M, Nuttall A, Ilsley JL, Ottersbach K, Tinsley JM, Sudol M, Winder SJ. Adhesion dependent tyrosine phosphorylation of (beta) dystroglycan regulates its interaction with utrophin. J Cell Sci. 2000;113(Pt 10):1717–26. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- 56.Pistilli EE, Bogdanovich S, Garton F, Yang N, Gulbin JP, Conner JD, Anderson BG, Quinn LS, North K, Ahima RS, Khurana TS. Loss of IL 15 receptor alpha alters the endurance, fatigability, and metabolic characteristics of mouse fast skeletal muscles. The Journal of clinical investigation. 2011;121(8):3120–32. doi: 10.1172/JCI44945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinn LS, Anderson BG. Interleukin 15, IL 15 Receptor Alpha, and Obesity: Concordance of Laboratory Animal and Human Genetic Studies. Journal of obesity. 2011;2011:456347. doi: 10.1155/2011/456347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire type actin nucleators cooperate with Formin 2 to drive asymmetric oocyte division. Current biology: CB. 2011;21(11):955–60. doi: 10.1016/j.cub.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.