Abstract
Bacteria must segregate their DNA and position a septum to grow and divide. In many bacteria MinD is involved in spatial regulation of the cytokinetic Z ring and ParAs are involved in chromosome and plasmid segregation. The use of the MinD/ParA family to provide positional information for spatial organization continues to expand with the recognition that orphan ParAs are required for segregating cytoplasmic protein clusters and the polar localization of chemotaxis proteins, conjugative transfer machinery, type IV pili and cellulose synthesis. Also, some bacteria lacking MinD use orphan ParAs to regulate cell division. Positioning of MinD/ParA proteins is either due to self-organization on a surface or reliance on a landmark protein which functions as a molecular beacon.
Positioning cellular components
Early microscopic observation of bacteria revealed the spatio-temporal regulation that must be operating within. Visible clues included a readily observable septum and, in some bacteria, the presence of a unipolar flagellum or polar type IV pili. Immunoelectron microscopy revealed the Z ring underlying the septum [1] and the advent of fluorescent technologies including GFP-fusions revealed more clues, chromosomal origins and plasmids duplicating and segregating to discrete cellular locations [2, 3]. More recently the distribution of some cytoplasmic protein clusters, carboxysomes and chemotaxis clusters was found to mimic that of plasmids [4, 5]. Investigation into these processes made it clear that members of the ParA/MinD family of proteins are involved in positioning large structures or regulating the position where large structures assemble. Here we look at how the MinD/ParA family of proteins are involved in these processes.
The MinD/ParA family
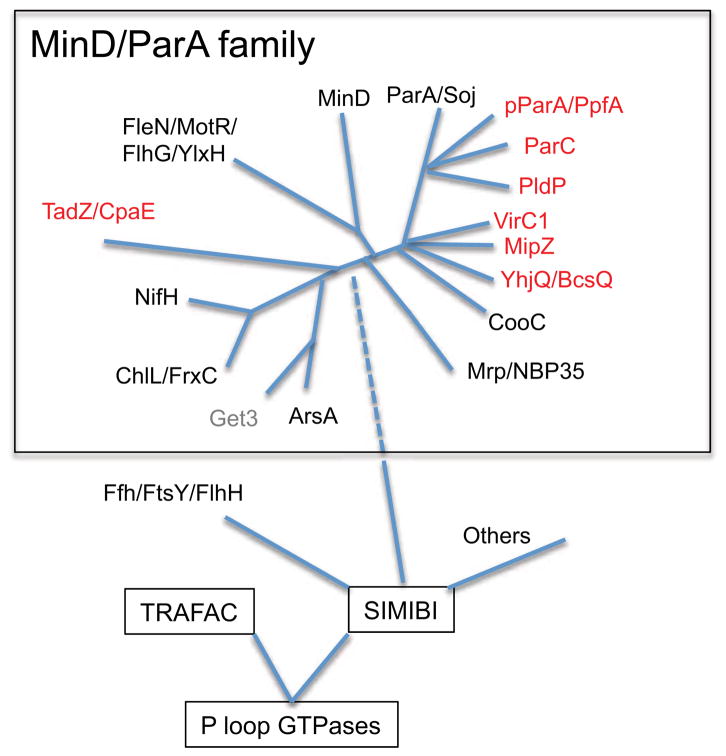
ParA and MinD are ATPases but members of the large P loop GTPase superfamily [6]. This superfamily has two classes, TRAFAC and SIMIBI, and MinD/ParA are members of the latter class along with other related ATPases. Sequence and structure analysis allowed Liepe et al. [6] to identify eight subfamilies (Figure 1, black). Two of these, MinD and ParA, have been studied for some time and are fairly widely disseminated among bacteria whereas most of the other related ATPases display a more limited distribution. An exception is the Mrp/ApbC/NBP35 subfamily, which is widely distributed in all domains of life and is involved in metabolism of Fe-S clusters [7, 8]. The remaining members are involved in a variety of cellular processes including nitrogen fixation (NifH), arsenite expulsion (ArsA), regulation of flagella (FlgG, but has many alternative names), and membrane insertion of proteins in eukaryotes with a C-terminal hydrophobic transmembrane domain (Get3). More recently, additional subfamilies or distinct clades within the MinD/ParA family have been identified that, like MinD and ParA, are involved in positioning protein structures (Figure 1, red). They have been referred to as orphan ParAs (not associated with the usual partner ParB) and have a limited distribution among bacteria. They include ParC (associated with positioning polar chemotaxis proteins [9]), VirC1 (required for the polar localization of conjugative transfer machinery [10]), TadZ/CpaE (involved in the polar positioning of type IV pili [11, 12]), YjhQ/BcsQ (required for polar synthesis of cellulose [13]) and plasmid related ParAs (associated with segregation of cytoplasmic protein clusters [4, 5]). Other members, MipZ and PdlP, are involved in spatial regulation of Z ring positioning in bacteria that lack MinD [14, 15].
Figure 1.
Phylogenetic tree displaying the position of MinD/ParA family members. The MinD/ParA family is a part of the SIMIBI class of P loop GTPases. Leipe et al. (6) recognized 8 subfamilies of the MinD/ParA/Mrp family (names in black). Get3 is closely related to ArsA. More recently recognized members are colored red. This diagram was based upon Leipe et al. (6) with additional information obtained from Perez-Cheeks et al. (11) and Ringgaard et al. (36).
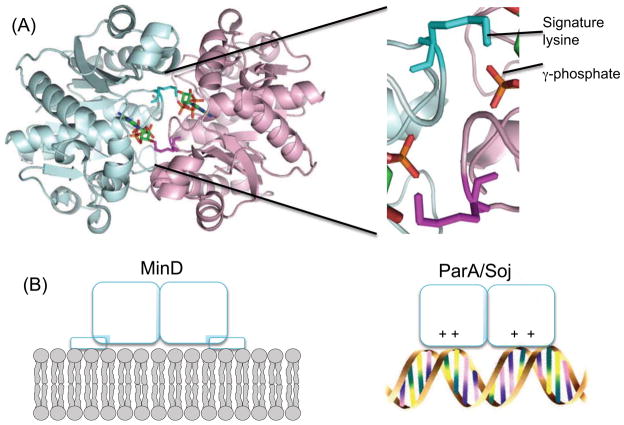
The hallmark of these ATPases is a ‘deviant Walker A motif – KGGXXGKT’ containing two conserved lysines [6, 16]. The second lysine is common to all Walker A motifs and is involved in the binding and hydrolysis of ATP. The amino terminal lysine, called the ‘signature’ lysine, mediates dimerization by binding to the phosphates of ATP bound to the other subunit (Figure 2a). This was first demonstrated for NifH [17] but has been subsequently shown for ParA (Soj), MinD, Get3 and others [18–20]. This lysine is also essential for ATP hydrolysis and is the functional equivalent of the arginine finger found in GAP proteins that activate the GTPase activity of Rho and Ras (members of the TRAFAC division). This lysine is missing in some TadZ/CpaE family members but, in the one case examined, its function is supplied by another lysine located elsewhere in the primary sequence [12].
Figure 2.
MinD/ParA proteins undergo ATP dependent dimerization to bind to surfaces and other proteins. (a) MinD dimer (PDB 3Q9L), with ATP (phosphates in orange) and signature lysines highlighted. The expanded view on the right shows the signature lysine in proximity to the γ-phosphate of ATP bound to other subunit. (b) ATP dependent dimerization of MinD and ParA lead to binding to surfaces. The MinD dimer binds to membranes through a C-terminal amphipathic helix and the ParA/Soj dimer binds nonspecifically to DNA through positive charged residues. Although binding to different surfaces, the orientation of the proteins on the surface is the same.
MinD/ParA are regulated by a nucleotide dependent switch
The ATP form of MinD/ParA proteins is a dimer that usually binds to a surface that allows it to take up residency in the cell [18, 21, 22]. MinD binds to the membrane using a C-terminal amphipathic helix and ParA binds nonspecifically to DNA through positive charges located on one face of the protein [23–25] (Figure 2b). Even though these proteins bind chemically different surfaces the same face of the protein is used resulting in the same orientation on the surface. Dimerization brings together two half sites to form a binding site with increased affinity for the surface (Figure 2b) as well as for protein partners. For example, the dimerization of MinD results in increased affinity for the membrane but also generates binding sites for its partners MinE and MinC [19]. Since the binding sites for the proteins partners are at the dimer interface, they only come into existence when MinD dimerizes.
Importantly, ParA and MinD have a partner protein that acts as an ATPase activating protein (AAP); MinE for MinD and ParB for ParA [27, 28]. The presence of an AAP with a spatially restricted distribution ensures that the residency established on a surface by MinD/ParA is dynamic and self-organized. For some other members of the MinD/ParA extended family, ATP-dependent dimerization does not lead to binding to a matrix, only to other proteins. For such proteins residency in the cell depends upon a landmark protein that acts as a molecular beacon to recruit the MinD/ParA family member [29]. As we shall see, MinD/ParA family members may use either mechanism, self-organization on a surface or reliance on a landmark protein, to localize.
In contrast to the ATP form, the ADP form of the MinD/ParA member is usually a monomer that freely diffuses in the cell. In some cases the monomer also binds to specific partners that either sequester the monomer form to delay its return to the ATP form or provides for novel regulation by interacting with another protein. For example, the Soj monomer (a chromosomal ParA in Bacillus subtilis) binds to and inhibits DnaA, the initiator of DNA replication [30]. Useful in dissecting the functions of the two forms are mutations that prevent dimerization (K11A, G12V in the deviant Walker A motif) or block ATP hydrolysis (D40A) and lock the protein in the dimer form (residue numbers are from Escherichia coli MinD) [22, 25, 31, 32]. Mutations can also be introduced that prevent nonspecific DNA binding (ParAs) [25] or membrane binding (MinD) [24]. Such mutations also lock proteins in the dimer form since ATP hydrolysis is not stimulated by the AAP unless the protein is bound to its respective surface [21, 33].
Oscillation (self-organizing) versus landmark (molecular beacon)
Proteins in bacteria localize by diffusion and capture [34], and ParA/MinD family members use two variations on this theme. One is an oscillatory mechanism observed with the Min and the plasmid ParA systems in E. coli [9, 35, 36]. This mechanism is also likely used by some bacteria to position large protein structures (carboxysomes and large cytoplasmic chemotaxis clusters) [10]. The other is a landmark mechanism observed with the Min system in B. subtilis and with a chemotaxis system in Vibrio cholerae [9, 29]. The common feature of the oscillatory mechanism is that dynamic binding to a surface (membrane or DNA) fueled by nucleotide hydrolysis leads to pattern formation (self-organization) whereas in the landmark mechanism the nucleotide cycle regulates binding to a protein that is localized in the cell and serves as a molecular beacon. Thus, the first mechanism is self-organizing, relying only on the shape of the cell or nucleoid, whereas the second requires another protein to provide the spatial cue.
Min system: oscillation versus landmark
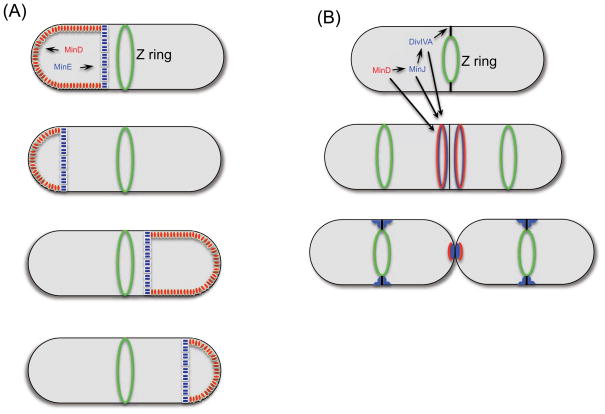
The Min system contributes to the spatial regulation of the Z ring in many bacteria by preventing its assembly away from midcell, especially near the poles [37]. The antagonist of FtsZ assembly, MinC is activated and positioned in the cell through its interaction with MinD. The E. coli Min system is a pure oscillatory system, getting its spatial cues directly from the shape of the cell. The system is very responsive to the cell’s shape and the pole-to-pole oscillation observed in wild-type cells (Figure 3a) shifts to a striped pattern in filamentous cells [38]. The oscillatory mechanism also finds the long axis in nearly round cells and establishes a rotating oscillation in Y-shaped cells [39, 40]. This ability to adapt to the shape of the cell without requiring protein landmarks is consistent with the ability of MinD and MinE to form dynamic patterns (travelling waves) on a planar lipid bilayer in vitro [41, 42].
Figure 3.
Localization behavior of MinD. MinD contributes to Z ring positioning in E. coli (a) and B. subtilis (b). In E. coli MinD and MinE oscillate between the poles to prevent Z ring assembly away from midcell. In B. subtilis DivIVA localizes to the incipient septum and recruits MinJ which recruits MinD and MinC. The decorated DivIVA ring is stable as the septum constricts. After the cells split, the Min proteins are released and DivIVA reorganizes.
The basis of the Min oscillation has been extensively explored and is due to pattern formation developing from a dynamic instability [43–45]. The ATP form of MinD (dimer) binds cooperatively to the membrane [46], and the ADP form (monomer) freely diffuses in the cytoplasm. MinE also exists in two conformations, a latent form that freely diffuses in the cytoplasm and an active form that binds MinD and the membrane [47]. MinE binding to MinD at the membrane stimulates the ATPase activity of MinD, releasing it from the membrane, but MinE is now associated with the membrane, and either dissociates or jumps to another membrane-bound MinD (the ‘Tarzan of the Jungle’ model). This latter feature appears necessary for robust pattern formation [48]. This interplay between MinD, MinE and the membrane leads to the observed dynamic pattern and has been modeled extensively [43].
In contrast to the pure oscillatory system in E. coli and many other Gram-negative organisms, the Min system in B. subtilis uses a landmark protein or molecular beacon to recruit the Min proteins [49] (Figure 3b). This protein, DivIVA, recognizes curved membranes at incipient septa [29]. In the absence of DivIVA, the Min inhibitor (MinC/MinD) is delocalized, blocking division throughout the cell causing a filamentous morphology. DivIVA forms a ring on either side of the invaginating septum and recruits MinJ which recruits MinD which recruits MinC [50, 51]. Once this DivIVA ring is decorated with the Min proteins it is able to prevent FtsZ released from the constricting Z ring from reassembling in the immediate vicinity [52]. This decorated ring maintains its diameter as septation ensues, but once cells separate the pole takes on a hemispherical shape and DivIVA is remodeled [29]. The Min proteins are eventually released and are able to join DivIVA assembling at the next incipient division site.
In B. subtilis, the binding of MinD to the membrane is ATP independent (likely due to a slightly longer amphipathic helix than E. coli MinD) [53]. Thus, the role of ATP binding is not to drive membrane binding but to drive interaction of MinD with its partners located at the pole. ATP binding also allows MinD to recruit MinC. How the MinD ATPase activity is regulated is not known. Thus, although this MinD binds to the membrane it is a landmark mechanism since the polar localization depends upon a molecular beacon, likely similar to other non-MinD members that are located at the cell’s pole (see below). Thus, the mechanism MinD uses depends upon its partner (MinE or DivIVA-MinJ), which varies among different bacteria [54].
Segregating plasmids and protein clusters: an oscillatory mechanism
Three systems for segregating low copy plasmids have been classified based upon the nucleotide hydrolyzing enzyme that powers segregation [55]. For type I the ATPase is a ParA type whereas for type II the ATPase (ParM) is related to actin. The third type employs a GTPase (TubZ) that it related to the FtsZ/tubulin family. Although the latter two employ dynamic filaments that push the plasmids apart, the ability to form cytoplasmic filaments is more controversial for the type I ParAs. Several of them have been shown to undergo ATP dependent polymerization in vitro, however, the physiological significance of this is still unclear. ParA binds nonspecifically to DNA forming a nucleoprotein filament, which is likely the relevant form of ParA [18].
Plasmid ParAs oscillate on the nucleoid due to reversible nonspecific DNA binding (Figure 4a) [36, 56]. The oscillation requires ParB bound to the centromere site ParS located on the plasmid. The oscillatory pattern is reminiscent of the Min system where MinE is chasing MinD. In this case, ParB with cargo attached (the plasmid) is chasing ParA that is spread over the nucleoid [36]. ParB bound to the ParS site stimulates the ATPase activity of ParA bound to nonspecific DNA (nucleoid). The simplest mechanism arising from these facts is a burnt bridge molecular ratchet mechanism [56]. In this model ParA dimerizes with ATP and binds nonspecifically to the nucleoid while ParB binds to ParS located on the plasmid. Interaction of the ParB/ParS complex with ParA leads to ATP hydrolysis and release of ParA from the DNA. Since the ParA is spread over the nucleoid it acts as a magnet to pull the ParB/ParS complex in one direction. As the complex moves, a ParA free region develops behind it, which in turn attracts ParA that is being released by ATP hydrolysis. Upon duplication of the ParB–ParS complex, one complex can be pulled in each direction leading to segregation. Modeling indicates that repeated cycles of ParA assembly and disassembly is sufficient for segregation of the duplicated plasmids [36].
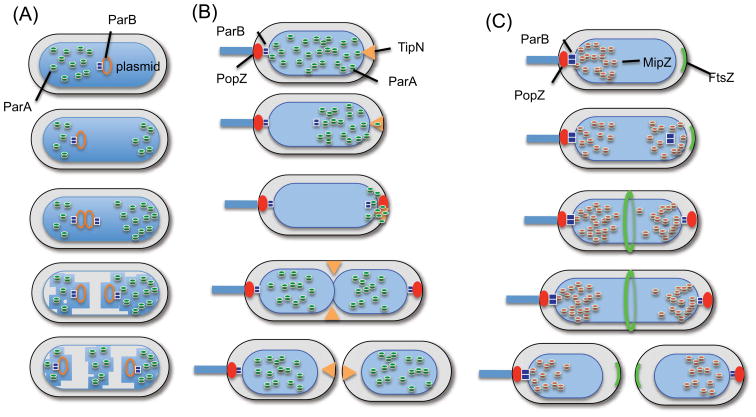
Figure 4.
Pattern formation involving ParA/MipZ and nonspecific DNA binding. ParA promotes the segregation of plasmids and cytoplasmic protein clusters (a) and the origin of chromosomes (b). (a) ParB bound to the parS site located on a plasmid displaces ParA that is bound nonspecifically to DNA. In some cases the cargo is not a plasmid but a cytoplasmic protein cluster (carboxysomes or chemotaxis clusters). (b) During chromosome segregation in C. crescentus the origin region is tethered to the polarity protein PopZ at the pole. Following duplication, one of the ParBs follows the receding ParA. The released ParA is maintained at the pole by TipN until late in the cell cyle when it is released and it spreads over the nucleoid. (c) MipZ, a distinct ParA protein, forms a bipolar gradient on the nucleoid to regulate the position of the Z ring in C. crescentus. The gradient emanates from ParB bound at the origin.
The oscillatory mechanism for segregating plasmids could be theoretically adapted to other cargo by simply having a component of the cargo interact with nucleoid associated ParA. This has apparently happened several times in evolution. In at least two cases orphan ParA proteins are required for segregation of large protein clusters, carboxysomes in one case [5] and large cytoplasmic chemotaxis clusters in another [4]. In the latter case a component of the chemostaxis cluster functions like ParB to tether the cargo to ParA and presumably stimulate ATP hydrolysis resulting in a molecular ratchet mechanism similar to that suggested for plasmids [57]. The orphan ParAs that function in this manner group with plasmid ParAs [9].
Segregation of chromosomal origins: oscillation with cues and aids
ParAs are also involved in chromosome segregation, at least in bacteria that do not undergo multifork replication. In comparison to plasmid segregation, however, several extrinsic proteins are required to ensure the unidirectional movement of the chromosomal origin (Figure 4b). The process appears similar among the two organisms studied, V. cholerae [58] and Caulobacter crescentus [32, 59, 60], although more details are known about the latter. The cell cycle starts with the origin sequestered at the old pole by ParB, which binds to centromere sites scattered around the origin and a polarly localized protein – PopZ, and ParA spread over the nucleoid [32]. As replication initiates, the ParB focus duplicates and one of them pursues the receding ParA. When delivered to the pole, the ParB-ParS complex binds to PopZ that has newly localized to this pole. During segregation the ParA released by ParB stimulation is sequestered by TipN, so that it cannot accumulate behind the migrating ParB which would cause it to stall or reverse direction (as seen with plasmids) [32]. At some point after segregation is completed, the ParA is released and spreads over the nucleoid to be in position for the next replication cycle. Thus, although both plasmid and chromosomal origin segregation rely on a ParB and nonspecific binding of ParA to the nucleoid, segregation of chromosomal origins requires additional proteins: PopZ, to tether ParB-origin complexes to the pole and TipN to ensure unidirectional movement.
Two proteins, two gradients, one regulator
In C. crescentus a distinct MinD/ParA member MipZ forms a bipolar gradient on the nucleoid to spatially regulate Z ring formation [14]. Like the ParA pattern in this organism, the bipolar MipZ gradient is dependent upon ParB and nonspecific DNA binding, however, its distribution on the nucleoid is temporally distinct from ParA (compare Figure 4, panels b and c). How can ParB cause two similar proteins to produce different patterns? The answer lies in the way ParB regulates the ATPase activity of the two proteins. For ParA, ParB functions as an AAP removing ParA from the DNA, whereas, for MipZ, ParB promotes dimerization and therefore DNA binding. Thus, the MipZ gradient emanates from ParB as MipZ monomers, recruited by ParB, undergo dimerization, diffuse away and bind nonspecifically to the nucleoid. So far MipZ is the only MinD/ParA member that has its ATPase regulated in this unusual manner.
A landmark mechanism for chemotaxis and type IV pili
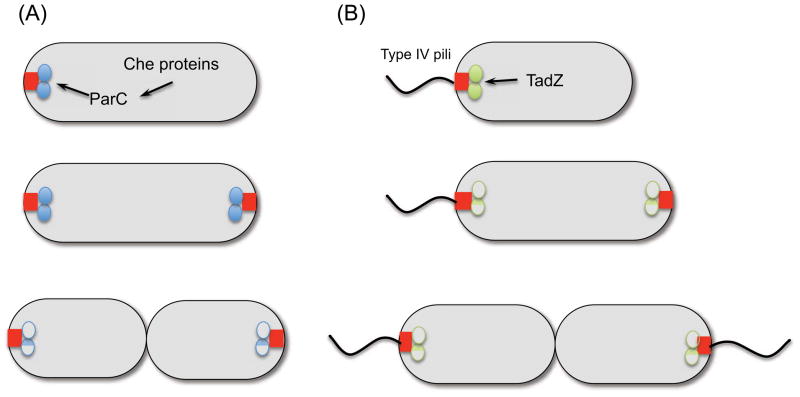
As mentioned above, some orphan ParAs are involved in the distribution of chemotaxis protein clusters by using an oscillating mechanism that mimics plasmid partitioning. However, other ParAs (designated ParCs) position chemotaxis proteins using a landmark mechanism [9]. A newborn V. cholerae cell contains a ParC focus at the old pole and as the cell cycle proceeds a new ParC focus develops at the new pole, so that following division each progeny cell inherits a cluster (Figure 5a). At no time is ParC localized on the nucleoid. Phylogenetically, ParC represents a new ParA clade whereas ParAs associated with cytoplasmic chemotaxis clusters fall with plasmid ParAs [9] (Figure 1). Although it is not clear how this ParA focus is recruited to the pole or how recruitment is regulated by nucleotide hydrolysis, it is clear that ParC is required for the polar recruitment of other chemotaxis proteins and that ATP hydrolysis is required. This mode of localizing proteins follows the landmark mechanism. Thus, there are two types of ParAs, one that oscillates on the DNA (and moves cargo) and one that does not bind DNA and residency depends on a preexisting polar determinant (and functions as a link to recruit other proteins).
Figure 5.
ParC and TadZ are involved in the polar localization of chemotaxis proteins and type IV pili respectively. (a) ParC is related to ParA but does not bind DNA. The polar determinant it binds to is unknown. (b) TadZ is a link between an unknown polar determinant and the type IV pilus machinery.
Type IV pili are found in many bacteria and are often localized to a single pole where they mediate adherence to surfaces (Figure 5b). There are at least three different subtypes, one of which is encoded by a widespread colonization island called tad. One of the dozen or so genes on this island, tadZ/cpaE encodes a fusion of a MinD/ParA-like protein with the receiver domain of a response regulator and both domains are needed for polar localization [11, 12]. Some TadZs are missing the signature lysine but a recent structure analysis of one of these revealed that this role is assumed by a lysine elsewhere in the structure [12]. It appears that TadZ is a link between a polar determinant functioning (PodJ-TipN in C. crescentus) as a beacon and other Tad components [62, 63].
Variations and questions
Investigation into the Min systems of E. coli and B. subtilis has already revealed that MinD can use either an oscillatory or landmark mechanism depending upon its protein partners. Also, one clade of orphan ParAs (ParC) does not bind DNA and uses a landmark mechanism to localize chemotaxis proteins, whereas other orphan ParAs are closer to plasmid ParAs and use an oscillatory mechanism to segregate non-DNA cargo, such as cytoplasmic chemotaxis clusters. Although there have been extensive studies of how oscillatory ATPases are regulated, little is known about the ATPases associated with the landmark mechanism. For example, MinE stimulation of MinD has been explored in some detail but how the ATPase activity of MinD in B. subtilis is regulated is unknown. Also, how the ATPase activity of VirC1 from A. tumefaciens is involved in localizing the transfer intermediate at the pole for conjugative DNA transfer is not clear [64].
Although some ParAs are clearly involved in segregation of chromosomal origins, for others it is not so clear. A well studied ParA from B. subtilis (Soj) and its ParB partner (SpoOJ) have long been thought to be involved in chromosome segregation, however, the evidence for direct involvement is not convincing [30]. SpoOJ does have a role in chromosome organization through recruitment of a DNA organizing protein SMC (structural maintenance of chomosomes) [61]. Also, SpoOJ and Soj cooperate to regulate the activity of DnaA; in the ADP form Soj prevents DnaA from assembling into an initiation helix at oriC [30]. Despite the rather widespread distribution of chromosomal ParA and ParB genes, they are not found in E. coli and closely related enterics. It may be that their involvement in chromosome segregation may be limited to bacteria (V. cholerae and C. crescentus) that do not carry out multifork replication (in contrast to E. coli and B. subtilis). However, this remains to be determined.
Bacteria have come up with a variety of mechanisms to spatially restrict assembly of the Z ring. E. coli and B. subtilis use a combination of Noc and Min to restrict Z ring assembly to midcell [37], whereas C. crescentus uses a single protein, MipZ, that seems to combine these functions [14]. Its ATPase activity is regulated in a very different manner. Rather than ParB stimulating ATP hydrolysis directly, ParB promotes dimerization of MipZ which undergoes ATP hydrolysis after binding nonspecifically to DNA. So far, this mechanism is unique to MipZ. Recently, the spatial regulation of cell division in Corynebacterium glutamicum (which also lacks MinD and Noc) was found to utilize another orphan ParA family member (PldP) [15]. Deletion of PldP causes aberrant divisions and its localization is similar to MinD of B. subtilis. How PldP works has not yet been investigated.
Clearly MinD/ParA proteins can be used as adapters in localizing proteins to the poles of cells. However, it is also clear that many other proteins can perform this task [65]. For example, although most type IV pili are found at the poles, only one of three identified type IV systems contains a MinD/ParA homologue [66].
Concluding remarks
The MinD/ParA family of proteins has evolved as a nucleotide dependent switch that regulates its affinity for a surface and other proteins. In the best studied examples, the protein bind to a surface and the switch is activated by a partner that is spatially restricted leading to an oscillatory pattern. Patterns have been observed on membranes that regulate the positioning of the Z ring and on the nucleoid leading to segregation of plasmids, oriC regions, protein clusters and positioning of the Z ring. In some cases the MinD/ParA members do not bind to a surface but to a landmark protein that act as a molecular beacon for recruitment. Such proteins are used to place a variety of structures at the poles of the cell. Clearly this family, which likely evolved from the more ancient Mrp chaperone [6], is quite adaptable and has been exploited over and over by bacteria to localize a variety of structures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–4. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.Webb CD, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–74. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 3.Gordon GS, et al. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–21. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 4.Thompson SR, et al. The positioning of cytoplasmic protein clusters in bacteria. Proc Natl Acad Sci U S A. 2006;103:8209–14. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage DF, et al. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–61. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 6.Leipe DD, et al. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 7.Boyd JM, et al. Archaeal ApbC/Nbp35 homologs function as iron-sulfur cluster carrier proteins. J Bacteriol. 2009;191:1490–7. doi: 10.1128/JB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hausmann A, et al. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc Natl Acad Sci U S A. 2005;102:3266–71. doi: 10.1073/pnas.0406447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ringgaard S, et al. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 2011;25:1544–55. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szardenings F, et al. ParA ATPases can move and position DNA and subcellular structures. Curr Opin Microbiol. 2011;14:712–8. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Cheeks BA, et al. The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol Microbiol. 2012;83:694–711. doi: 10.1111/j.1365-2958.2011.07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q, et al. Structure of the pilus assembly protein TadZ from Eubacterium rectale: Implications for polar localization. Mol Microbiol. 2011;83:717–27. doi: 10.1111/j.1365-2958.2011.07954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Quere B, Ghig JM. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol Microbiol. 2009;72:24–40. doi: 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- 14.Thanbichler M, Shapiro MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–62. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 15.Donovan C, et al. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J Bacteriol. 2010;192:3441–51. doi: 10.1128/JB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 17.Schindelin H, et al. Structure of ADP x AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–6. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 18.Leonard TA, et al. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EMBO J. 2005;24:270–82. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu W, et al. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Mol Microbiol. 2011;79:1515–28. doi: 10.1111/j.1365-2958.2010.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–6. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z, et al. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci U S A. 2002;99:6761–6. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou H, et al. Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction. J Bacteriol. 2005;187:629–38. doi: 10.1128/JB.187.2.629-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szeto TH, et al. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci U S A. 2002;99:15693–8. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Lutkenhaus J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol. 2003;47:345–55. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- 25.Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci U S A. 104:20326–31. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiekebusch D, et al. Localized dimerization and nucleoid binding drive fradient formation by the bacterial cell division inhibitor MipZ. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell. 2001;7:1337–43. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 28.Bouet JY, Funnell BE. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 1999;18:1415–24. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eswaramoorthy P. Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. MBio. 2011;2:e000257–11. doi: 10.1128/mBio.00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scholefield G, et al. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31:1542–55. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholefield G, et al. SpoOJ regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol. 2011;79:1089–100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- 32.Schofield WB, et al. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–81. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro L, et al. Why and how bacteria localize proteins. Science. 2009;326:1225–8. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–24. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringgaard S, et al. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci U S A. 2009;106:19369–74. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–62. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 38.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:4971–6. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varma A, et al. The Min system as a general cell geometry detection mechanism: branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J Bacteriol. 2008;190:2106–17. doi: 10.1128/JB.00720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corbin BD, et al. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 2002;21:1998–2008. doi: 10.1093/emboj/21.8.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loose M, et al. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–92. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 42.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci U S A. 2010;107:8071–8. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard M, Kruse K. Cellular organization by self-organization: mechanisms and models for Min protein dynamics. J Cell Biol. 2005;168:533–6. doi: 10.1083/jcb.200411122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loose M, et al. Protein self-organization: lessons from the Min system. Annu Rev Biophys. 2011;40:315–36. doi: 10.1146/annurev-biophys-042910-155332. [DOI] [PubMed] [Google Scholar]
- 45.Meinhardt H, de Boer PA. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci U S A. 2001;98:14202–7. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lackner LL, et al. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol. 2003;185:735–49. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park KT, et al. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146:396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loose M, et al. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat Struct Mol Biol. 2011;18:577–83. doi: 10.1038/nsmb.2037. [DOI] [PubMed] [Google Scholar]
- 49.Marston AL, et al. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–30. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patrick JE, Kearns DB. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol. 70:1166–79. doi: 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- 51.Bramkamp M, et al. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol Microbiol. 2008;70:1556–69. doi: 10.1111/j.1365-2958.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- 52.Gregory JA, et al. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 2008;22:3475–88. doi: 10.1101/gad.1732408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- 54.Rothfield L, et al. Spatial control of bacterial division-site placement. Nat Rev Microbiol. 2005;3:959–68. doi: 10.1038/nrmicro1290. [DOI] [PubMed] [Google Scholar]
- 55.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–42. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 56.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts MA, et al. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc Natl Acad Sci U S A. 2012;109:6698–703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–82. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shebelut CW, et al. Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci U S A. 2010;107:14194–8. doi: 10.1073/pnas.1005274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–8. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–96. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 62.Viollier PH, et al. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–8. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viollier PH, et al. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 99:13831–6. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atmakuri K, et al. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J. 2007;26:2540–51. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirkpatrick CL, Viollier PH. Poles apart: prokaryotic polar organelles and their spatial regulation. Cold Spring Harb Perspect Biol. 2011;3:a006809. doi: 10.1101/cshperspect.a006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayers M, et al. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 2010;5:1203–18. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]