Abstract
A novel protein identification framework, PILOT_PROTEIN, has been developed to construct a comprehensive list of all unmodified proteins that are present in a living sample. It uses the peptide identification results from the PILOT_SEQUEL algorithm to initially determine all unmodified proteins within the sample. Using a rigorous biclustering approach that groups incorrect peptide sequences with other homologous sequences, the number of false positives reported is minimized. A sequence tag procedure is then incorporated along with the untargeted PTM identification algorithm, PILOT_PTM, to determine a list of all modification types and sites for each protein. The unmodified protein identification algorithm, PILOT_PROTEIN, is compared to the methods SEQUEST, InsPecT, X!Tandem, VEMS, and ProteinProspector using both prepared protein samples and a more complex chromatin digest. The algorithm demonstrates superior protein identification accuracy with a lower false positive rate. All materials are freely available to the scientific community at http://pumpd.princeton.edu.
Introduction
Tandem mass spectrometry (MS/MS) has emerged as the premier tool for protein identification of cellular samples.1,2 Most large scale studies currently use a shotgun proteomics approach, where proteins are extracted from a living sample, enzymatically digested, and fractionated.3 The peptides are then fragmented using MS/MS and then typically analyzed using either database4–14 or hybrid de novo/database methods.15–24 There is currently no automated high-throughput pure de novo approach for protein identification in the literature. Protein identifications can then be inferred based on the individual peptide sequences. This final protein list reported by an algorithm is the prime objective of a large-scale proteomics experiment, so it is imperative that the methods used to generate the list can predict a high number of correct protein identifications with a low number of false positives.3,25
As large-scale cellular samples have a diverse population of proteins, it is difficult to quantify the protein identification accuracy of an algorithm exclusively by the total number of correct protein “hits”. The false-discovery rate (FDR) has been used as an additional metric for the quality of a protein identification list. Statistical methods based on parametric distributions,26 hierarchical modeling,27,28 cumulative scoring,29 or target-decoy strategies30 have been developed to use the evidence for peptide identifications for computing protein identification probabilities. The FDR for protein identification is typically higher than that for peptide identification since any errors in peptide identification will propagate to the protein level. For example, false identifications of proteins are typically the result of the annotation of a single incorrect peptide while correctly identified proteins are often labeled on the basis of many peptides. Though many instruments are available for MS/MS analysis, the most accurate measurements come from using high-resolution detector types including time-of-flight, Orbitrap, and ion cyclotron resonance. The high accuracy of these instruments can yield better peptide and protein identification and can help reduce the number of false positive results reported.
Though these methods can provide very reasonable assessments for the protein identification FDR of a particular tool, it is not possible to exactly quantify the number of false positives reported for a large-scale sample. That is, due to the unknown size and types of proteins that are in a typical shotgun proteomics experiment, the number of “true” proteins cannot be identified. To address this issue, a standard protein mixture31 can be developed where the protein list is known a priori. Using such a mixture, the number of protein “hits” can be quantified based solely on the proteins used to construct the sample and any common contaminants that are found throughout the samples. Though such a protein mixture may have an order of magnitude less proteins than a typical cellular sample, it can provide critical insight and serve as a test bed for the predictive capability of an algorithm on both the peptide and protein level.
Determination of an accurate protein list is also crucial for the identification of post-translational modifications (PTMs). Several algorithms have been developed for sample analysis that are capable of protein identification and post-translational modification (PTM) search.4–7,9–17,20–24,32–41 Typically, an initial analysis of cellular data will have a limited number of variable PTMs due to the exponential increase in database search time that result in enumeration of all combinations of modified peptides. This is typically resolved by implementing a two-pass approach33,42,43 where the database is initially scanned either with no modifications or a small subset of variable modifications to eliminate proteins that did not score above a given threshold (based on the peptide hits). In the second pass, the protein list found from the first search can be used in an untargeted search that contains a larger variety of variable modifications or other unusual digestion/fragmentation information. The inaccuracies in the protein list from the first pass may transfer to the second pass if a peptide from an incorrect protein is used to identify potential PTMs.
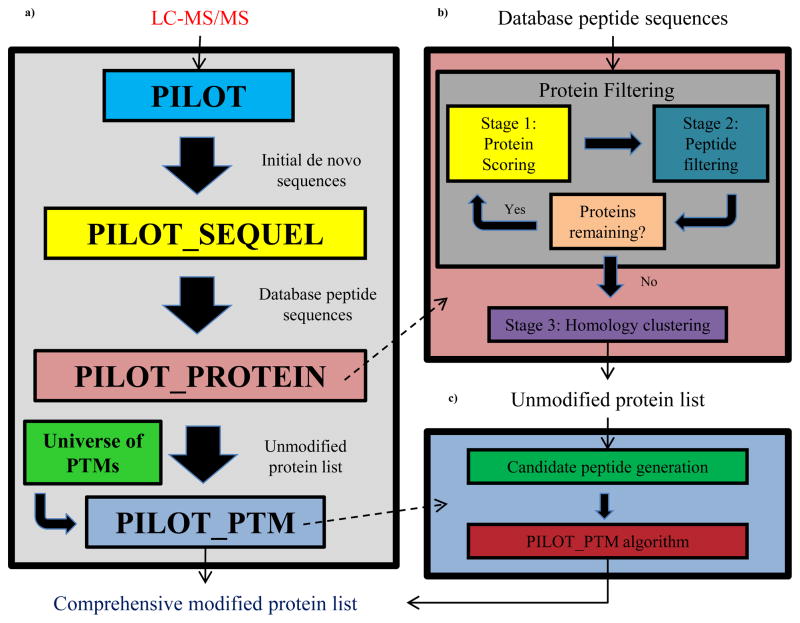
In this paper, a complete proteomics workflow method (Figure 1a) is introduced to identify a comprehensive list of unmodified and modified proteins using high-resolution MS/MS. LC-MS/MS data is initially analyzed using the PILOT44,45 algorithm to find a rank-ordered list of unmodified de novo peptide sequences. These sequences are subsequently analyzed using PILOT_SEQUEL46 to find unmodified database peptides that closely match the de novo sequences. A novel protein identification method, PILOT_PROTEIN, has been developed to predict a comprehensive list of unmodified proteins (Figure 1b) from the peptide list generated by PILOT_SEQUEL. PILOT_PROTEIN combines the scores of the de novo sequences and the database peptides to score all of the possible proteins and outputs an unmodified protein list with a minimal number of false positives. Using this output unmodified protein list, the PILOT_PTM47 algorithm will perform a second pass over the LC-MS/MS data and perform an untargeted PTM search and identify any modification types and sites that are present on a sample protein (Figure 1c). The final result will be a comprehensive protein list that contains the types and sites of all modifications present in the data. The novel aspects of this work include (i) the development of a unmodified protein identification algorithm that produces a competitive number of protein “hits” with respect to state-of-the-art algorithms, (ii) the utilization of a rigorous biclustering algorithm to identify peptide homologues that are incorrectly labeled and therefore reduce the false positive output, (iii) the generation of template amino acid sequences from the unmodified protein list that can serve as good inputs so the PILOT_PTM algorithm can identify a good list of modified spectra, and (iv) the development of a completely integrated webtool (http://pumpd.princeton.edu) that allows free access to the PILOT, PILOT_SEQUEL, PILOT_PROTEIN, and PILOT_PTM algorithms for identification of all sample proteins along with all corresponding types and sites of PTMs.
Figure 1.
(a) Overall framework for LC-MS/MS analysis. LC-MS/MS data is sent to PILOT algorithm for de novo sequence generation. The de novo sequences are compared against a protein database using PILOT_SEQUEL to extract database peptides and their corresponding proteins. This information is passed to PILOT_PROTEIN for generation of an unmodified protein list. The unmodified protein list is used with PILOT_PTM to annotate all protein modification types/sites. (b) Framework for PILOT_PROTEIN. Proteins are initially filtered based on PILOT and PILOT_SEQUEL peptide scores. Peptide homologues are identified using OREO to help increase protein identification specificity. (c) Framework for PILOT_PTM. Using the smaller unmodified protein list, candidate peptides for PILOT_PTM are generated for each MS/MS spectrum after a sequence tag search. PILOT_PTM will output the highest-scoring modification set for annotation.
Methods
PILOT_PROTEIN Algorithm for Unmodified Protein Identification
The framework for PILOT_PROTEIN consists of three distinct stages (Figure 1b). The input to the algorithm is a complete list of proteins, each of which is assigned a rank-ordered list of peptides from the PILOT_SEQUEL algorithm. The first stage scores all proteins in the current protein list. Scoring of each protein uses the individual scores of each rank-one peptide that is found within the protein. Bias toward redundant sequences is reduced by considering only the top score for a given peptide sequence. The top scoring protein is retained and is annotated with all corresponding peptides. The second stage analyzes all remaining proteins to filter out any spectra that may contain a rank-two or higher peptide that can be associated with the protein found in stage one. Any proteins that no longer have a peptide are removed from consideration while all others are analyzed using stage one. This iterative procedure continues until no proteins remain. The third stage of PILOT_PROTEIN consists of a peptide clustering approach using OREO.48,49 Using the Smith-Waterman50 alignment score as a distance metric, all peptides within the filtered protein list are clustered together to identify any homologous sequences that have sequence mass differences within the parent mass tolerance. This stage of the algorithm helps to identify sequences that are incorrectly annotated and would otherwise lead to a lower protein identification specificity. The algorithm is described in full detail below.
Input/Output
Input to the PILOT_PROTEIN algorithm consists of a list of MS/MS spectra, each of which has been analyzed with the hybrid de novo/database method PILOT_SEQUEL. From PILOT_SEQUEL, each MS/MS spectrum is assigned a list of scored peptides that are directly derived from a protein database. Each peptide corresponds to a list of all proteins in the database that contain the amino acid sequence as an enzymatically cleaved peptide. The output to the user is a rank-ordered list of scored proteins with each protein containing (i) the peptide score, (ii) a list of peptides found, and (iii) a list of MS/MS spectra that contain the peptides.
Stage 1: Protein Scoring
Initially given is a set of MS/MS spectra (t ∈ MSMS), each of which will have a rank-ordered list of peptides p ∈ Pept with a score . Each peptide p is a theoretically digested sequence from a list of proteins r ∈ Prp. The complete list of peptides for analysis is given by Equation (1).
| (1) |
Using this peptide list, the complete list of proteins for analysis is then defined as in Equation (2).
| (2) |
Using the complete protein list ProtList, all proteins are scored using their corresponding peptides (p ∈ Pror) as shown in Equation (3). Bias toward redundant sequences is reduced by considering only the top score for a given peptide sequence (Eqn. 4).
| (3) |
| (4) |
The protein rk in ProtList with the highest score is removed from the set and added to the filtered protein list FilProt.
Stage 2: Peptide Filtering
A given MS/MS scan t is annotated if t contains at least one peptide p with score greater than a threshold ( ) that is part of the protein’s theoretical peptide list (p ∈ Prork). The threshold value is representative of a PILOT_SEQUEL peptide that is a direct match to a de novo sequence without the potential rewards of high-confidence residues.46 The value is defined as the score for which a 2% FDR is achieved for the LC-MS/MS data set. If possible, the scan t is annotated with the highest-scoring peptide. All annotated spectra are removed from the set MSMS and added to the set FilMSMS. If there are no MS/MS spectra remaining in the set MSMS, then the protein filtering terminates. Otherwise, the stage 1 process is repeated beginning at Equation (1).
If multiple proteins match a single peptide, this peptide will be annotated to the protein that has the strongest set (i.e., highest scoring) of additional peptides in the data. This is a consequence of the scoring methodology of the algorithm since the method (i) scores all proteins using any peptide information, (ii) retains the highest scoring protein, and finally (iii) removes all peptides from consideration that are associated with the protein found in (ii). This three stage process repeats itself until the number of remaining peptides is equal to zero. If during part (ii), any two (or more) proteins have the exact same list of supporting peptides (either one or more peptides), all proteins in this list are considered as equally valid and are reported to the user as a valid “match”. Note that without further peptide information, it is not possible to distinguish between the proper annotation of any one of these proteins.
Stage 3: Sequence Based Clustering
Once the unmodified protein list (FilProt) has been filtered and scored, an analysis of the low-rank proteins can help to eliminate false positives. Many of the incorrectly annotated proteins will be associated with peptides that are only found once or twice within the full LC-MS/MS scan. Therefore, it is critical that any peptides with low spectral counts be further analyzed to validate the assignment. The PILOT_PROTEIN algorithm uses the biclustering method OREO48,49 to group together peptides that have sequence similarity and identify potential homologues for peptide sequences. The mathematical model for biclustering is detailed below.
Scoring Matrix
To score a pair of distinct peptides, a FASTA alignment matrix is traditionally used.46 Matrices based upon evolutionary distances between amino acids are not used because conservation of mass between the peptide sequences is very important. Specifically, for two peptides to be considered as homologues, they must not differ by more than twice the threshold tolerance for the parent mass. This criterion is imposed because the difference between the actual parent mass and the experimental parent mass can be at most the threshold parent error tolerance for any peptide. Thus, if two peptides differ by more than twice the threshold tolerance, then it is not possible to re-assign either of the peptides to the alternate spectrum. Additionally, it is anticipated that an incorrectly annotated peptide will have a sequence that is very similar to another sequence that was annotated in the LC-MS/MS analysis or that is a theoretically digested peptide of a high-rank protein but not found in a MS/MS spectrum annotation. Thus, a scoring matrix is used that rewards exact residue matches with a score of +5 and penalized incorrect matches with a score of −5. Leucine and isoleucine matches are given a score of +5 and lysine/glutamine matches are given a score of +5 if the fragment ion tolerance is greater than 0.03 Da. These scoring values have been chosen based on successful application of the PILOT_SEQUEL algorithm on high resolution test data sets.46
Isobaric Residues
It is important to consider the treatment of isobaric residues during the alignment procedure. Isobaric residues can exist on either of two compared peptide sequences, and are generally present due to incorrect de novo sequence predictions. This can possibly reduce the overall alignment score between two peptides and prevent the identification of a homologous pair. To compensate, the Smith-Waterman50 alignment routine in the FASTA algorithm is altered to replace the penalty for sequence mismatch and gap insertion with a reward (i.e., +5) for isobaric alignment.
Biclustering Mathematical Model
The complete mathematical model for biclustering utilizes an input matrix of values to identify cluster boundaries within a re-ordered matrix.48,49 However, PILOT_PROTEIN uses an input vector of peptides for cluster determination, so a reduced form of the model is required. A full description of re-ordering over a matrix is presented elsewhere.48,49
All distinct peptide sequences are sorted by increasing total mass and several vectors are created. The mass-sorted array is decomposed into smaller vectors at every point where two adjacent peptide sequences have total masses that differ by more than twice the parent mass tolerance. All vectors that contain low-confidence sequences are then analyzed with the biclustering model to identify any homologous high-scoring peptides. For a given vector, the index i represents a specific element in the vector whose peptide sequence is given as Si.
Binary variables (yi,i′) are defined that represent the position of the peptide sequences in the final re-ordering of the vector.
| (5) |
That is, if y3,9 is equal to one, then element 3 is immediately above element 9 in the final arrangement of the vector. If y3,9 is equal to zero, this implies that element 3 is not immediately above element 9 in the final ordering, but does not reveal any additional information.48,49
The objective function maximizes the total alignment score between adjacent peptide sequences (Eqn. 6).
| (6) |
Equation (6) will identify all peptides that have high sequence similarity to the query sequence Siq. The cluster boundaries of the optimal re-ordering are found when the normalized alignment score (Ai,i′ divided by length of sequence Si) of adjacent elements is less than 2. The cluster of peptides containing the query sequence Siq is subsequently analyzed to determine the validity of the query sequence.
Homology Labeling
For each low-scoring spectral assignment q, a cluster of homologous sequences Cq is defined by OREO.48,49 The spectral assignment is considered to be invalid if there exists another sequence q′ in the cluster that has been annotated in at least three MS/MS spectra and at least one of those MS/MS spectra has a PILOT_SEQUEL score above a given threshold. The threshold value is set to 6.5, which is indicative of a database peptide that has both (i) high sequence similarity to the de novo sequence and (ii) is matched to high-confidence de novo sequence residues.46 Beginning with the cluster sequence that has the highest alignment score, the above criterion is checked. If the two conditions are met, then the low-scoring sequence q is assumed to be a homologue of q′ and the MS/MS scans annotated with sequence q are re-annotated with sequence q′. If the conditions are not met, then this process is repeated using the sequence with the second-highest alignment score and will iteratively proceed through the entire cluster until a homologue is found or all sequences fail the criterion. If all sequences within the cluster fail the criterion, then the original annotation is maintained.
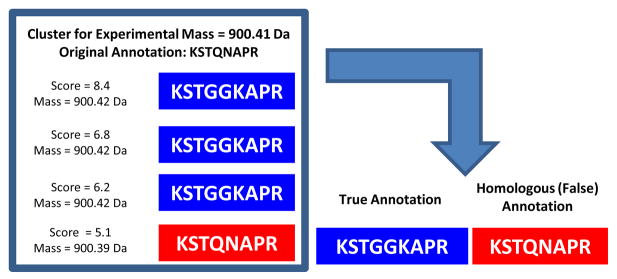
As an example, Figure 2 shows the clustering approach applied to a MS/MS spectrum with experimental parent mass 900.41 Da. The original annotation (in red) is KSTQNAPR, which is an invalid assignment and is the only peptide assignment in the LC-MS/MS associated with the corresponding protein. Without the clustering approach, this false-positive protein will be output by the PILOT_PROTEIN algorithm if the minimum protein score threshold is lower than 5.1 (the score of the sole peptide annotation). However, the clustering method identifies several candidate peptides (in blue) with sequence similarity to the incorrect peptide. Note that this example shows a peptide sequence (KSTGGKAPR) that has been assigned to three distinct MS/MS spectra, and the top scoring assignment has a score of 8.4. Thus, the clustering approach will label the KSTQNAPR assignment as a homologous (false) annotation and re-assign the spectrum to the KSTGGKAPR sequence.
Figure 2.
Example of peptide homology clustering. The original annotation (KSTQNAPR; red) to a spectrum with experimental mass 900.41 Da can be corrected using the proper assignment of KSTGGKAPR (blue) to three other distinct spectra.
Note that the possibility of removing a peptide homologue for which a sequence differs by only one amino acid depends on the tolerance of the parent mass error. For high resolution instruments, this particular case should happen very infrequently since two peptides that differ by one amino acid will generally have parent masses that are beyond the allowable threshold tolerance. If a lower resolution instrument is used, then it is possible that a proper homologous peptide would be removed if the single different amino acid is similar for each peptide (e.g., I and N). However, this analysis has been restricted to high-resolution parent mass detectors so the only ambiguities that may arise are between I and L or between K and Q. The authors note that the assignment of I or L is impossible for any algorithm to distinguish and that the K/Q homology was not detected in this study.
Once the biclustering/homology labeling routine is completed for all peptides with low spectral counts, then all proteins are re-scored using the new peptide annotations. Any peptide that was considered a homologue is not used for protein scoring, but is still associated with the protein as a redundant sequence. All proteins that do not pass the scoring threshold ( ) are eliminated from consideration. This threshold criterion directly builds off of the threshold cutoff for PILOT_SEQUEL and implies that a protein would either require one assigned database peptide that had a good match to the de novo sequence or else a combination of peptides would be necessary. Whenever a set of peptides that identifies a particular protein may also correspond to another protein, PILOT_PROTEIN will report a “protein group” for that group of peptides to allow the user to resolve any ambiguity. All remaining peptides are output to the user.
Search for Protein Modifications using PILOT_PTM
Upon generation of all unmodified proteins in a protein sample, a targeted or untargeted modification search can be performed. The PILOT_PTM algorithm47 will utilize the smaller list of unmodified proteins on a second-pass of the LC-MS/MS data to determine all modification sites and types for any peptide that is an enzymatic fragment from these proteins. The comprehensive list of modifications used for the search is constructed using all known PTMs, chemical derivatives, and artifacts found in the UniMod,51 RESID52 and Delta Mass53 databases. Each modified peptide corresponding to the modified protein will also be annotated with the modification information. PILOT_PTM will be run on every MS/MS spectrum that is not annotated with a peptide that is assigned to a protein by PILOT_PROTEIN. To run the PILOT_PTM algorithm, a list of test peptides must be generated by in silico digestion of the unmodified protein list. It is assumed that any modified peptide found in the sample must be part of an unmodified protein found from PILOT_PROTEIN. The following sections detail the approach for test peptide generation for PILOT_PTM.
Candidate Peptide Generation
Using the maximum number of missed cleavages, the number of specific termini, and the digestion enzyme input by the user, the unmodified proteins from PILOT_PROTEIN are theoretically digested to generate a “candidate peptide” list. When analyzing a MS/MS spectrum for PTMs, the first step is to identify a three amino acid sequence tag that will isolate one or more “test peptides” that may be found in the MS/MS spectrum. The mathematical model for sequence tag generation is outlined below.
Sequence Tag Generation
After the MS/MS spectrum is preprocessed, there exists a list of filtered ion peaks, (p ∈ P), each of which is associated with a given mass (Mp) and intensity (Ip). A complete list of three amino acid sequences q is input and used to generate appropriate sequence tags t as follows. Given a base peak pb (mass Mpb) and an amino acid sequence q, a sequence tag t is found if there exists an end peak pe (mass Mpe) such that |Mpe − Mpb − Mq| < tolfrag where Mq is the mass of the three amino acid sequence q. A peak p is considered part of the sequence tag set Pt if there exists a N-terminal subsequence of q such that where is the mass of the subsequence. Note that the null subsequence and the full subsequence are considered, so pb and pe are in the set Pt. Complementary peaks may be added to the set Pt if they exist in P. The full set of sequence tags is labeled as T.
Sequence Tag Scoring
Each sequence tag t is scored using the objective function in Equation (7). The sum of the intensities of the peaks that comprise the tag (Ip) is premultiplied by a weighting coefficient (Ct) that is generally equal to 1, but can be reduced as follows. If the mass error between two consecutive peaks is greater than 40% of the nominal user input fragment tolerance, then Ct is reduced by 0.2. If two consecutive peaks represent an amino acid doublet or triplet (i.e., one or two missing peaks, respectively), then Ct is reduced by 0.2 or 0.4, respectively.
| (7) |
The top 5 sequence tags are scanned against the candidate peptide list to extract out the test peptides for the MS/MS spectrum. A candidate peptide is retained if an exact match to the sequence tag is found within the amino acid sequence and if the mass gaps on the N-terminal and C-terminal sections are within −50 Da and 250 Da. The mass gap limitation is imposed to select peptides that will ultimately have a modification mass within the given mass window. This analysis generally retains 5–10 test peptides for use as input to the PILOT_PTM algorithm.
PILOT_PTM
All MS/MS spectra not annotated with PILOT_PROTEIN are analyzed using PILOT_PTM47 for an untargeted PTM search (Figure 1c). All unmodified proteins identified by PILOT_PROTEIN are theoretically digested to generate a candidate peptide list. MS/MS spectra are analyzed for the existence of a sequence tag15 which generates a list of test peptides, as outlined above. Each test peptide is analyzed with PILOT_PTM and the peptide with the highest cross-correlation score is retained along with the corresponding set of PTMs. Completion of this approach for all MS/MS spectra yields a comprehensive list of modified and unmodified proteins, with all PTM sites, PTM types, and supporting peptides output to a user. If the PILOT_PTM method is unable to fully resolve the particular amino acid site for a given modification type due to incomplete fragmentation, then the method will output a list of amino acid sites for which the modification type may exist. In the analysis below, PILOT_PTM was able to localize the expected site for a modification for each annotated spectrum. Though the top scoring peptide from PILOT_PTM is reported in this manuscript, PILOT_PTM will also output a rank-ordered list of all modification sets for a peptide47 through the use of integer cuts.54 Note that the ILP model used for the PILOT_PTM algorithm can be formulated using network based constraints.44,45,55–59 Some annotations that correspond to the same amino acid sequence, the same set of modifications, and different site assignment for the modifications may be reported as a lower-rank sequence. All such annotations are assumed to be inferior to the top-rank sequence and are not included as part of the analysis.
Sample Preparation
Test Set A - QTOF Peptides
These spectra were derived from a publicly available data set.60 The spectra were collected with Q-TOF2 and Q-TOF-Global mass spectrometers using a mixture of alcohol dehydrogenase (yeast), myoglobin (horse), albumin (bovine, BSA), and cytochrome c (horse). A test set of 37 spectra was obtained using only “ acceptable spectra” as previously defined.44
Test Set B - Orbitrap Peptides
Stock solutions of a 16 protein mixture were prepared containing equal amounts of each protein as previously described.46 The proteins were digested with trypsin and analyzed by automated microcapillary liquid chromatography and a LTQ-Orbitrap hybrid mass spectrometer (ThermoFinnagin, San Jose, CA). Both MS and MS/MS spectra were recorded on the instrument and a test set of 401 spectra was annotated using the SEQUEST algorithm.4
Test Set C - Standard Protein Mix Peptides
Six 18-protein mixtures were preparted for LC-MS/MS analysis as previously described.31 The proteins were digested with trypsin and analyzed using either QTOF (QSTAR), LTQ-FT, or Orbitrap mass spectrometers.
Test Set D - Total Chromatin Fraction
HeLa S3 cells were cultured and harvested as recently described.61,62 5 distinct chromatin fraction samples from the HeLa cells were prepared using either a salt extraction, a micrococcal nuclease (MNase) digestion, or a total extraction. The salt extraction and MNase digestion provided both a pellet and a supernatant extraction.62 For each sample, extracted protein was separated using 1D-SDS-PAGE and in gel digested by trypsin following treatment with iodoacetamide. Peptide digests were then analyzed by nanoflow LC-MS/MS on an Orbitrap mass spectrometer as previously described.63
PILOT_PROTEIN Scoring Method
PILOT_PROTEIN is benchmarked against several state-of-the-art algorithms using the standard protein mix database (data set C) and the total chromatin fraction (data set D). Data set C is comprised of a known mixture of 18 sample proteins, so the list of correct protein hits is known a priori. For this data set, the accuracy of an algorithm is measured using two metrics: (a) protein identification sensitivity and (b) protein identification specificity. The definition of each accuracy metric for each algorithm is given below. Data set D is prepared by extracting a chromatin fraction and will therefore contain a large amount of proteins that cannot be comprehensively annotated a priori. Therefore, the identification accuracy of an algorithm will be measured using a reverse sequence decoy database64 and the number of peptide spectrum matches (PSMs) will be analyzed at various levels of false discovery rate. A PSM is defined as the peptide-spectrum pair associated with the assignment of a peptide sequence to a particular MS/MS spectrum. For a LC-MS/MS run, a peptide identification algorithm can report both a list of unique peptides and a list of PSMs. The unique peptide list gives an indication of how many distinct peptides were identified throughout the experiment while the PSM list provides an indication of how many MS/MS spectra were identified with a peptide. Note that the number of PSMs must be higher than the number of unique peptides because the same peptide may be found in different MS/MS spectra. Though a peptide sequence may be repeated in the PSM list, the peptide-spectrum pair will be unique in the PSM list because only one peptide will be annotated for each MS/MS spectrum. For a benchmark false discovery rate of 2%, a comparison of the number of unique peptides, PSMs, and identified proteins will be reported for all algorithms (see Table 2 and Table 4, Figure 5 and Figure 6).
Table 2.
Summary of identified unique peptides, peptide spectrum matches (PSMs), and proteins for each chromatin extraction methodology. The total number of unique annotations across all six algorithms is given, and the percentage of unique identifications is listed for each algorithm in parenthesis. All results are based on a 2% false discovery rate that was calculated using a reverse sequence decoy database. The cumulative sum of all five extraction methods is reported in the bottom right of the table
| Salt Pellet Extraction | Salt Supernatant Extraction | ||||||
|---|---|---|---|---|---|---|---|
| Algorithm | Peptides | PSMs | Proteins | Algorithm | Peptides | PSMs | Proteins |
| PILOT_PROTEIN | 3164 (0.844) | 6707 (0.759) | 1262 (0.887) | PILOT_PROTEIN | 4107 (0.588) | 5993 (0.481) | 1807 (0.752) |
| SEQUEST | 2476 (0.660) | 4635 (0.525) | 1058 (0.744) | SEQUEST | 4480 (0.642) | 6692 (0.535) | 1784 (0.744) |
| InsPecT | 2136 (0.569) | 3750 (0.425) | 941 (0.662) | InsPecT | 3367 (0.482) | 4592 (0.367) | 1474 (0.614) |
| X!Tandem | 2008 (0.535) | 3457 (0.391) | 904 (0.636) | X!Tandem | 2929 (0.420) | 3867 (0.309) | 1348 (0.562) |
| ProteinProspector | 1476 (0.393) | 2268 (0.257) | 732 (0.515) | ProteinProspector | 2674 (0.383) | 3436 (0.275) | 1295 (0.540) |
| VEMS | 1044 (0.278) | 1399 (0.158) | 565 (0.397) | VEMS | 3110 (0.446) | 4118 (0.329) | 1419 (0.591) |
| Total | 3751 | 8832 | 1422 | Total | 6980 | 12509 | 2399 |
| MNase Pellet Extraction | MNase Supernatant Extraction | ||||||
| Algorithm | Peptides | PSMs | Proteins | Algorithm | Peptides | PSMs | Proteins |
| PILOT_PROTEIN | 3035 (0.766) | 6750 (0.544) | 898 (0.833) | PILOT_PROTEIN | 2573 (0.889) | 6401 (0.755) | 714 (0.930) |
| SEQUEST | 2212 (0.558) | 3836 (0.309) | 731 (0.678) | SEQUEST | 2006 (0.693) | 4022 (0.474) | 610 (0.794) |
| InsPecT | 3262 (0.823) | 7581 (0.610) | 960 (0.891) | InsPecT | 2035 (0.703) | 4080 (0.481) | 616 (0.802) |
| X!Tandem | 2183 (0.551) | 3736 (0.301) | 724 (0.672) | X!Tandem | 1549 (0.535) | 2575 (0.304) | 514 (0.669) |
| ProteinProspector | 1822 (0.460) | 2768 (0.223) | 642 (0.596) | ProteinProspector | 1419 (0.490) | 2335 (0.275) | 500 (0.651) |
| VEMS | 1868 (0.471) | 2874 (0.231) | 659 (0.611) | VEMS | 1137 (0.393) | 1708 (0.201) | 438 (0.570) |
| Total | 3962 | 12418 | 1078 | Total | 2895 | 8477 | 768 |
| Total Extraction | Cumulative Total | ||||||
| Algorithm | Peptides | PSMs | Proteins | Algorithm | Peptides | PSMs | Proteins |
| PILOT_PROTEIN | 7861 (0.867) | 32033 (0.757) | 3183 (0.898) | PILOT_PROTEIN | 14011 (0.807) | 58784 (0.693) | 4519 (0.884) |
| SEQUEST | 8729 (0.963) | 39125 (0.924) | 3413 (0.961) | SEQUEST | 13975 (0.805) | 58310 (0.690) | 4493 (0.878) |
| InsPecT | 6707 (0.740) | 23532 (0.556) | 2634 (0.742) | InsPecT | 12103 (0.697) | 43535 (0.515) | 3751 (0.733) |
| X!Tandem | 5680 (0.627) | 17485 (0.413) | 2266 (0.638) | X!Tandem | 10206 (0.588) | 31120 (0.368) | 3312 (0.647) |
| ProteinProspector | 5234 (0.578) | 14787 (0.349) | 2113 (0.595) | ProteinProspector | 9239 (0.532) | 25594 (0.303) | 3075 (0.601) |
| VEMS | 3666 (0.405) | 8250 (0.195) | 1571 (0.443) | VEMS | 7957 (0.458) | 18349 (0.217) | 2696 (0.527) |
| Total | 9062 | 42326 | 3550 | Total | 17367 | 84562 | 5119 |
Table 4.
Summary of modified peptide and protein identification for a total chromatin extraction. The total number of all peptides, modified peptides, all PSMs, modified PSMs, all proteins, modified proteins, and all modifications for 50 LC-MS/MS data sets are shown for a 2% false discovery rate. The total amount of unique entries across all algorithms is also displayed for each column. The percentage of unique entries for a given algorithm is listed in parenthesis
| Algorithm | Peptides | Modified Peptides | PSMs | Modified PSMs | Proteins | Modified Proteins | Total Modifications |
|---|---|---|---|---|---|---|---|
| PILOT_PROTEIN | 14,011 (0.807) | 633 (0.617) | 58,784 (0.693) | 1,251 (0.475) | 4,519 (0.883) | 336 (0.713) | 3,572 |
| InsPecT | 12,103 (0.697) | 592 (0.577) | 43,535 (0.515) | 1,119 (0.425) | 3,751 (0.733) | 312 (0.662) | 3,508 |
| X!Tandem | 10,206 (0.588) | 415 (0.404) | 31,120 (0.368) | 690 (0.262) | 3,312 (0.647) | 246 (0.522) | 3,444 |
| Total | 17,367 | 1026 | 25,594 | 2,633 | 5,119 | 471 | - |
Figure 5.
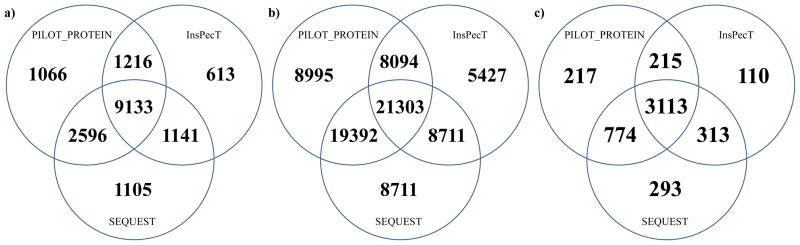
Venn diagram for the unique unmodified peptides (a), PSMs (b), and proteins (c) identified by PILOT_PROTEIN, SEQUEST, and InsPecT. The diagram shows the number of annotations by one or a combination of the three algorithms for a false discovery rate of 2%.
Figure 6.
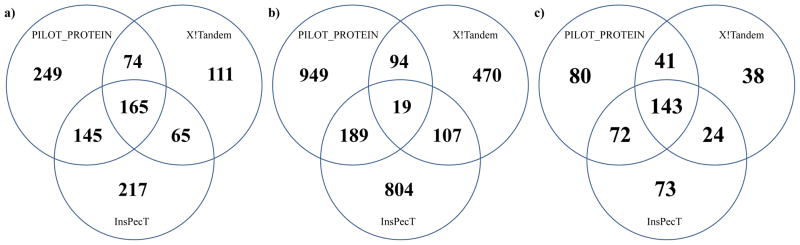
Venn diagram for modified peptide (a), PSM (b), and protein (c) identifications for the chromatin data. The diagram shows the number of modified peptide, PSM, or protein identifications that were annotated by one or a combination of the three algorithms for a false discovery rate of 2%.
Protein Identification Sensitivity
Each LC-MS/MS run in the standard protein mix database was derived from an 18-protein mixture. The sensitivity of a given algorithm is defined as the total number of these 18 proteins that are output from the algorithm. When an algorithm reports a list of homologues for a certain protein, this list will be examined for the presence of a sample protein. If one of these proteins is found, that protein will be marked and added to the list of correct proteins.
Protein Identification Specificity
Along with the 18 proteins that are used to generate the stock solutions, 15 other contaminant proteins were commonly found in the experimental results.31 The specificity is measured using the number of predicted proteins that are not part of either the 18 sample proteins or the 15 contaminants. When a homologue list is found, the list is checked for either a sample protein or a contaminant. If found, no false positives are reported for the homologue list. If not, then only one false positive is reported for the algorithm.
PILOT_PROTEIN Parameters
The following section discusses the parameters used for each algorithm for each test set. For all sets, a maximum of three missed cleavages and two specific termini were required. Carbamidomethylated cysteine was used as a fixed modification while no variable modifications were allowed. The protein database used was the NCBI non-redundant database (Sept. 19, 2011 release; 12,679,685 entries). The complete list of taxonomies was used for data sets A, B, and C while the homo sapiens taxonomy was used for data set D. The set of absolute tolerance parameters for each data set are listed in Table 1. Note that the fixed parameters used to analyze PILOT_PROTEIN (i.e., reward/penalty for sequence matching, weighting constraints, homology labeling) were chosen by training the algorithm on data sets A and B. The parameter values were then fixed for use in the analysis of data sets C and D.
Table 1.
Search tolerance parameters. Parent and fragment ion search tolerances for each data set
| Data Set | Instrument | Parent Tolerance (Da) | Fragment Tolerance (Da) |
|---|---|---|---|
| A | QTOF | 0.2 | 0.2 |
| B | Orbitrap | 0.1 | 0.1 |
| C | Orbitrap | 0.2 | 0.5 |
| C | QTOF | 0.2 | 0.2 |
| C | QSTAR | 0.5 | 0.5 |
| C | QTOF1 | 0.5 | 0.5 |
| C | QTOF2 | 1.0 | 0.5 |
| C | LTQ-FT | 0.2 | 0.5 |
| D | Hybrid Orbitrap/Ion trap | 0.2 | 0.5 |
Results
The protein identification accuracy of PILOT_PROTEIN was initially tested on two small data sets consisting of (a) 36 QTOF spectra from 9 proteins (data set A) and (b) 701 Orbitrap spectra from 12 proteins (data set B).46 PILOT_PROTEIN was able to identify 100% of the proteins from the two data sets while reporting no false positives. Further, all peptides that were validated with SEQUEST4 were correctly assigned to each protein. To test PILOT_PROTEIN on more comprehensive data, data set C derived from all Orbitrap, QTOF, and LT-FTQ LC-MS/MS files in the Standard Protein Mix Database31 and data set D derived from a chromatin extraction are utilized. The capability of the method was benchmarked with five state-of-the-art algorithms VEMS,32 SEQUEST,4 InsPecT,,16 X!Tandem,33 and ProteinProspector.41 The 112 LC-MS/MS files from data set C contain 18 known proteins along with 15 possible contaminant proteins while the 50 LC-MS/MS files from data set D contain a more complex array of proteins that are associated with chromatin. Both data sets were analyzed using the NCBI non-redundant database. The complete list of taxonomies was used for data set C while the homo sapiens taxonomy was used for data set D. Note that while SEQUEST was previously used to analyze the information from data set C,31 both the database (nr vs. swissprot) and the fragment/parent tolerances are different in this study. The peptide and protein identification results for each tested algorithm and each LC-MS/MS run are presented as Supplementary Material.
Data Set C: Standard Protein Mix Database
The comparative results for all three instruments from data set C are shown in Figure 3. Within each graph, the change in protein identification accuracy (sensitivity) with respect to changing false discovery rate (specificity) is shown. The graphs are generated from a total of 112 LC-MS/MS runs. The curves can be reconstructed using the comprehensive protein list reported by each algorithm in the Supplementary Material. For a given score cutoff value, the resulting number of true proteins (hits) and false proteins (misses) are reported and are used to construct the curves in Figure 3. The score cutoff for PILOT_PROTEIN is the exact value listed in the table while the score cutoff for the other algorithms is derived using the negative log of the protein probability (SEQUEST, VEMS, InsPecT) or expectation value (ProteinProspector, X!Tandem). To generate the ROC curves from protein identifications (see Supplementary Material), the lowest protein score threshold was chosen such that no false positives are reported. This cutoff value defines the left-hand side of each ROC curve and represents the maximum number of proteins that can be reported with no false positives. This cutoff score was then incrementally decreased and the number of false positives and protein hits were reported at each iteration. The iterations were terminated when all true protein hits that were reported by an algorithm were above the threshold score. Further reduction of the score threshold for the algorithm will add false positives and no true hits. Note that the protein identification rate (PIR) is defined as:
| (8) |
where is the number of “true” proteins found and is the total number of possible true proteins (i.e., 18 times the number of LC-MS/MS runs). The false discovery rate (FDR) is defined as:
| (9) |
where FP is the number of incorrect proteins (i.e., not a true protein or a known contaminant) found and is the total number of proteins found. For each of the three instruments (Orbitrap, QTOF, and LTQ-FT), PILOT_PROTEIN consistently demonstrates enhanced sensitivity (higher PIR) at each of the specificity (FDR) levels. Each of the instruments will be discussed briefly to highlight the key findings for each set of data.
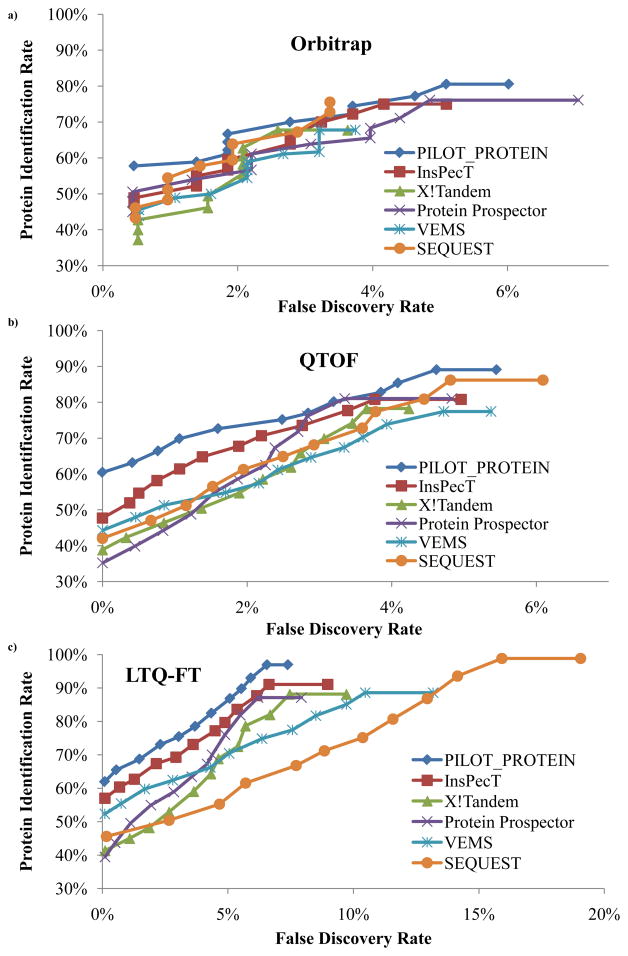
Figure 3.
Standard protein mix database ROC curves. The graphs represent the change in the protein identification accuracy with changing false-discovery rate. (a) 10 Orbitrap LC-MS/MS runs. (b) 68 QTOF runs. (c) 38 LTQ-FT LC-MS/MS runs.
Orbitrap
The 18 proteins were repeatedly identified for the 10 Orbitrap LC-MS/MS runs, leading to a total of 180 correct protein identifications that could be reported by each algorithm. The right-most points for each curve in Figure 3 represent the sensitivity/specificity when the protein score cutoff threshold is set to the minimum value such that all correct protein hits will be reported. This gives an indication of how many possible protein hits can be reported by a given algorithm. Note that the quantification of the protein sensitivity (number of false positives) is representative of the minimum false positive rate that can be expected by an algorithm if true protein hits are reported. Over all data sets, PILOT_PROTEIN annotates the highest amount of proteins (145 hits; 80.6% PIR) correctly, while reporting only 11 false positives (6.0% FDR). The next highest total is found by Protein Prospector, which annotated 137 proteins correctly (76.1% PIR) with 16 false positives (7.0% FDR). SEQUEST annotates 136 proteins (75.6% PIR) and 7 false positives (3.4% FDR) while InsPecT annotates 135 proteins (75.0% PIR) and 11 false postivies (5.1% FDR). X!Tandem and VEMS both report the least amount of proteins (122 hits; 67.8% PIR) and the same amount of false positives (7 FP).
To obtain a more accurate representation of the protein specificity at a given protein sensitivity, the PIR is analyzed for each algorithm when the FDR is set to a target level. Note from Figure 3a that PILOT_PROTEIN maintains a higher PIR than all competing algorithms for all target FDR levels with the exception of a small region between 3–4% FDR. Specifically, if a maximum target level of 2.5% FDR is selected, then PILOT_PROTEIN reports 120 protein hits (66.7% PIR) while the next highest algorithm, SEQUEST, reports 115 protein hits (63.9% PIR). At this benchmark FDR level, X!Tandem, Protein Prospector, InsPecT, and VEMS report protein hits of 113 (62.8% PIR), 110 (61.1% PIR), 107 (59.4% PIR), and 106 (58.9% PIR), respectively.
QTOF
For the 68 QTOF LC-MS/MS runs, the sensitivity vs. specificity for each protein identification algorithm is displayed graphically in Figure 3b. PILOT_PROTEIN annotated a total of 1090 correct proteins out of 1224 possible with a FDR of 5.4%. SEQUEST annotated the next highest amount of proteins with 1055 true hits, though the FDR increased to 6.1%. Protein Prospector reported the next highest total number of proteins, followed by InsPecT, X!Tandem, and VEMS.
PILOT_PROTEIN consistently maintains a higher protein identification rate than all other algorithms for each given false discovery rate. While the protein sensitivity for PILOT_PROTEIN is only slightly higher than InsPecT, Protein Prospector, or SEQUEST for higher levels of FDR (> 3%), the difference in sensitivity begins to increase as the FDR is decreased below 3%. This is extremely important as PILOT_PROTEIN demonstrates a significant enhancement in the true protein identification rate. Specifically, at 1.5% FDR, PILOT_PROTEIN reports 855 true hits while the next highest algorithm, InsPecT, reports 793 true hits. The number of identified proteins for each of the additional four algorithms decreases by at least 99 from InsPecT, with SEQUEST reporting the highest total of the four.
LTQ-FT
For the 38 LTQ-FT LC-MS/MS runs, a total of 684 proteins could be identified, and the resulting data is shown in Figure 3c for each algorithm. At the minimum score cutoff, PILOT_PROTEIN identified a total of 663 proteins correctly (96.9% PIR) with 80 false positivies (7.4% FDR). Only SEQUEST was able to annotate more proteins (676 hits; 98.8% PIR), though the amount of false positives increased substantially to 237 (19.1% FDR). InsPecT, VEMS, X!Tandem, and Protein-Prospector all report a similar amount of true hits (596 – 623) and all have between 78 – 141 false positives (7.9% – 13.2% FDR).
Similar to the two previous data sets, PILOT_PROTEIN has a higher protein identification rate (sensitivity) for each level of false discovery rate (specificity) than any other algorithm. The difference in sensitivity between PILOT_PROTEIN and InsPecT remains relatively constant while the difference between VEMS or SEQUEST increases at higher values of FDR and the difference between X!Tandem or Protein Prospector increases at lower values of FDR. Though the range of FDR for this particular data set (0 – 20%) is wider than the QTOF or Orbitrap data (0 – 7%), this increase is largely due to the high FDR of VEMS and SEQUEST. The other four algorithms had ranges of FDR that were consistent with previous data. Using a benchmark level of 1.5% FDR as a basis for comparison, PILOT_PROTEIN is able to annotate 470 proteins correctly (68.7% PIR) which is 41 more proteins then the next best method, InsPecT (429 hits; 62.7% PIR). Using a 1.5% FDR, VEMS and Protein Prospector report 379 and 339 proteins (55.4% and 49.6% PIR, respectively), SEQUEST reports 312 true hits (45.6% PIR), and X!Tandem reports 308 hits (45.0% PIR).
Note that Figure 3a and Figure 3c represent ROC curves for instruments operating with a high resolution MS1 and a low accuracy MS2 (via the LTQ). The similarity of these data sets is reflected in the two ROC curves. At a false discovery rate of 2%, the six algorithms identify between 50%–65% of the proteins for the Orbitrap data and 45%–67% of the proteins for the LTQ-FT data. The range of protein identification rate for this FDR level and the range of FDR levels across the data is likely higher for the LTQ-FT data because there was 3.8 times as much data present for that instrument (38 LC-MS/MS runs) as opposed to the Orbitrap (10 LC-MS/MS runs).
Data Set D: Chromatin Extraction
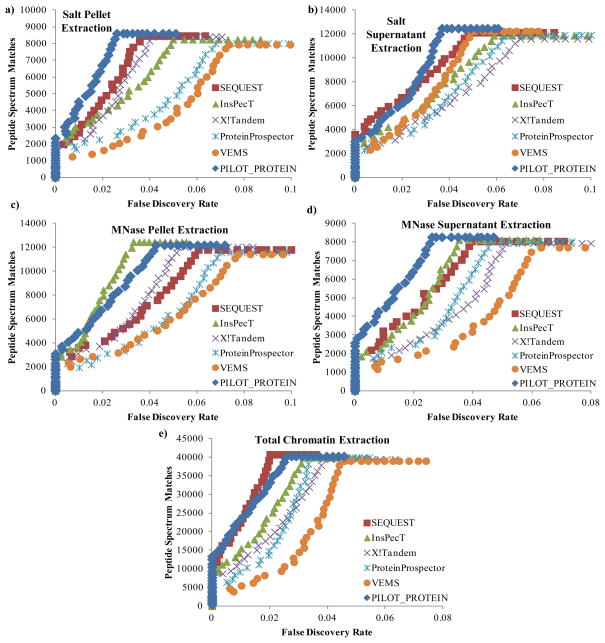
The plot of peptide spectrum matches (PSMs) compared with false discovery rate for data set D is shown in Figure 4 for each algorithm. Each graph is generated by analyzing the 10 repeat injections of a different chromatin extraction technique. Each mixture has extracted and purified chromatin from HeLa H3 cells using either a total extraction, a salt extraction, or a micrococal nuclease (MNase) digestion. The salt and MNase extractions produced both supernatant and pellet chromatin fractions.62 The plots in Figure 4 can be reproduced from the peptide identification data reported for each algorithm in the Supplementary Material.
Figure 4.
Chromatin fraction ROC curves. The graphs represent the change in the number of peptide spectrum matches with changing false-discovery rate. (a) Salt pellet extraction. (b) Salt supernatant extraction. (c) MNase pellet extraction. (d) MNase supernatant extraction. (e) Total extraction.
For the salt pellet extraction, PILOT_PROTEIN reported a higher number of PSMs than all of the other algorithms for a variety of false discovery rate (FDR) levels. SEQUEST tends to report the second highest amount of PSMs followed closely by X!Tandem and InsPecT. At lower levels of FDR near 1%, InsPecT begins to identify more PSMs than either X!Tandem or SEQUEST.
The salt supernatant extraction plot in Figure 4b shows that SEQUEST reports the highest number of PSMs at lower levels of FDR (less than 2%) and is followed closely by PILOT_PROTEIN, though at a FDR level of approximately 2.5%, PILOT_PROTEIN begins to report more PSMs than SEQUEST. InsPecT, X!Tandem, ProteinProspector, and VEMS all report a similar number of PSMs at a 2% FDR, though the gap between the four algorithms begins to widen at FDR levels between 2% – 5% as VEMS increased the number of reported PSMs relative to the other three algorithms.
The MNase pellet extraction data shows that PILOT_PROTEIN reports the highest number of PSMs at FDR levels below 1% with InsPecT and SEQUEST reporting the next highest. At a FDR level of approximately 1.5%, InsPecT begins to report the highest number of PSMs with PILOT_PROTEIN ranking second. Both SEQUEST and X!Tandem consistently rank third and fourth for this data set, though SEQUEST reports more PSMs than X!Tandem at low FDR levels and the reverse is true at higher FDR levels.
For the MNase supernatant data, PILOT_PROTEIN consistently annotates a higher number of PSMs than each of the other five algorithms for all of the relevant FDR levels. SEQUEST and InsPecT report the second and third most PSMs, with SEQUEST ranking second until at FDR level of approximately 2.5% where InsPecT begins to rank second. X!Tandem and ProteinProspector report the next highest amount of PSMs and are followed by VEMS.
The total chromatin extraction shows both SEQUEST and PILOT_PROTEIN reporting a superior number of PSMs. PILOT_PROTEIN annotates a slightly higher number for FDR levels below 1.5%, while SEQUEST begins to increase the annotations relative to PILOT_PROTEIN above this level. InsPecT reports the third highest number of PSMs across many FDR levels followed by ProteinProspector and X!Tandem.
For two of the five data sets (salt pellet and MNase supernatant), PILOT_PROTEIN outperforms all other algorithms for each level of false discovery rate (FDR) from the range of 1% – 3%. In both of these data sets, PILOT_PROTEIN reports over 2,000 more peptide spectrum matches (PSMs) than any other algorithm at a 2% FDR. Additionally, PILOT_PROTEIN reports at least 500–700 more unique peptides and 100–200 more proteins than any other algorithm at this FDR. For two other data sets (MNase pellet and total extraction), PILOT_PROTEIN annotates more PSMs at lower FDR levels (< 1%), though InsPecT (MNase pellet) and SEQUEST have more annotations at higher FDR levels. Note that PILOT_PROTEIN performed the second best to each of these algorithms for these data sets and the gap between the PSMs reported by the first and second algorithm is significantly smaller than for the first two data sets. That is, at a 2% FDR, PILOT_PROTEIN only reported about 800 fewer PSMs, 200–300 fewer unique peptides, and 60–75 fewer unique proteins than the top scoring algorithm, (i.e., InsPecT for MNase pellet or SEQUEST for total extraction). The fifth data set (salt supernatant) shows SEQUEST performing the best at low FDR levels and PILOT_PROTEIN performing the best at higher FDR levels.
Breaking down each PSM graph into two regions (FDR ≥1% and FDR < 1%), it is seen that PILOT_PROTEIN is the top scoring algorithm in 7 of the 10 regions considered (i.e., all regions except low FDR salt supernatant, high FDR MNase pellet, and high FDR total extraction). Additionally, PILOT_PROTEIN is the second best scoring algorithm for the remaining three regions. Alternatively, SEQUEST is the top scoring algorithm for two regions (i.e., low FDR salt supernatant and high FDR total extraction), the second best for four regions, the third best for three regions, and the fourth best for one region. InsPecT is the top algorithm for one region (i.e., high FDR MNase pellet), the second algorithm for three regions, the third for four regions, and the fourth for two regions.
As an illustrative example, the results for a FDR of 2% are shown in detail in Table 2. The number of distinct peptides, PSMs, and proteins are reported for each algorithm along with the percentage of unique entries across all six algorithms in parenthesis. Across all five data sets (50 LC-MS/MS runs and approximately 450,000 MS/MS spectra), PILOT_PROTEIN reports a total number of 58,784 PSMs consisting of 14,011 distinct peptides and 4,519 distinct proteins. SEQUEST reports the next highest numer of PSMs with 58,310 matches coming from 13,795 distinct peptides and 4,493 proteins. InsPecT annotated 12,103 peptides and 3,751 proteins using 43,535 PSMs and X!Tandem reports 10,206 peptides and 3,312 proteins from 31,120 PSMs. Protein-Prospector and VEMS were able to annotate 3,075 proteins, 9,239 peptides, and 25,594 PSMs and 2,696 proteins, 7,957 peptides, and 18,349 PSMs, respectively.
The unique peptide, PSM, and protein identification crossover for the three algorithms (PILOT_PROTEIN, SEQUEST, and InsPecT) that identified the most number of PSMs is shown in Figure 5 for a 2% FDR. In each panel, note that a majority of the unique PSMs (21,303), peptides (9133), and proteins (3,113) were found by all of the three algorithms. Additionally, the number of identifications for each panel in Figure 5 reported by exactly two algorithms is generally higher than that reported for any single algorithm. However, there does exist a fraction of PSM, peptide, or protein identifications (around 6–10% for each algorithm) that are reported by only one of the three algorithms. PILOT_PROTEIN (8,995 individual PSMs), SEQUEST (8,711 individual PSMs), and InsPecT (5,427 PSMs) report 23,133 PSMs that are not found by either of the other two algorithms. These identifications represent 28.7% of the total amount of PSMs found by the three algorithms (Figure 5b). For the unique peptides (Figure 5a) and proteins (Figure 5c), the number of identifications reported by only one algorithm reduces to 16.5% and 12.3% of the total, respectively. This provides evidence that the use of multiple identification algorithms to verfiy the assignments made for an LC-MS/MS data set can enrich the quantity of peptide and protein identifications output to a user.
Sequence Based Clustering Results
For each of the LC-MS/MS runs in data sets C and D, the sequence based clustering algorithm was able to remove several peptides that were incorrectly assigned by the PILOT_SEQUEL hybrid de novo/database identification algorithm. Table 3 shows the total number of peptides that were labeled incorrectly along with the fraction of those peptides that were re-annotated with a correct peptide assignment. The remainder of the peptides were simply removed from the list of annotations. For the standard mix proteins (data set C), 217 PSMs were identified by the clustering algorithm for the Orbitrap data, 773 PSMs for the QTOF data, and 830 for the LTQ-FT data. Of these spectra, 59 were re-assigned with another peptide for the Orbitrap data, 387 for the QTOF data, and 398 for the LTQ-FT data. Table 3 also shows the decrease in identified proteins that came directly as a result of the clustering algorithm. For data set C, the true positive proteins consist of the 18 sample mixture proteins and the 15 contaminants while any other protein is considered a false positive protein. At a protein false-discovery rate level of 2%, this led to a decrease of 23 false positive proteins for the Orbitrap data, 47 for the QTOF data, and 111 for the LTQ-FT data. Many of these false positive proteins would have been annotated solely by the peptide spectrum match that was eliminated via the clustering algorithm. The decrease in the number of true positive proteins for PILOT_PROTEIN was 6 for the Orbitrap data, 6 for the QTOF data, and 25 for the LTQ-FT data.
Table 3.
Summary of sequence based clustering results for PILOT_PROTEIN. For each data set, the total number of peptide spectrum matches (PSMs) identified by the clustering routine is listed along with the fraction of those spectrum that were reassigned to another peptide. The decrease in the number of both true positive and false positive proteins reported by the algorithm is also listed for a 2% false discovery rate
| Data Set | Clustered PSMs | Reassigned PSMs | Missed Proteins | |
|---|---|---|---|---|
| True | False | |||
| Data Set C - Orbitrap | 217 | 59 | 6 | 23 |
| Data Set C - QTOF | 773 | 387 | 6 | 47 |
| Data Set C - LTQ-FT | 830 | 398 | 25 | 111 |
| Data Set D - Salt Pellet | 115 | 47 | 1 | 46 |
| Data Set D - Salt Supernatant | 211 | 76 | 29 | 40 |
| Data Set D - MNase Pellet | 123 | 38 | 17 | 66 |
| Data Set D - MNase Supernatant | 123 | 50 | 45 | 80 |
| Data Set D - Total Extraction | 134 | 34 | 38 | 40 |
For the chromatin data (set D), the number of PSMs identified by the clustering algorithm was 115 for the salt pellet extraction, 123 for the MNase pellet extraction, 123 for the MNase supernatant extraction, 134 for the total extraction, and 211 for the salt supernatant. The total number of spectra that were reassigned was 34 for the total extraction, 38 for the MNase pellet, 47 for the salt pellet, 50 for the MNase supernatant, and 76 for the salt supernatant. For data set D, the true positive proteins are proteins that is contained in the NCBI nr database while a false positive protein is part of the reverse sequence decoy database. Using a 2% false discovery rate for the PSMs as defined earlier, this led to a reduction of 272 false positive proteins and 130 true positive proteins across all five data sets. Many of the true positve proteins and all of the false positive proteins would have only been annotated using a single PSM that was removed via the clustering algorithm.
Data Set D: PTM Identification
To demonstrate the capability of PILOT_PROTEIN and PILOT_PTM, the chromatin data set was used for an untargeted post-translational modification analysis. A universal list of modifications comprised of information from several databases was used as an input to PILOT_PTM.47 Generally, five to ten peptide sequences were used as candidate template sequences for PILOT_PTM. The modified template sequence that had the highest cross-correlation score was retained. To help reduce the number of false positive modifications, a reverse sequence decoy database was used to establish a false discovery rate for each cross-correlation score. The decoy database was constructed from the smaller list of unmodified proteins output by the PILOT_PROTEIN algorithm. A threshold score cutoff was utilized that corresponded to a 2% false discovery rate.
To benchmark the capability of PILOT_PROTEIN, the algorithm was compared against the methods InsPecT and X!Tandem. Both methods employ a two-pass variable modification search method that initially generates a small protein list in the first pass of the method and then search for an expanded set of variable modifications in the second stage of the method.16,33 A previous study compared the residue and peptide prediction accuracy of all three methods47 and demonstrated that PILOT_PTM has a superior prediction accuracy for both modified and unmodified spectra. In this study, the variable modification search results are presented for each of the three algorithms to determine the number of modified peptides, the number of modified proteins, and the counts for each type of modification. Search criteria for all three methods included a 0.2 Da parent tolerance, a 0.5 Da fragment tolerance, 2 tryptic termini, 2 maximum missed cleavages, and 531 variable post-translational modifications, chemical derivatives, and artifacts.47 A false discovery rate of 2% was imposed using a reverse sequence decoy database based off of the small list of unmodified proteins generated by each algorithm. The resulting peptide and protein identifications for all three algorithms are reported in the Supplementary Material.
The summary of all peptide and protein identifications for all 50 LC-MS/MS data sets is shown in Table 4 for a 2% FDR. The total number of peptides, PSMs, and proteins reported by each algorithm is reported in Table 2 along with the percentage of unique identifications compated to the total in parenthesis. PILOT_PROTEIN identified 3,572 modifications on 633 distinct peptides, indicating that 4.57% of the peptides were modified throughout all 50 data sets. InsPecT identified 3,508 modifications on 592 distinct peptides while X!Tandem identified 3,444 modifications on 415 distinct peptides. PILOT_PROTEIN was able to identify 4,519 distinct proteins across all data sets, of which 336 (7.50%) were labeled with at least one modification. X!Tandem reported a total of 3,312 proteins including 246 modified proteins and InsPecT reported a total of 3,751 proteins including 312 modified proteins.
The histogram of all modifications identified over all 50 LC-MS/MS data sets is shown in Table 5. Modifications are annotated in Table 5 based on the amino acid location along the peptide. Due to the ambiguity between an amino acid acetylation and an acetylation of the N-terminus, all acetylations that occur on an N-terminal residue will be labeled as an N-terminal acetylation. To distinguish between their relative locations on the peptide, all methylations or di-methylations that occur on a C-terminal residue have been specifically marked (see Table 5). This distinction is intended to provide information about modification frequency at the terminal position and does not imply that these modifications are on the C-terminus. Information regarding frequency with respect to sequence position will be helpful for the classification of PTMs within a proteome.65 The vast majority of modifications identified by PILOT_PROTEIN, X!Tandem, and InsPecT was oxidized methionine with 2,938 hits, 2,950 hits, and 2,875 hits, respectively. After oxidation, the most common modification reported by PILOT_PROTEIN was methylation with 363 counts found on residues located at the C-terminus, 65 counts for lysine, and 29 counts for arginine. The next most abundant modifications included dimethylation (56 on the C-terminus, 21 on lysine, and 29 on arginine) and acetylation (51 on the N-terminus, 21 on lysine, and 9 on arginine). For all algorithms, the majority of the modifications (except oxidation) are found on the C-terminus (419 modifications for PILOT_PROTEIN, 390 for InsPecT, and 372 for X!Tandem) and lysine was the amino acid with the most abundant amount of modifications (103 modifications for PILOT_PROTEIN, 78 for InsPecT, and 98 for X!Tandem). Arginine also contained several modifications, with 51 total reported from PILOT_PROTEIN, 46 total from InsPecT, and 45 total from X!Tandem.
Table 5.
Post-translational modification identification (PTM) results. The total number of each PTM reported over all 50 LC-MS/MS runs is reported for each algorithm for a 2% false discovery rate. The format for a modification name is Am where A is the amino acid residue and m is the modification. Note that CT and NT refer to a modification that is located at the C-terminus and N-terminus, respectively. The labels for the modifications are: a - Acetylation; d - Dimethylation; m - Methylation; t - Trimethylation; o - Oxidation.
| Modification | Algorithm | ||
|---|---|---|---|
| PILOT_PROTEIN | InsPecT | X!Tandem | |
| CT-Kd | 33 | 30 | 22 |
| CT-Km | 208 | 194 | 185 |
| CT-Rd | 23 | 18 | 19 |
| CT-Rm | 155 | 148 | 146 |
| Ka | 8 | 8 | 11 |
| Kd | 21 | 20 | 16 |
| Km | 65 | 45 | 60 |
| Kt | 9 | 5 | 11 |
| Mo | 2938 | 2950 | 2875 |
| NTa | 51 | 35 | 45 |
| Ra | 9 | 3 | 8 |
| Rd | 13 | 12 | 12 |
| Rm | 29 | 31 | 25 |
The crossover for unique peptides, PSMs, and protein identifications for the three algorithms is shown in Figure 6 for a 2% FDR. The peptides shown in Figure 6a include all modifications that are assigned to that peptide. If two identified peptides have the same sequence, but a different combination of modification types or sites, then they are designated as distinct peptides. Similarly, two PSMs are said to be equivalent between two algorithms if both the peptide and modification set are equal for a given spectrum. The proteins reported in Figure 6c represent only the protein identification and are not indicative of the quantity or location of modifications found on that protein by a particular algorithm. Note that the number of peptides and PSMs that are identified by all three algorithms is not a dominant fraction of the total, as it was in Figure 5. The additional complexity with localization of modifications provides an enhanced layer of complexity that makes it difficult for certain spectra to be properly identified by multiple algorithms. This is clearly evident in Figure 6b, where a majority of the unique PSMs are identified by only one of the three algorithms. This result occurs because it was common for one MS/MS spectrum to be annotated by only one of the three algorithms. Further, when two or more algorithms annotated a spectrum with a different modified peptide, it was often found that the peptide sequence and modification types were similar, but the localization of the modifications was different (see Supplementary Material). Note that the lack of overlap at the PSM level for the three algorithms diminishes slightly at the peptide level and more significantly at the protein level. That is, the relative amounts of unique peptides identified by only one algorithm diminishes (see Figure 6a). While certain MS/MS spectra may be difficult to annotate for a given algorithm, this does not imply a 1:1 loss of unique peptide data. The protein data in Figure 6c shows an overlap between three algorithms that has the highest number of annotations. Further, the overlap between PILOT_PROTEIN and InsPecT is almost as high as the number of individual identifications by either of those two algorithms alone.
Discussion
A novel mixed integer linear optimization framework for the identification of all unmodified and modified proteins in a cellular sample was developed. PILOT_PROTEIN used the results of the peptide identification algorithm PILOT_SEQUEL to initially generate a list of all unmodified proteins in the sample. Using a biclustering approach, all peptide homologues are identified and incorrectly assigned peptide sequences are identified and removed from consideration. This helps reduce the number of false positives by eliminating low-scoring peptide identifications. The protein identification accuracy of the PILOT_PROTEIN algorithm was clearly demonstrated using data31 taken from several MS/MS instruments. The biclustering approach was very effective in identifying peptide similarities and reducing false positive reporting compared to competing methods.
The results of PILOT_PROTEIN for unmodified protein identification were benchmarked using SEQUEST, InsPecT, X!Tandem, ProteinProspector, and VEMS. Overall, PILOT_PROTEIN reported superior results against each algorithm. Given an unmodified protein list from PILOT_PROTEIN, the PILOT_PTM algorithm can use an untargeted search to generate a modified protein list that contains all modification types and sites for each protein in the sample. The PILOT, PILOT_SEQUEL, PILOT_PROTEIN, and PILOT_PTM algorithms represent a complete package for identification of all sample proteins along with all corresponding PTMs and are capable of analyzing an LC-MS/MS data set with a computation run time of approximately 8 CPU seconds per MS/MS spectrum. The package is run on a Beowulf cluster with 24 Intel Xeon 2.83 GHz processors and utilized a message passing interface to parallelize the data processing. Full utilization of all processors for one LC-MS/MS experiment significantly reduces the computational run time to approximately 0.6 CPU seconds per MS/MS spectrum. The algorithms are currently available as a singular webtool free of charge at http://pumpd.princeton.edu. A successful large-scale application of the aforementioned suite of algorithms for the elucidation of gingival crevicular fluid has been recently reported.66
Supplementary Material
Acknowledgments
CAF and BAG acknowledge financial support from the National Science Foundation (CBET-0941143). CAF acknowledges financial support from the National Institute of Health (R01LM009338). Although the research described in the article has been funded in part by the U.S. Environmental Protection Agency’s STAR program through grant (R 832721-010), it has not been subjected to any EPA review and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred. BAG acknowledges support from Princeton University and the American Society for Mass Spectrometry Research award.
Footnotes
Supporting Information Available
The peptide and protein identification results for the standard protein mixture and the chromatin data are available as supplementary material.
This material is available free of charge via the Internet at http://pubs.acs.org/.
Competing Interests Statement
The authors declare no competing financial interests.
Author’s contributions
R.C.B developed model, interpreted data, and wrote paper; P.A.D. developed model, interpreted data, and wrote paper; M.D.P. designed experiments, conducted experiments, and interpreted data; B.A.G. designed experiments, conducted experiments, interpreted data, and wrote paper; C.A.F. developed model, interpreted data, and wrote paper.
References
- 1.Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 2.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 3.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eng JK, McCormack AL, Yates JR., III An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 5.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Lu B, Chen T. A suffix tree approach to the interpretation of tandem mass spectra: applications to peptides of non-specific digestion and post-translational modifications. Bioinformatics. 2003;19:ii113–ii121. doi: 10.1093/bioinformatics/btg1068. [DOI] [PubMed] [Google Scholar]
- 7.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open Mass Spectrometry Search Algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 8.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 9.Shadforth I, Xu W, Crowther D, Bessant C. GAPP: A Fully Automated Software for the Confident Identification of Human Peptides from Tandem Mass Spectra. J Proteome Res. 2006;5:2849–2852. doi: 10.1021/pr060205s. [DOI] [PubMed] [Google Scholar]
- 10.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth Analysis of Tandem Mass Spectrometry Data from Disparate Instrument Types. Mol Cell Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Aebersold R, Schwilkowski B. ProbID: a probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectrometry. Proteomics. 2002;2:1406–1412. doi: 10.1002/1615-9861(200210)2:10<1406::AID-PROT1406>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Algient, Spectrum Mill for MassHunter Workstation. http://www.chem.algient.com/
- 13.Colinge J, Masselot A, Giron M, Dessingy T, Magnin J. OLAV: Towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3:1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 14.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 16.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: Identification of Posttranslationally Modified Peptides from Tandem Mass Spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 17.Searle BC, Dasari S, Wilmarth PA, Turner M, Reddy AP, David LL, Nagalla SR. Identification of Protein Modifications Using MS/MS de Novo Sequencing and the OpenSea Alignment Algorithm. J Proteome Res. 2005;4:546–554. doi: 10.1021/pr049781j. [DOI] [PubMed] [Google Scholar]
- 18.Han Y, Ma B, Zhang K. SPIDER: Software for Protein Identification from Sequence Tags with De Novo Sequencing Error. 2004. [DOI] [PubMed] [Google Scholar]
- 19.Matthiesen R, Trelle MB, Højrup P, Bunkenborg J, Jensen ON. VEMS 3.0: Algorithms and Computational Tools for Tandem Mass Spectrometry Based Identification of Post-translational Modifications in Proteins. J Proteome Res. 2005;4:2338–2347. doi: 10.1021/pr050264q. [DOI] [PubMed] [Google Scholar]
- 20.Kim S, Na S, Sim JW, Park H, Jeong J, Kim H, Seo Y, Seo J, Lee KJ, Paek E. Modi: a powerful and convenient web server for identifying multiple post-translational peptide modifications from tandem mass spectra. Nucleic Acids Res. 2006;34:W258–W263. doi: 10.1093/nar/gkl245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a New Proteomic Tool for Mapping Substoichiometric Post-translational Modifications, Finding Novel Types of Modifications, and Fingerprinting Complex Protein Mixtures. Mol Cell Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL. Prosight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 2007;35:W701–W706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez P, Gras R, Frey J, Appel RD. Popitam: towards new heuristic strategies to improve protein identification from tandem mass spectrometry data. Proetomics. 2003;3:870–878. doi: 10.1002/pmic.200300402. [DOI] [PubMed] [Google Scholar]
- 24.Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Genome Res. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI. Protein Identification by Tandem Mass Spectrometry and Sequence Database Searching. Method in Molecular Biology. 2007;367:87–119. doi: 10.1385/1-59745-275-0:87. [DOI] [PubMed] [Google Scholar]
- 26.Sadygov RG, Liu H, Yates JR. Statistical models for protein validation using tandem mass spectral data and protein amino acid sequence databases. Anal Chem. 2004;76:1664–1671. doi: 10.1021/ac035112y. [DOI] [PubMed] [Google Scholar]
- 27.Gerster S, Qeli E, Ahrens CH, Buhlmann P. Protein and gene model inference based on statistical modeling in k-partite graphs. Proc Natl Acad Sci. 2010;107:12101–12106. doi: 10.1073/pnas.0907654107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen C, Wang Z, Shankar G, Zhang X, Li L. A hierarchical statistical model to assess the confidence of peptides and proteins inferred from tandem mass spectrometry. Bioinformatics. 2008;24:202–208. doi: 10.1093/bioinformatics/btm555. [DOI] [PubMed] [Google Scholar]
- 29.Li YF, Arnold RJ, Li Y, Radivojac P, Sheng Q, Tang H. A bayesian approach to protein inference problem in shotgun proteomics. J Comput Biol. 2009;16:1183–1193. doi: 10.1089/cmb.2009.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein Identification False Discovery Rates for Very Large Proteomics Data Sets Generated by Tandem Mass Spectrometry. Mol Cell Prot. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilmek J, Eddes JS, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken PR, Katz JE, Mallick P, Lee H, Schmidt A, Ossola R, Eng JK, Aebersold R, Martin DB. The Standard Protein Mix Database: A Diverse Data Set To Assist in the Production of Improved Peptide and Protein Identification Software Tools. J Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthiesen R, Lundsgaard M, Welinder KG, Bauw G. Interpreting peptide mass spectra by VEMS. Bioinformatics. 2003;19:792–793. doi: 10.1093/bioinformatics/btg059. [DOI] [PubMed] [Google Scholar]
- 33.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commum Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 34.Baumgartner C, Rejtar T, Kullolli M, Akella LM, Karger BL. SeMoP: A New Computational Strategy for the Unrestricted Search for Modified Peptides Using LC-MS/MS Data. J Proteome Res. 2008;7:4199–4208. doi: 10.1021/pr800277y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Yan B, Song Y, Xu Y, Cai L. Peptide sequence tag-based blind identification of post-translational modifications with point process model. Bioinformatics. 2006;22:e307–e313. doi: 10.1093/bioinformatics/btl226. [DOI] [PubMed] [Google Scholar]
- 36.Seo J, Jeong J, Kim YM, Hwang N, Paek E, Lee KJ. Strategy for Comprehensive Identification of Post-translational Modifications in Cellular Proteins, Including Low Abundant Modifications: Application to Glyceraldehyde-3-phosphate Dehydrogenase. J Proteome Res. 2008;7:587–602. doi: 10.1021/pr700657y. [DOI] [PubMed] [Google Scholar]
- 37.Hansen BT, Davey SW, Ham AJL, Lieber DC. P-Mod: An Algorithm and Software To Map Modifications To peptide Sequences Using Tandem MS Data. J Proteome Res. 2005;4:358–368. doi: 10.1021/pr0498234. [DOI] [PubMed] [Google Scholar]
- 38.DiMaggio PA, Jr, Young NL, Baliban RC, Garcia BA, Floudas CA. A Mixed-Integer Linear Optimization Framework for the Identification and Quantification of Targeted Post-translational Modifcations of Highly Modifed Proteins using Multiplexed ETD Tandem Mass Spectrometry. Mol Cell Proteomics. 2009;8:2527–2543. doi: 10.1074/mcp.M900144-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havilio M, Wool A. Large-Scale Unrestricted Identification of Post-Translational Modifications Using Tandem Mass Spectrometry. Anal Chem. 2007;79:1362–1368. doi: 10.1021/ac061515x. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Chen W, Cobb MH, Zhao Y. PTMap–A sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. PNAS. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clauser KR, Baker PR, Burlingame AL. Role of accurate mass measurement (+/−, 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 42.Creasy DM, Cottrell JS. Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics. 2002;2:1426–1431. doi: 10.1002/1615-9861(200210)2:10<1426::AID-PROT1426>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 43.Tanner S, Bafna V, Pevzner PA. Unrestrictive identification of post-translational modifications through peptide mass spectrometry. Nat Protoc. 2006;1:67–72. doi: 10.1038/nprot.2006.10. [DOI] [PubMed] [Google Scholar]
- 44.DiMaggio PA, Jr, Floudas CA. A Mixed-Integer Optimization Framework for De Novo Peptide Identification. AIChE J. 2007;53:160–173. doi: 10.1002/aic.11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiMaggio PA, Jr, Floudas CA. De Novo Peptide Identification via Tandem Mass Spectrometry and Integer Linear Optimization. Anal Chem. 2007;79:1433–1446. doi: 10.1021/ac0618425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiMaggio PA, Jr, Floudas CA, Lu B, Yates JR., III A Hybrid Method for Peptide Identification Using Integer Linear Optimization, Local Database Search, and Quadrupole Time-of-Flight or OrbiTrap Tandem Mass Spectrometry. J Proteome Res. 2008;7:1584–1593. doi: 10.1021/pr700577z. [DOI] [PubMed] [Google Scholar]
- 47.Baliban RC, DiMaggio PA, Plazas-Mayorca MD, Young NL, Garcia BJ, Floudas CA. A novel approach for untargeted post-translational modification identification using integer linear optimization and tandem mass spectrometry. Mol Cell Proteomics. 2010;9:764–779. doi: 10.1074/mcp.M900487-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiMaggio P, McAllister S, Floudas CA, Feng XL, Rabinowitz J, Rabitz H. A network flow model for biclustering via optimal re-ordering of data matrices. J Glob Opt. 2010;47:343–354. doi: 10.1186/1471-2105-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiMaggio PA, McAllister SR, Floudas CA, Feng XL, Rabinowitz JD, Rabitz HA. Biclustering via optimal re-ordering of data matrices in systems biology: rigorous methods and comparative studies. BMC Bioinformatics. 2008;9:458. doi: 10.1186/1471-2105-9-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 51.Creasy DM, Cottrell JS. Unimod: Protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 52.Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 53.Mitchelhill K. Delta Mass: A Database of Protein Post Translational Modifications. http://www.abrf.org/index.cfm/dm.home.
- 54.Floudas CA. Nonlinear and Mixed-Integer Optimization. Oxford University Press; New York: 1995. [Google Scholar]
- 55.Floudas CA, Paules GE., IV A Mixed-Integer Nonlinear Programming Formulation for the Synthesis of Heat-Integrated Distillation Sequences. Comp Chem Eng. 1988;12:531–546. [Google Scholar]
- 56.Kokossis AC, Floudas CA. Synthesis of Isothermal Reactor-Separator-Recycle Systems. Chem Eng Sci. 1991;46:1361–1383. [Google Scholar]
- 57.Kokossis AC, Floudas CA. Optimization of Complex Reactor Networks-II: Nonisothermal Operation. Chem Eng Sci. 1994;49:1037–1051. [Google Scholar]
- 58.Floudas CA. Anastasiadis Synthesis of General Distillation Sequences with Several Multicomponent Feeds Products. Chem Eng Sci. 1988;43:2407–2419. [Google Scholar]
- 59.Ciric AR, Floudas CA. A retrofit approach for heat exchanger networks. Comp Chem Eng. 1989;13:703–715. [Google Scholar]
- 60.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commum Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 61.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Meth. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 62.Torrente MP, Zee BM, Baliban RC, Young NL, LeRoy G, Floudas CA, Hake SB, Garcia BG. Proteomics Interogation of Human Chromatin. PLoS ONE. 2011;6:e24747. doi: 10.1371/journal.pone.0024747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-Specific Remodeling by HMGB1 and Linker Histone H1 Silences Proinflammatory Genes during Endotoxin Tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 65.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Nat Sci Rep. 2011;1:1–5. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baliban RC, Sakellari D, Li Z, DiMaggio PA, Garcia BA, Floudas CA. Novel protein identification methods for biomarker discovery via a proteomic analysis of periodontally healthy and diseased gingival crevicular fluid samples. J Clinical Periodontol. 2011;39:203–212. doi: 10.1111/j.1600-051X.2011.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.