Abstract
In the developing CNS, unique functional identities among neurons and glia are, in part, established as a result of successive transitions in gene expression programs within neural precursor cells. One of the temporal-identity windows within Drosophila CNS neural precursor cells or neuroblasts (NBs) is marked by the expression of a zinc-finger transcription factor (TF) gene, castor (cas). Our analysis of cis-regulatory DNA within a cas loss-of-function rescue fragment has identified seven enhancers that independently activate reporter transgene expression in specific sub-patterns of the wild-type embryonic cas gene expression domain. Most of these enhancers also regulate different aspects of cas expression within the larval and adult CNS. Phylogenetic footprinting reveals that each enhancer is made up of clusters of highly conserved DNA sequence blocks that are flanked by less-conserved inter-cluster spacer sequences. Comparative analysis of the conserved DNA also reveals that cas enhancers share different combinations of sequence elements and many of these shared elements contain core DNA-binding recognition motifs for characterized temporal-identity TFs. Intra-species alignments show that two of the sub-pattern enhancers originated from an inverted duplication and that this repeat is unique to the cas locus in all sequenced Drosophila species. Finally we show that three of the enhancers differentially require cas function for their wild-type regulatory behavior. Cas limits the expression of one enhancer while two others require cas function for full expression. These studies represent a starting point for the further analysis of cas gene expression and the TFs that regulate it.
Keywords: Drosophila CNS development, cis-regulatory DNA, sub-pattern enhancers, DNA sequence conservation, castor gene regulation
Introduction
During Drosophila neurogenesis, neural precursor cells or neuroblasts (NBs) are singled-out from adjacent neuroectodermal cells by a cascade of regulatory events involving both cell intrinsic and extrinsic instructive signals (for reviews see Campos-Ortega, 1995 and Skeath, 1999). Commencing soon after gastrulation, NBs exit the neuroectoderm and initiate lineage development in a sub-ectodermal proliferative zone by cycling through a series of asymmetric divisions, producing a ganglion mother cell (GMC) with each event. Each GMC divides to yield either neurons or glia (for review see Lin and Lee, 2012). During CNS NB lineage development a temporal network of cell-fate programs, distinguished by the sequential expression of the temporal-identity transcription factors (TFs) [Hunchback (Hb) -> Krüppel (Kr) -> POU domain proteins 1 and 2 (Pdm) -> Castor (Cas) -> Grainyhead (Grh)], collectively function over the course of several hours to generate multilayered basal (inner or dorsal) to apical (outer or ventral) uniquely fated neuronal subpopulations (review by Brody and Odenwald, 2002; Pearson and Doe, 2004; Lin and Lee, 2012).
The layered sub-populations of neurons and glia in both the ventral cord neuromeres and cephalic lobes can be identified by the expression of the temporal-identity TF that is transiently expressed in NBs during the generation of their GMCs. First-born, deeper neuronal subpopulations within all ganglia express hb while more superficial layers of later-born cells are marked by their expression of cas (Kambadur et al., 1998). In addition to playing a role in developing embryonic CNS lineages, most of the temporal-identity TFs are expressed postembryonically during larval and adult CNS development. For example, cas is expressed in larval ventral cord NBs, in linearly organized NB clusters on both sides of the interhemispheric brain junction and in adult neurons of the pars intercerebrallis, ellipsoid body and fan-shaped body fibers (Hitier et al., 2001). cas loss-of-function mutations are embryonic lethal (Mellerick et al., 1992) and mosaic mutant analysis has also demonstrated that cas is required for adult protocerebrum development (Hitier et al., 2001).
The availability of sequenced genomes from multiple species of different phyla has galvanized the development of phylogenetic footprinting tools to discover and compare cis-regulatory DNA (Wasserman et al., 2000; Odenwald et al., 2005; Visel et al., 2007; Loots and Ovcharenko, 2007; Yavatkar et al., 2008; and Brody et al., 2012). For example, the EvoPrinter comparative genomics tool combined with the cis-Decoder conserved DNA sequence database search and alignment algorithms have facilitated the discovery of functionally related Drosophila enhancers (Yavatkar et al., 2008; Brody et al., 2012). Analysis of known, in vivo tested cis-regulatory DNA reveals that individual enhancers can be identified by their conserved DNA sequence clusters (CSCs). Alignments of the multiple conserved sequence blocks (CSBs) within a CSC reveal repeat and unique sequence elements that can be used to identify functionally related enhancers (Brody et al., 2008 and 2012). Adjacent independent enhancers can now be resolved from one another by the lack of sequence conservation within the intervening spacer DNA and by the greater variability in spacer lengths observed among multiple species, relative to the more evolutionary constrained size of orthologous CSCs (Kuzin et al., 2009).
To increase our understanding of the molecular details that control the synchronized NB expression of the temporal-identity TFs and ultimately gain insight into mechanisms of neuronal diversification, we have undertaken the identification and cis-regulatory analysis of enhancers that regulate the dynamic expression of cas. As an initial step to locate essential cas enhancers, we identified a 17.5 kb genomic fragment that rescues an embryonic lethal casnull allele to full-viability. This study reports on the in vivo cis-regulatory analysis of multiple shorter regions that encompass the rescue construct, demonstrating that CSCs within the 13.1 kb of flanking DNA and the cas 5’UTR function as independent enhancers that regulate different aspects of cas gene expression. Intra-genomic searches also reveal that sub-regions of two enhancers originated from an inverted duplication, and the extent of sequence identity between the repeat halves differs among 12 Drosophila species. Similar to the cis-regulatory studies of other dynamically expressed genes during embryonic development such as nerfin-1 (Kuzin et al., 2011), hb (Hirono et al., 2012) and odd-paired (Fujioki and James, 2012), our analysis of cas cis-regulation reveals that multiple independent enhancers are responsible for different aspects of cas spatial/temporal expression. We also show that three of the enhancers that contain conserved Cas DNA-binding sites require cas for their proper regulation. Collectively, these studies (and those of others) indicate that multiple independent enhancers appear to be a general regulatory strategy used to control dynamic gene expression in heterogeneous populations of cells undergoing a diversity of developmental programs.
1. Results and discussion
1.1. Comparative genomic analysis of the castor locus
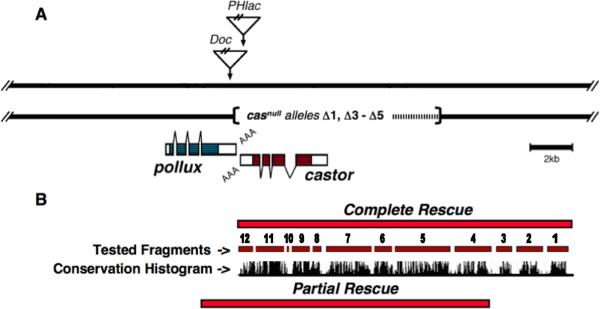
To locate cis-regulatory DNA essential for cas function, we first identified two overlapping genomic fragments that when independently inserted on the 2nd chromosome rescued the embryonic lethality of a 3rd chromosome casnull imperfect P-element excision allele (Fig. 1; the H23A 1 casnull allele is described in Mellerick et al., 1992). Although both rescue fragments restored embryonic viability and no discernable differences between Cas expression in the H23A 1/rescue-fragment transformant and wild-type embryonic backgrounds were detected, only one rescue fragment, which had an additional 4.5 kb of upstream DNA, rescued the H23A 1 allele to full-viability (Fig. 1 and data not shown). H23A 1 mutant larvae that were homozygous for the shorter partial rescue fragment died during the first or second instar developmental phases. A 12 species relaxed EvoPrint of the cas genomic region, including the complete rescue fragment, identified multiple CSCs both in the transcribed sequence and within its 5’ flanking region (Fig. 2). The analysis also revealed that the full rescue fragment contained three additional CSCs not present in the partial rescue fragment (Fig. 1 and 2). cis-Decoder CSB alignments of the cas CSCs also revealed that many of the sequence elements that are shared among the CSCs contain identifiable DNA-binding sites for characterized TFs, including POU-domain, HOX, bHLH, Pbx and Ets TFs (Fig. 2).
Fig. 1.
Genomic rescue fragments delimit essential castor cis-regulatory DNA. A) The genomic organization of the pollux (plx) and castor (cas) genes located on the 3rd chromosome at the cytological position 83C. The linear representation spans ~28 kb of the locus showing the integration site positions of a Doc retrotransposon and PHlac P-element enhancer-trap vector within the H23A enhancer-trap line. Aligned with the map are casnull deletion alleles created by imperfect excisions of the PHlac P-element (Mellerick et al., 1992 and Zhang et al., 1996). The dashed line within the deleted region indicates breakpoint uncertainties. The plx and cas genes are transcribed from opposite strands with the blue (plx) and red (cas) shaded transcribed regions representing their ORFs and angle bars indicating introns. B) Aligned to the map in panel A are genomic rescue fragments that, when inserted on the 2nd chromosome, either fully or partially rescue the embryonic lethality of the casnull H23A 1 PHlac P-element imperfect excision allele (see Mellerick et al., 1992 for imperfect excision details). The partial rescue fragment, lacking the 4.5 kb of the distal portion of the complete rescue fragment, rescues embryonic lethality, however larva die during the 1st and 2nd instar developmental phases. Numbers and red bars aligned under the rescue fragment indicate DNA fragments that were tested for cis-regulatory activity. Also shown is a 12 Drosophila species conservation histogram obtained from the UCSC Genome Browser that highlights the presence of multiple clusters of conserved DNA sequences within the tested fragments.
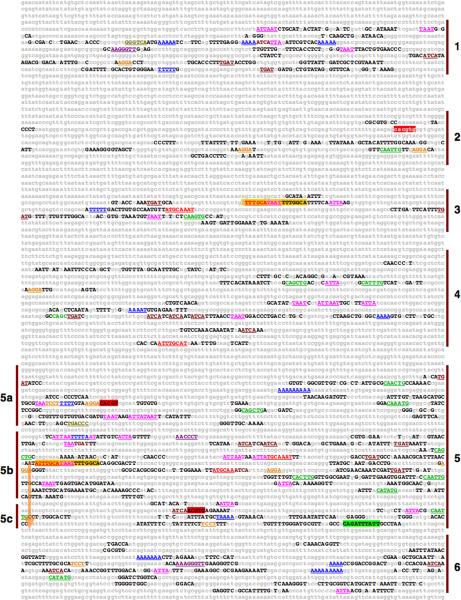
Fig. 2.
EvoPrint analysis of the complete rescue fragment (17.55 kb) identifies multiple conserved sequence clusters (CSCs) and an inverted repeat. Capital letters represent bases in the D. melanogaster reference sequence that are conserved in all, or all but one, of the following orthologous regions within D. simulans, D. sechellia, D. erecta, D. yakuba, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. virilis, D. mojavensis and D. grimshawi. The numbered vertical bars in the margins indicate the CSCs that were tested for in vivo cis-regulatory activity (summarized in Table 1; 5’ and 3’ ends of the tested fragments are listed in the Supplemental data table 1). The cas transcribed region is identified by highlighted sequences and represent its 5’ UTR (yellow), ORF (blue) and 3’ UTR (green). The gold colored arrows indicate the positions and orientation of inverted repeat sequences identified by the EvoPrinter repeat finder program and the green and gold highlighted sequences indicate the 5’ and 3’ boundaries of the inverted repeat described in Fig. 3. Also highlighted in gold is a 18 bp conserved sequence within the cas-3 CSC that matches the gold highlighted 18 bp sequence within cas-5 and cas-7. Different font-colored-underlined or highlighted DNA sequences correspond to core transcription factor DNA-binding sites (homeodomain, ATTA-pink; POU domain, ATGCAAAT-red; bHLH, CANNTG-green: Hunchback/Castor, TTTTT/AT-blue; Krüppel, AACCCT-violet; Tramtrack, TCCT-orange; PBX sites, TGAT-dark red; Seven-up sites, GGGTCA-gold; and Single-minded/Tango sites, ACGTG-red highlight). Note that the three most distal CSCs (cas-1 -> cas-3) are not present in the partial rescue fragment.
Using the EvoPrinter repeat finder search program and composite eBLAT intragenomic alignments (described in Yavatkar et al., 2008), we identified an inverted repeat that partially overlaps two of the upstream CSCs and is present once in each of the 12 Drosophila genomes examined (Fig. 2 and 3). In D. melanogaster, the proximal half of the inverted repeat is located 680 bp upstream of the predicted start of transcription and is separated from the distal half by an 1,881 bp spacer that spans the cas-6 CSC (Fig. 2 and 3A). Both distal and proximal halves contain CSBs that are part of the cas-5 and cas-7 CSCs respectively, and both of these CSCs contain unique CSBs that flank the repeat halves (Fig. 2). Analysis of the inverted duplication in different Drosophila species revealed that all sequenced species have the inverted repeat, but the size, extent of repeat sequence identity and the orientation of its intervening region vary among different species (Fig. 3). For example, the sequence identity between proximal and distal repeat halves is not complete in D. melanogaster, as there are base-pair differences within the central region of each repeat and larger identity gaps in their outer ends (Fig. 3a). In addition, the repeats within the D. virilis and D. mojavensis species both have differences when compared to D. melanogaster (Fig. 3B and C). In contrast to D. melanogaster, the D. virilis repeat is larger (distal half 1,587 bp vs. 1,188 bp) and the central regions of the repeat halves are nearly identical with only a single base-pair difference between them, while the D. mojavensis repeat sequence identity is significantly lower than that observed in the repeat of other species (Fig. 3 and data not shown). Comparison of the D. mojavensis repeat halves reveals that the central portion of the repeats has not been conserved (Fig. 3).
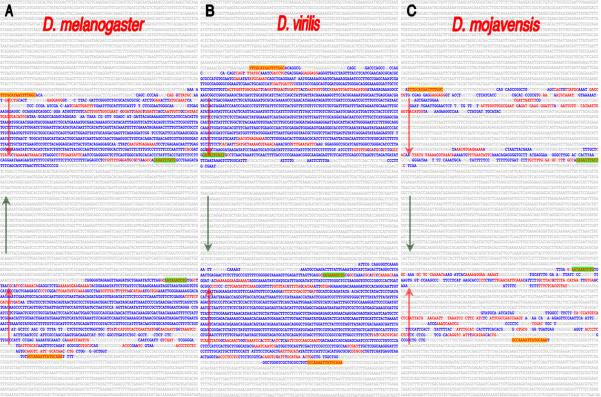
Fig. 3.
Intra-genomic alignments revealed an inve rted repeat that is present once in each of the Drosophila genomes and located upstream of the cas transcribed sequence. Shown are the inverted repeats within the D.melanogaster (A), D.virilis (B) and D.mojavensis (C) cas orthologous sequences. Uppercase blue- and red-colored sequences are identical (but inverted) within the repeats. The red-typeface sequences highlight CSBs that are common among the different orthologous regions in the repeat sequences and the yellow and green highlighted CSBs identify the outer-most repeat sequence blocks present in all species (orientation of the repeats is illustrated by the arrows). The repeats were initially identified via the EvoPrinter repeat finder program and their inverted orientation revealed by subsequent composite eBLAT analysis. The D. melanogaster inverted repeat extends from position -0.68 to -4.31 kb above the predicted cas transcription start site. Note the species-specific differences in the length and extent of sequence identity between the inverted halves. Conserved sequences flanking the inverted repeats are not highlighted.
The inter-species alignments also revealed that the cas-6 CSC (located within the intervening spacer between the repeat halves) had flipped its orientation in five of the Drosophila species (D. persimilis, D. willistoni, D. virilis, D. mojavensis and D. grimshawi) relative to the repeat-spacer-repeat arrangement present in D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae and D. pseudoobscura (Fig. 3 and data not shown). The intra-species alignments also revealed that, unlike any of the other species examined, the cas locus within D. grimshawi is duplicated, and one of the duplicates does not include the adjacent pollux gene (data not shown).
cis-Decoder analysis of the conserved sequences (including both intra- and inter-CSC alignments) reveals that many of the CSCs share different combinations of conserved sequence elements, and many of these elements are repeated within the individual CSCs (Fig. 2, Supplemental data Fig. 1 and data not shown). As discussed above, cas-5 and cas-7 are structurally related to one-another (Fig. 2 and 3). The shared repeat and unique elements within the cas-5 and -7 CSCs have been used to discover other late temporal network Drosophila NB enhancers (Brody et al. 2012). In addition, the inter-CSC alignments also revealed that the cas-3 CSC shares an 18 bp sequence (ATTTGCATAATTTTGGCA) with both the cas-5 and -7 CSCs with all but the first base conserved in the cas-3 CSC (Fig. 2; gold-colored highlighted sequence). The shared sequence contains a POU-homeodomain TF DNA-binding octamer sequence (ATTTGCAT) and the core DNA-binding motif for Antennapedia class homeodomain TFs (TAAT). Unlike cas-5 and cas-7, which both activate expression in the embryo, cas-3 drives expression in a set of larval brain neurons (see below). Future experiments are required to delineate function of the 18 bp sequence.
The presence of shared conserved sequence elements also suggests that cas-1 and -6 CSCs may belong to a structurally related family of enhancers. Alignments revealed that the two CSCs share the following sequences GCAAGGGTT, TTGGGTTG and TCAAAGGGT (Supplemental data Fig. 1). These elements all contain Kr consensus DNA-binding sites (Schroeder et al., 2004). Although it is beyond the scope of this paper to examine the role of Kr in the regulation of these enhancers, our result is suggestive of a role for this temporal-identity TF in the regulation of cas expression. Previous work of others has shown that other CSCs in the cis-Decoder database contain clusters of conserved Kr binding sites including a number of segmentation and neural enhancers that are either known or putative targets of Kr regulation, including the hb HZ enhancer driving expression in the embryonic ectoderm (Berman, et al. 2004), the pdm-2 CE8012 neuroblast enhancer (Berman et al, 2004) and a hairy stripe 1 enhancer (Riddihough, 1991).
The comparative analysis also highlighted conserved amino acid codons within the ORF (Fig. 2). Invariant codon nucleotide positions for the poly-glutamine (first exon) and the Zn-fingers (exons 2 and 3) domains were the most prominent while the 3’ end of the ORF and the 3’ UTR lacked sequence conservation. Unlike the Zn-finger domain codons, that show evolutionary divergence in many of their wobble positions, most of the codons for the conserved poly-glutamine tracks within the first exon showed significantly fewer substitutions in their wobble positions, most likely reflecting the fewer nucleotide options for the glutamine codon wobble position. To test if any of the ORF CSCs may also function as enhancers, we examined their potential to activate transgene reporter expression during different phases of development, and did not detect any cis-regulatory activity (discussed below).
1.2. cas CSCs activate transgene expression in sub-regions of cas gene expression
To determine the cis-regulatory behavior(s) of cas enhancers, we tested each of the CSCs contained in the complete rescue fragment using gypsy-insulated enhancer/reporter transgenes (Kuzin et al., 2011; Brody et al., 2012). To further control for chromosomal integration-specific events that could possibly affect reporter expression, we employed the phiC31 integrase mediated site-specific integration system to insure that all reporter transgenes were inserted into the same chromosomal environment (Groth et al., 2004; Markstein et al., 2008). Multiple independent transformant lines were tested for each CSC reporter transgene (see Table 1), and no significant variability was detected among the independent lines for each construct. Mutational analysis of enhancers has revealed that when CSBs are removed or altered, the enhancer often becomes unstable, triggering a high degree of cis-regulatory variability. For example, functional analysis of conserved sequences within the nerfin-1 NB enhancer reveal that its bilateral expression symmetry within ventral cord NBs that flank the midline is dependent on maintaining its CSBs and their individual sequences (Kuzin et al., 2011).
Table 1.
Summary of CSC cis-regulatory activity
| CSC | Transformant linesa | Embryo Expression | Larva Expressionb | Adult Expression | Figure |
|---|---|---|---|---|---|
| #1 | 4 | Brain and VNC NBs | NBs and neurons of the brain, thorax, and optic lobe | Olfactory lobe neurons | 4A, 6A, 7A |
| #2 | 4 | Ventral cord midline mesectodermal cells | NBs and neurons of the brain & thorax | Small subset of brain neurons | 4B, 6C |
| #3 | 4 | No expression | Small subset of CNS neurons | Small subset of brain neurons | 6B |
| #4 | 3 | Intermediate row VNC neurons | Small subset of CNS neurons | Small subset of brain neurons | 4C, 7B |
| #5 | 3 | Subset Brain and VNC NBs | Small subset of CNS neurons | No expression | 4D, 6D |
| #6 | 3 | Subset Brain and VNC late stage NBs | CNS Neurons | Small subset of brain neurons | 4H, 6F, 7C |
| #7 | 3 | Subset Brain and VNC NBs | No expression | No expression | 4I |
| #8 | 3 | Brain and VNC GMCs | CNS Neurons | Olfactory lobe neurons | 4L, 6E, 7D |
| #9 | 2 | No expression | No expression | No expression | not shown |
| #10 | 2 | No expression | No expression | No expression | not shown |
| #11 | 5 | No expression | No expression | No expression | not shown |
| #12 | 5 | No expression | No expression | No expression | not shown |
Number of independent transformant lines examined.
Excluding vector-dependent salivary gland expression (see Zhu and Halfon, 2007).
Whole-mount embryo in situ mRNA localization of transgene reporter expression revealed that all but four of the tested CSCs activate expression in different or overlapping regions of the cas gene embryonic expression window. Examples of peak CSC-reporter expression for the embryonic enhancers are shown in Fig. 4. Reporter expression was not detected in tissues outside of the CNS for any of the CSCs tested nor was expression observed outside the cas late temporal window within the embryonic CNS. One of the four CSCs that did not activate embryo reporter expression, the cas-3 CSC, was found to direct transgene expression in the larval CNS (described below) while the other three fragments that span the ORF, introns and 3’ UTR (fragments 9–12) were not active during any of the developmental windows examined in this study (Fig. 6 and data not shown).
Fig. 4.
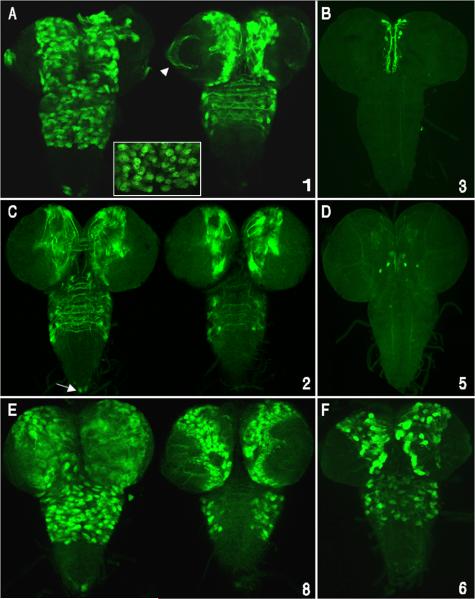
cas CSCs function as cis-regulatory enhancers that control different aspects of embryonic CNS expression. Shown are enhancer-reporter transgene embryo expression patterns for seven CSCs and sub-fragments from cas-5 and -7 CSCs. Whole-mount stained embryos (staging according to Hartenstein and Campos-Ortega, 1984; ventral, lateral and dorsal views are given from left to right, with embryos oriented with anterior up) reveal peak transgene reporter mRNA expression detected by digoxigenin labeled RFP riboprobes for each of the enhancer-reporter constructs. The numbers in the lower right corner of each panel correspond to the CSCs shown in Fig. 2 with the red and green colored numbers indicating the different sub-fragments of CSCs 5 and 7, respectively. A) The cas-1 CSC activates reporter expression in a large subset of CNS NBs (both ventral cord and cephalic lobes) during stage 11. B) cas-2 CSC directs reporter expression in a subset of cells within the ventral cord midline mesectodermal cells and in neural precursor cells that line the anterior midgut primordium (arrow) during stage 9, similar to the onset of endogenous cas gene expression (described in Mellerick et al., 1992 and Kambadur et al., 1998). C) cas-4 CSC activates expression in intermediate column ventral cord NBs and in a sub-set of cephalic lobe NBs, during stage 11. D) Similar, but not identical to cas-1, the cas-5 CSC drives expression in a wide-range of NBs at stage 11. E - G) cas-5 CSC sub-fragments (5a, b and c) activate reporter expression in different subsets of NBs but not in all NBs that express the full cas-5/reporter transgene. E) During late stage 11, the 5a sub-fragment activates expression in a subset of ventral cord midline NBs and in cephalic lobe NBs. F) The central region of cas-5 CSC (5b) that spans the distal part of one of the inverted repeats and flanking CSBs (outside of the repeat) regulates expression in intermediate row ventral cord NBs during stage 12, and the proximal inverted repeat sub-region (5c) shown in panel G activates expression in a single medial row NB and adjacent intermediate row NB per ventral cord hemisegment. Note, the full cas-5 CSC (D) activates expression in more NBs than the sum of its sub-fragments. H) Peak expression of the cas-6 reporter/transgene occurs in NBs during stage 13. I) Similar to cas-1 and cas-5, cas-7 CSC reporter expression was detected in subsets of NBs during stage 11. J and K) cas-7 sub-regions (7a and 7b) that span the proximal half of the upstream inverted repeat and flanking CSBs. J) Like cas-5c, cas-7a activates reporter expression in a single medial row NB per hemisegment and in an adjacent intermediate row NB during late stage 11. K) cas-7b activates expression in ventral cord midline NBs and in a small subset of cephalic lobe NBs during early stage 12. Like cas-5 CSC and its sub-regions, cas-7 sub-regions do not drive reporter expression in all cas-7 CSC positive cells. L) Reporter expression driven by the cas-8 CSC, the 5’UTR, is detected in most GMCs and/or nascent neurons throughout the CNS during stage 11.
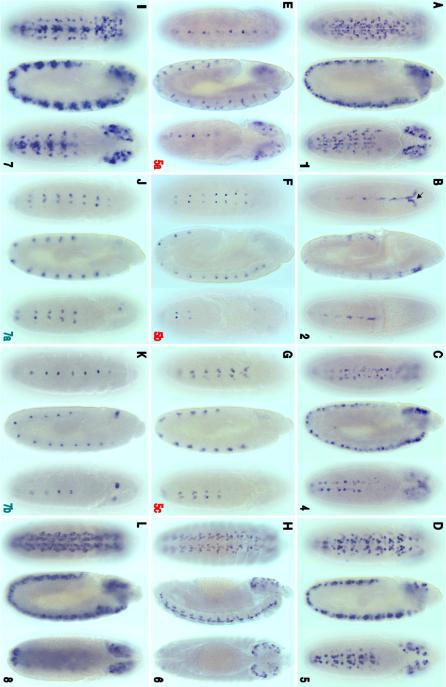
Fig. 6.
Many cas embryonic enhancers also direct gene expression during larval CNS development. CSC reporter Gal4 expression was detected using a Gal4 activated UAS/GFP-mCD8 transgene followed by anti-GFP fluorescent immunostaining. Images show dissected third-instar larval brains and ventral cords (anterior up) with panels A, C and E showing optical sections from the ventral (left) and dorsal (right) regions of the CNS. Panels B, D and F show stacked Z-series optical sections of the whole CNS. Numbers on the lower right side of each panel represent CSC regions shown in Fig. 2. A) cas-1 enhancer/reporter is expressed in many putative type II NBs and their lineages, including precursors and neurons of the central brain, thorax, and optic lobe neurons (arrowhead). cas-1 also directs expression in a cluster of abdominal ventral cord neurons located at the posterior tip (arrow). B) cas-3 enhancer/reporter expression in a subset of neurons within the medial brain hemispheres. Given their position and size, these medial-anterior neurons may be part of the neurosecretory cell group in the pars intercerebralis (de Valasco et al., 2007). C) cas-2 reporter is predominantly expressed in subsets of brain and thoracic neurons whose axons cross the midline, and in two neurons located at the posterior tip of the ventral cord (arrow). There are fewer GFP positive NBs compared with cas-1, suggesting that cas-2 activates expression predominantly in neurons. D) cas-5 enhancer/reporter activity was detected in 2 central brain neurons. E) cas-8 CSC activated reporter expression in a large subset of central brain lineages including optic lobe medullary NBs and their progeny, many thoracic lineages that project across the midline and posterior tip ventral cord neurons. F) cas-6 enhancer/reporter is expressed in a subset of Type II NB lineages.
During embryonic stage 9, cas gene expression is first detected in segmental clusters of ventral midline mesectodermal cells and in cells that line the anterior midgut primordium (Mellerick et al., 1992; Kambadur et al., 1998). At this stage, no expression is detected in either cephalic lobe or ventral cord NBs. The expression pattern observed with the cas-2 CSC/reporter transgene matches the early onset of the endogenous cas expression (Fig. 4B). Located 10.3 kb 5’ to the predicted transcription start site, the cas-2 enhancer contains 10 CSBs of 6 bp or greater in length that span 557 bp of genomic DNA (Fig. 2). cis-Decoder intra-CSC alignments of the CSBs identified both repeat and palindromic conserved sequences, including two copies of a 6 bp repeat (CCCTTT) and the palindromes (TTATAA and CAATTG) (Fig. 2 and data not shown). Core DNA-binding motifs for bHLH and Tramtrack TFs (Fig. 2; green and orange colored font, respectively) are present in three of the CSBs. Also present, but only partially conserved in the 12 species that were included in the EvoPrint (not present in D. virilis, D. mojavensis and D. grimshawi) is a docking site for the Single-minded (Sim) and Tango (Tgo) dimer (CACGTG, red highlighted sequence in Fig. 2). The role and expression of Sim/Tgo bHLH TFs in midline development is discussed in Freer et al. (2011) and in Fulkerson and Estes (2011).
Expression analysis of cas-1, -4, -5, -7 and -8 CSC/reporter transgenes suggest that these enhancers participate in regulating different or overlapping aspects of the spatial and temporal cas embryonic NB expression dynamics (Fig. 4). Analysis of cas gene expression has shown that subsets of cephalic lobe and ventral cord NBs initiate cas expression during embryonic stage 10 (Kambadur et al., 1998). The first ventral cord NBs to express Cas are the late delaminating medial row NB6-1 NBs located on the posterior edge of the gnathal, thoracic and abdominal segments. Soon after this onset, additional medial, intermediate and lateral row NBs initiate expression, and by stage 12 most ventral cord and cephalic lobe NBs have detectable levels of cas mRNA and/or protein. Cas expressing NBs, identified based on their large cell body diameters and their position underlying the ectoderm, and their Cas positive descendants (GMCs, nascent neurons and glia) are positioned on the outer surface of the cephalic lobes and on the ventral/ventral-lateral surfaces of the developing subesophageal ganglion and ventral cord neuromeres (see Fig. 2 and 3 in Kambadur et al., 1998). Similar to wild-type cas expression, cas-1, -4, -5, -7 and -8 CSCs activate reporter expression in overlapping patterns of ventral cord and cephalic lobe NBs (Fig. 4 panels A, C, D and F), while cas-8 directs expression predominantly in GMCs and nascent neurons (Fig. 4L).
The less-conserved central regions of the inverted repeat within the cas-5 and -7 CSCs indicate that these relatively large CSCs might span multiple independent enhancers (Fig. 2 and 3). For example in D. mojavensis, which has the poorest conservation within the inverted repeat, the distal half of the repeat within the cas-5 CSC contains a 600 base-pair gap of less-conserved sequence between CSBs. To test if the different sub-regions of cas-5 and -7 could function independently as enhancers and if the combined expression patterns of the sub-fragments recapitulate the full expression domains of the CSCs, we generated three smaller enhancer/reporter transgenes for the cas-5 CSC (cas-5a, b and c) and two that cover subsets of the cas-7 CSBs (cas-7a and b) (Fig. 2). Although each sub-region of both the cas-5 and -7 CSCs activated reporter expression within different sub-patterns of their full CSCs expression domain, collectively the sub-fragments of each CSC did not recapitulate the full embryonic expression of the cas-5 or cas-7 enhancers (compare Fig. 4D to panels E - G and Fig. 4I to panels J and K). Both full enhancers activate reporter expression in greater numbers of NBs, with the most significant difference between complete CSCs and their sub-regions being detected in the cephalic lobes and lateral ventral cord NBs. Corresponding sub-regions of the inverted repeat halves exhibited similar cis-regulatory behaviors. For example, both the cas-5c and cas-7a sub-regions activate reporter expression in medial row ventral cord NBs. The sub-fragments that span both repeat and unique flanking CSBs (cas-5b and cas-7b) do not overlap in their expression patterns, suggesting that the unique CSBs flanking the repeats harbor spatial cis-regulatory information for these regions. Interestingly, although both cas-5 and -7 CSCs direct expression within subsets of ventral midline cells and both contain highly conserved Sim/Tgo DNA-binding sites, one of the cas-7 sub-regions, cas-7a, includes one of the conserved Sim/Tgo core docking site flanked by other conserved sequences (TTTCTCACGTT), but does not activate midline expression (Fig. 4J).
Near the end of the cas temporal NB expression window (starting at the end of stage 12 and progressing through stage 13), the cas-6 enhancer activates reporter expression in a subset of NBs positioned along the outer edges of the cephalic lobes and along the ventral/ventral-lateral edges of the ventral cord neuromeres (Fig. 4H). As with endogenous cas expression, cas-6 enhancer/reporter expression diminishes during stage 14 until only a small subset of ventral cord cells have detectable levels of expression (data not shown).
1.3. cas enhancers alter their cis-regulatory behavior in casnull mutant embryos
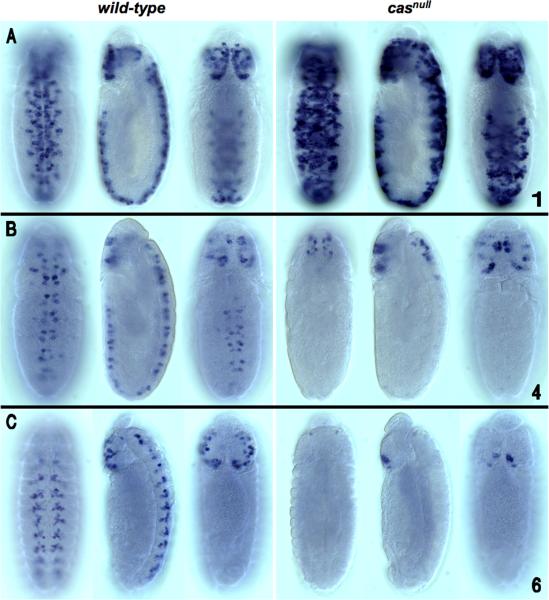
As an initial step toward identifying the TFs that are required for cas expression, we examined the reporter expression patterns of the cas embryo enhancers in H23A 1 casnull mutant embryos. EvoPrint analysis revealed that all but one of the cas CSCs (cas-2) have multiple CSBs that contained core recognition motifs for Cas DNA-binding (Fig. 2, blue colored font). Three of the enhancers were found to require cas for wild-type behavior (Fig. 5 and data not shown). One of the enhancers, the cas-1 CSC, which activates reporter expression in both cephalic lobe and ventral cord NBs, was hyper-expressed in casnull embryos (Fig. 5A). Both extra cells and higher levels of expression were observed. cas-4 and cas-6 exhibited diminished expression in a casnull background. For both constructs, only brain expression remained; for cas-4, expression in the brain was not impaired, while for cas-6, a reduced number of cells expressing the reporter was also apparent in the brain cephalic lobes. It is unclear whether the effects of cas mutant background on cas enhancer/reporter expression are direct or indirect, since many regulatory events have been observed to be downstream of cas (Baumgardt et al., 2009). The Baumgardt study suggested that the broad temporal window regulated by cas could be subdivided into multiple feed-forward loops. Absence of cas function had no detectable effect on the expression of cas-2, cas-5 or cas-7. Interesting two of the non-responsive enhancers, cas-5 and cas-7 both contain multiple Cas binding sites. Future studies are required to understand the mechanism of cas self-regulation.
Fig. 5.
Loss of cas function differentially affects cas CSC/reporter transgene expression in the developing embryonic CNS. Shown are ventral (left), lateral (center) and dorsal (right) views of individual transformant embryos from wild-type and casnull backgrounds (anterior up). CSC reporter/transgene expression was detected by whole-mount embryo in-situ mRNA hybridization using a digoxigenin labeled Gal4 riboprobe for the cas-1 reporter/transgene and a RFP riboprobe for the cas-4 and cas-6 reporter/transgenes. A) Compared to its wild-type expression dynamics, loss of cas function triggered heightened expression levels of the cas-1/reporter transgene throughout the CNS in addition to its expression in a greater number of NBs. B and C). cas-4 (stage 12 embryo) and cas-6 (stage 13 embryo) reporter/transgenes, respectively, exhibited diminished expression in a casnull background. For both constructs, loss of transgene expression was most significant in the ventral cord; cas-4/reporter transgene expression in the cephalic lobes was not significantly impaired, while for cas-6, reduced number of transgene expressing NBs was more significant in the subesophageal ganglion.
1.4. cas enhancers are also active in the larva and adult CNS
Previous studies have shown that cas is expressed in the larval ventral nerve cord and in the larval brain (Hitier et al., 2001). cas enhancer-trap expression studies have also revealed that cas is most likely expressed in a subset of ellipsoid body and fan-shaped body fibers, and in the pars intercerebralis (Hitier et al., 2001). Given these observations, we sought to determine if any, or all, of the cas CSCs function as enhancers in larvae or adults (Fig. 6 and 7).
Fig. 7.
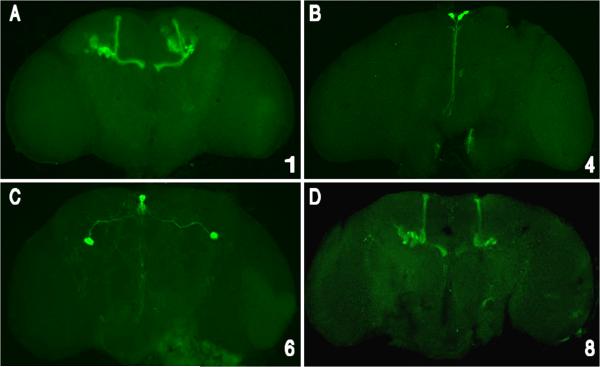
cas CSCs also function in the adult brain as enhancers that drive expression in limited numbers of neural lineages. Gal4 transgene expression was visualized by crossing the transformants with a UAS/GFP-mCD8 transgene containing reporter line and anti-GFP immuno-flourescent localization. Expression of each construct is represented by stacked optical confocal images (frontal views) of adult brains. A & D) cas-1 and cas-8 express in the olfactory lobe and the membrane-tagged GFP-CD8 marks the projection neurons that project to the mushroom body. B) cas-4 is expressed in an anterior cell from the medial neurosecretory group. C) cas-6 is expressed in two anterior medial cells with posterior projecting axons and a pair of large cell-body lateral neurons that project axons towards the midline and dendrites extending throughout the adult brain.
In the 3rd instar larvae, cas-1, -2 and -8 CSCs drove reporter expression within ventral nerve cord and brain NB lineages (Fig. 6A, C and E respectively; each is represented by two z-series at different focal planes). Many larval NB lineages are made up of coherent clusters of cells and a projecting axonal fascicle, as described by Pereanu and Hartenstein (2006), and reviewed by Ito and Awasaki (2008) (also see Fig. 6A inset). cas-1 enhancer/reporter activity was detected in many NB lineages of the central brain and thorax, and also in a two terminal abdominal neurons whose axonal projections did not cross the midline. The cas-2 enhancer/reporter drove expression in sets of brain and thoracic neurons that cross the midline (Fig. 6C). Compared to cas-1 fewer cells expressed the reporter, suggesting that cas-2 regulates expression predominantly in neurons. cas-8 (the 5’UTR) drives reporter expression in a large set of central brain lineages, including medullary NBs and their progeny, and many neurons project between the hemispheres (Fig 6E). The reporter is also expressed in lamina and medulla neurons of the optic lobe. cas-8 is expressed in lateral thoracic neurons that cross the midline and in many ventral lineages.
The expression patterns of three other CSCs (cas-3, -5 and -6) are represented by a single z-series representing the full dorsal/ventral extent of the third-instar larva (Fig. 6B, D and F). cas-3 enhancer/reporter, which was silent in the embryo, is expressed in a small set of larval brain neurons (Fig. 6B). Given their position and size, the anterior cells are probably the median neurosecretory cell group in the pars intercerebralis (Hitier et al., 2001; de Valasco et al., 2007). cas-4 enhancer/reporter drove expression in a single pair of medial brain neurons that project posterior, again corresponding to a putative neurosecretory cell (data not shown). cas-5 enhancer/reporter, which includes the distal inverted repeat, expressed in several more posterior central brain neurons. cas-6 enhancer/reporter, consisting of the spacer between the inverted repeats, expressed in fewer lineages, comparable in number to cas-2 (Fig. 6C), and like cas-2 fewer NBs are labeled. Positive thoracic neurons are arrayed laterally and project axons across the midline. cas-7 enhancer/reporter was completely inactive in early 3rd instar larvae (data not shown). Additional co-labeling studies are required to identify specific NBs and the identities of the post-mitotic cells within their lineages.
In the adult, both cas-1 and cas-8 CSCs activate expression in the olfactory lobe. The membrane-tagged GFP-mCD8 marks the projection neurons that project to the mushroom body (Figs 7A and D). This expression could be related to a previously documented requirement of cas for axon pathfinding within the mushroom body (Hitier et al., 2001). The cas-4 CSC regulates reporter expression in an anterior cell from the medial neurosecretory group (Fig 7B), while cas-6 drives expression in two anterior medial cells that project axons posteriorly and a pair of large lateral cells that project axons towards the midline, and neurites throughout the adult brain (Fig 7C).
2. Summary
This paper describes the comparative genomic and expression analysis of enhancers that regulate the late temporal network determinant castor. Our analysis reveals that the embryonic expression of cas is controlled by multiple enhancers, termed here sub-pattern enhancers, that activate expression in overlapping similar but non-identical subsets of cells in the brain and ventral cord. Even the CSCs that include the inverted repeats regulate expression in non-identical subsets of cells, as double labeling experiments revealed that the patterns overlap but were not identical (data not shown). Given that the first three distal-most enhancers direct expression to different spatial/temporal windows of cas expression, the absence of these enhancers in the partial rescue fragment most likely results in its inability to completely rescue the casnull mutant.
The inverted repeat upstream of the cas transcribed sequence partially constitute two different but functionally related enhancers. The selective pressures that maintain the inverted repeat in the different Drosophila species are currently unknown. Models for the evolution and maintenance of the inverted duplication can be built on the following assumptions: First, having repeat copies of a regulatory sequence must be advantageous since both sets are highly conserved; and second, the species-specific nature and extent of inverted sequence identity between the two repeat halves indicates that each may play a role in maintaining repeat identity with its partner. For example, one half of the repeat could possibly serve as a template for rectifying differences between the two. Given that the extent of sequence identity between the two halves, varies between species, sequence corrections (rectification) most likely occurred at different times during species divergence. For example, the relatively high sequence identity between the D. virilis repeat halves suggests that its putative sequence rectification was more recent compared to any corrections that may have occurred in D. mojavensis.
Analysis of the cas enhancer cis-regulatory activity in larvae and adults shows that many of these enhancers are multi-functional; that is, they direct gene expression in embryos, larvae and/or adults. cis-Decoder analysis reveals that some of the enhancers fall into two structurally related families based on the sharing of conserved sequence elements; one class, represented by the cas-3, cas-5 and cas-7 CSCs are characterized by the presence of multiple POU-homeodomain TF DNA-binding sites. The second class is represented by cas-1 and cas-6, which each share many sequence elements including multiple Kr binding sites but lack POU TF docking sites.
The existence of cas sub-pattern enhancers can be compared to recent studies of segmentation enhancers in Drosophila. Multiple enhancers function to drive gap gene expression in similar non-identical fashion acting to ensure the full pattern of expression of the regulated gene (Perry et al., 2011). Similarly, transgenic rescue experiments suggest that most of the partially redundant enhancers associated with the sloppy-paired locus of Drosophila are required for full gene function in maintaining wingless expression and parasegment boundaries throughout embryogenesis (Fujioka and Jaynes, 2012). We have also discovered that multiple enhancers associated with the nerfin-1 gene can drive expression in different subsets of neural precursor cells and neurons (Kuzin et al., 2009). Finally, the presence of sub-pattern enhancers is not confined to Drosophila. In vertebrates, expression of Ngn1 in the midbrain, hindbrain, trigeminal ganglia, and ventral–neural tube appear to be due to sub-pattern enhancers that are located both 5′ and 3′ of the Ngn1 coding sequence (Nakada et al., 2004). Our studies and those of others indicate the existence of sub-pattern enhancers is likely a general phenomenon required by the different regulatory environments in different tissues and lineages generated during development. These insights into cas regulation provide the basis for further, more detailed analysis of the molecular events that enable NBs to transition from one temporal gene expression program to the next.
3. Experimental procedures
3.1. Genomic rescue of a casnull mutation
Essential genomic DNA that constitutes the cas locus at cytological position 83C on the 3rd chromosome was delineated by rescuing the embryonic lethal cas loss-of-function P-element imperfect excision allele H23A 1 (see Mellerick et al., 1992 for details) with a cas genomic fragment inserted on the 2nd chromosome. As shown in Fig. 1, two genomic fragments that overlap in their central regions but have different 5’ and 3’ extended sequences were obtained from overlapping lambda genomic clones isolated from a D. melanogaster genomic library using a cas cDNA probe and standard library screening methods. After sequencing, the genomic fragments were cloned into the Not-I restriction site of the pCasper4 P-element transformation vector. Full details of the cloning steps, P-element transformation protocol and selection for 2nd chromosome insertions are available upon request.
3.2. Enhancer-reporter transgenes
CSC fragments were generated by standard PCR protocols. Primer sequences are provided in the Supplemental data Table 1. PCR-amplified genomic fragments were cloned into Invitrogen pCRII-TOPO vector for sequence verification. To test the cis-regulatory function of the CSC fragments, they were inserted into a modified pCa4B site-specific integration vector (Markstein et al., 2008) that we call pBullfinch-Gal4 (Brody et al., 2012). The genomic fragments were additionally inserted into pCa4B-RFP and pCa4B-GFP for testing enhancer activity (Kuzin et al., 2011). The pBullfinch-Gal4 vector was generated by inserting into pCa4B vector with following fragments: a polylinker site, the Hsp70 minimum promoter (from the pRed H-Stinger vector; Barolo et al., 2004), the Gal4 ORF (from S. cerevisiae) and the SV40 3'UTR (from the pRed H-Stinger vector; Barolo et al., 2004). Details of the cloning steps and vector sequence are available upon request. Third chromosome site-specific P-element integration transformants were generated using the site-specific integration vectors (described above) with the y, w; y+[attp2] transformant (Groth et al., 2004; Markstein et al., 2008). For expression analysis in cas mutants, transgenes were inserted into the second chromosome landing pad of the y, w; y+[attp16] transformant (Groth et al., 2004; Markstein et al., 2008).
3.3. Embryo expression analysis
Embryo collection and fixation were performed according to the procedures described by Patel (1994). For in situ hybridization detection of reporter expression, we used the Berkeley Drosophila Genome project embryo in situ hybridization protocol (http://www.fruitfly.org/about/methods/RNAinsitu.html) adapted for 1.6 ml Eppendorf tubes. Riboprobes were prepared using the Roche (Indianapolis, IN) DIG RNA Labeling kit. For simultaneous co-localization studies, we used the FISH protocol developed in the Krause Lab (Lécuyer et al., 2008). All details are available upon request. After whole-mount in situ hybridization, embryos were viewed in 70% glycerol/30% phosphate-buffered saline (PBS), and photographed using a Nikon microscope equipped with Nomarski (DIC) optics. Embryo developmental stages were determined by morphological criteria (Campos-Ortega and Hartenstein, 1985).
3.4. Immunohistochemistry and confocal imaging of larval and adult brain transgene expression patterns
Gal4 reporter expression in the larval brain and CNS of wandering larvae was analyzed using UAS/GFP-mCD8 (Lee and Luo, 1999) as the reporter. In this analysis, we crossed cas enhancer/Gal4 transgene containing males to UAS/GFP-mCD8 virgin females and independently examined 10-12 third instar larva CNS. Brain dissection, immunohistochemistry, and confocal imaging were performed as described previously (Lee and Luo, 1999). For immunohistochemistry, rabbit anti-GFP (1:1,500, Invitrogen, San Diego, CA) and Alexa 488 goat anti-rabbit IgG (1:1,000, Invitrogen) were used to enhance the GFP signal. The confocal image stacks were analyzed using ImageJ software.
For each genotype, at least ten adult flies of mixed genders were collected 1 days after eclosion and used for immunohistochemistry and imaging. Brain dissection, immunohistochemistry, and confocal imaging were performed as described previously (Gao et al., 2008). For immunohistochemistry, rabbit anti-GFP (1:1,500, Invitrogen, San Diego, CA) and Alexa 488 goat anti-rabbit IgG (1:1,000, Invitrogen) were used to enhance the GFP signal. The confocal image stacks were analyzed using ImageJ software.
Supplementary Material
Highlights.
castor gene expression pattern is regulated by 7 independent sub-pattern enhancers
Each cas enhancer consists of clusters of conserved DNA sequences
The multifunctional enhancers regulate expression in embryos, larvae and/or adults
cas enhancers combinatorially share conserved TF binding sites and novel sequences
The Castor transcription factor differentially regulates three of the cas enhancers
Acknowledgements
The authors would like to thank Takeshi Awasaki for his comments, Antonios Ekatomatis for technical assistance and Judith Brody for editorial expertise. This research was supported by the Intramural Research Program of the NIH, NINDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
References
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–42. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Díaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–82. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biology. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development. 2002;129:3763–70. doi: 10.1242/dev.129.16.3763. [DOI] [PubMed] [Google Scholar]
- Brody T, Rasband W, Baler K, Kuzin A, Kundu M, Odenwald WF. cis-Decoder discovers constellations of conserved DNA sequences shared among tissue-specific enhancers. Genome Biol. 2007;8:R75. doi: 10.1186/gb-2007-8-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Rasband W, Baler K, Kuzin A, Kundu M, Odenwald WF. Sequence conservation and combinatorial complexity of Drosophila neural precursor cell enhancers. BMC Genomics. 2008;9:371. doi: 10.1186/1471-2164-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Yavatkar AS, Kuzin A, Kundu M, Tyson LJ, Ross J, Lin TY, Lee CH, Awasaki T, Lee T, Odenwald WF. Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers. Dev Dyn. 2012;241:169–89. doi: 10.1002/dvdy.22728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA. Genetic mechanisms of early neurogenesis in Drosophila melanogaster. Mol Neurobiol. 1995;10:75–89. doi: 10.1007/BF02740668. [DOI] [PubMed] [Google Scholar]
- Cui X, Doe CQ. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116:943–52. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302:309–23. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Freer SM, Lau DC, Pearson JC, Talsky KB, Crews ST. Molecular and functional analysis of Drosophila single-minded larval central brain expression. Gene Expression Patterns. 2011;11:533–56. doi: 10.1016/j.gep.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerstenberg S, Broadus J, Doe CQ. Asymmetry and cell fate in the Drosophila embryonic CNS. Int J Dev Biol. 1998;42:379–83. [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB. Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev. Biol. 2012;362:309–19. doi: 10.1016/j.ydbio.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson E, Estes PA. Common motifs shared by conserved enhancers of Drosophila midline glial genes. J Exp Zool B Mol Dev Evol. 2011;316:61–75. doi: 10.1002/jez.b.21382. [DOI] [PubMed] [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, Wang JW, Odenwald WF, White BH, Meinertzhagen IA, Lee CH. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–42. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 2006;20:2618–27. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono K, Margolis JS, Posakony JW, Doe CQ. Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons. Gene Expr Patterns. 2012;12:11–17. doi: 10.1016/j.gep.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitier R, Chaminade M, Préat T. The Drosophila castor gene is involved in postembryonic brain development. Mech Dev. 2001;103:3–11. doi: 10.1016/s0925-4773(01)00312-4. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–21. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Adv Exp Med Biol. 2008;628:137–58. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–60. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A, Kundu M, Brody T, Odenwald WF. Functional analysis of conserved sequences within a temporally restricted neural precursor cell enhancer. Mech Dev. 2011;128:165–77. doi: 10.1016/j.mod.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A, Kundu M, Ekatomatis A, Brody T, Odenwald WF. Conserved sequence block clustering and flanking inter-cluster flexibility delineate enhancers that regulate nerfin-1 expression during Drosophila CNS development. Gene Expr Patterns. 2009;9:65–72. doi: 10.1016/j.gep.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, Necakov AS, Cáceres L, Krause HM. High-resolution fluorescent in situ hybridization of Drosophila embryos and tissues. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot5019. doi:10.1101/pdb.prot5019. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies, sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lin S, Lee T. Generating neuronal diversity in the Drosophila central nervous system. Dev Dyn. 2012;241:57–68. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Loots G, Ovcharenko I. ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics. 2007;23:122–24. doi: 10.1093/bioinformatics/btl546. [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–83. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Parab P, Simmons A, Omer-Abdalla A, Johnson JE. Separable enhancer sequences regulate the expression of the neural bHLH transcription factor neurogenin 1. Dev Biol. 2004;271:479–87. doi: 10.1016/j.ydbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–36. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc. Natl. Acad. Sci. 2005;102:14700–5. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–87. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–47. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108:13570–5. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–53. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddihough G, Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 1991;5:840–54. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB. At the nexus between pattern formation and cell-type specification: the generation of individual neuroblast fates in the Drosophila embryonic central nervous system. Bioessays. 1999;21:922–31. doi: 10.1002/(SICI)1521-1878(199911)21:11<922::AID-BIES4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341:331–35. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Tautz D, Lehmann R, Schnuerch H, Schuh R, Seifert E, Kienlin A, Jones K, Jaeckle H. Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature. 1987;327:383–89. [Google Scholar]
- Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–9. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J, Desplan C. The products of the Drosophila gap genes hunchback and Kruppel bind to the hunchback promoters. Nature. 1989;341:335–37. doi: 10.1038/341335a0. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–8. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- Yang X, Yeo S, Dick T, Chia W. The role of a Drosophila POU homeo domain gene in the specification of neural precursor cell identity in the developing embryonic central nervous system. Genes Dev. 1993;7:504–16. doi: 10.1101/gad.7.3.504. [DOI] [PubMed] [Google Scholar]
- Yavatkar AS, Lin Y, Ross J, Fann Y, Brody T, Odenwald WF. Rapid detection and curation of conserved DNA via enhanced-BLAT and EvoPrinterHD analysis. BMC Genomics. 2008;9:106. doi: 10.1186/1471-2164-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Halfon MS. Vector-dependent gene expression driven by insulated P-element reporter vectors. Fly (Austin) 2007;1:55–6. doi: 10.4161/fly.3892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.