Abstract
Influenza is a major cause of morbidity and mortality in the United States. Studies have shown that excessive T cell activity can mediate pneumonitis in the setting of influenza infection, and data from the 2009 H1N1 pandemic indicate that critical illness and respiratory failure following infection was associated with greater infiltration of the lungs with CD8+ T cells. T cell immunoglobulin and mucin domain 3 (Tim3) is a negative regulator of Th1/Tc1-type immune responses. Activation of Tim3 on effector T cells has been shown to down-regulate proliferation, cell-mediated cytotoxicity, and IFNγ production as well as induce apoptosis. Here, we demonstrate that deletion of the terminal cytoplasmic domain of the Tim3 gene potentiates its ability to down-regulate Tc1 inflammation and that this enhanced Tim3 activity is associated with decreased phosphorylation of the TCR-CD3ζ chain. We then show that mice with this Tim3 mutation infected with influenza are protected from morbidity and mortality without impairment in viral clearance or functional heterotypic immunity. This protection is associated with decreased CD8+ T cell proliferation and decreased production of inflammatory cytokines, including IFNγ. Furthermore, the Tim3 mutation was protective against mortality in a CD8+ T cell-specific model of pneumonitis. These data suggest that Tim3 could be targeted to prevent immunopathology during influenza infection and demonstrate a potentially novel signaling mechanism utilized by Tim3 to down-regulate the Tc1 response.
INTRODUCTION
Influenza causes a highly contagious respiratory disease among humans (1) and is a major cause of morbidity and mortality, accounting for up to 150,000 hospitalizations and 20,000 deaths in the United States annually (2, 3). Accumulating data suggests that excessive T cell activity can mediate pneumonitis in the setting of influenza infection (4–7). Indeed, recent data from the 2009 H1N1 pandemic indicate that critical illness and respiratory failure following infection was associated with higher circulating levels of cytokines including TNF, IL-6 and IFNγ, and greater infiltration of the lungs with CD8+ T cells (8–11). Additionally, cytokine levels correlated positively with severity of illness scores (8). In this situation, inhibition of T cell activity leads to mitigation of the lung inflammation (12–14). However, it also can lead to profound immunosuppression, which in the case of viral pneumonitis can reduce viral clearance (12–15). It follows that a therapeutic strategy that could control excessive T cell-mediated injury without significant immunosuppression or impairment in the ability to clear infections would be an ideal treatment for severe influenza pneumonitis.
T cell immunoglobulin and mucin domain 3 (Tim3) is a type-1 transmembrane receptor with immunoregulatory properties on effector T cells and antigen presenting cells (APCs) (16). Tim3 is expressed on Th1 and Tc1 cells and has been shown to be a negative regulator of the Th1/Tc1 response (17–19). On these cells, activation of Tim3 via ligand binding results in decreased T cell proliferation (20), decreased T cell-mediated cytotoxicity (21, 22), decreased IFNγ production (23, 24) and induction of apoptosis (23). However, Tim3 is also expressed on natural killer (NK) cells and APCs, and on APCs activation of Tim3 has a pro-inflammatory role (25). One of the Tim3 ligands has been identified as galectin-9 (gal-9) (23), a β-galactoside-binding lectin that is induced by IFNγ and has a number of immunoregulatory functions (26–28). Binding of Tim3 by gal-9 has been shown to result in phosphorylation of a highly conserved tyrosine residue (human Y265; murine Y256) (29), but the overall mechanisms by which Tim3 signals are relatively undefined. Furthermore, there are multiple other tyrosine residues in the Tim3 cytoplasmic domain with relatively unknown signaling functions (30). Overall, these data suggest that the signaling pathways of Tim3 are highly complex and likely depend on the cell type on which it is expressed.
Animal models of immunoinflammatory disease including experimental autoimmune encephalitis (17, 19, 23, 31), experimental autoimmune arthritis (32) and transplantation (33, 34) have suggested that the predominant effect of Tim3 activation in vivo is anti-inflammatory. Studies suggest that Tim3 plays a similar role during acute viral infection of the eye, by limiting the antigen-specific CD8+ effector T cell response and thereby immune-mediated bystander injury (21, 22). We therefore hypothesized that Tim3 would play an important role in regulating the immune response to influenza infection. In an effort to explore both the functional role of Tim3 in influenza pathogenesis as well as mechanisms of Tim3 signaling, we generated a Tim3 mutant mouse (Tim3mut) with preservation of a highly conserved tyrosine kinase motif (Y256) and deletion of a portion of the remaining distal cytoplasmic domain. Here, we demonstrate that this deletion enhances the negative regulatory activity of Tim3 on CD8+ T cells and is associated with decreased phosphorylation of CD3ζ, a potentially novel mechanism for the inhibitory activity of Tim3. Furthermore, the enhanced ability of Tim3 to down-regulate the Tc1 response in Tim3mut mice is associated with reduced morbidity and mortality during influenza infection. Importantly, mutation of Tim3 had no adverse effect on viral clearance or the development of cellular immunity.
MATERIALS AND METHODS
Generation of Tim3 Mutant Mice
Mice with a deletion of the terminal cytoplasmic domain of the Tim3 gene were generated using modified bacterial artificial chromosome (BAC) technology as previously described (35). Briefly, BAC clones spanning the Tim3 locus were obtained from Research Genetics (Invitrogen Life Technologies, Carlsbad, CA). Two sequences flanking exon 7 were cloned into the 5’ and 3’ insertion sites of the selection cassette of the pSKY replacement vector. BAC host cells were transformed with the pBADλredαβ plasmid, which helped produce electroporation-competent cells. The linear fragment released from the pSKY backbone was then electroporated into the BAC host, and the transformants were selected for simultaneous resistance to chloramphenicol (from the BAC backbone) and zeomycin (from the insert). After selection, homologous recombinants were identified by PCR. Tim3.5/Pzeo primers identify a predicted 440-bp PCR product in the mutant BAC confirming 5’ targeting. To confirm 3’ targeting, an additional primer Tim3.2 was included in the PCR. As predicted, the PSV/Tim3.2 primers identify a 490-bp PCR product in the mutant BAC and 326-bp product in the wild-type (WT) BAC. The modified BAC was electroporated into embryonic stem cells and fluorescence in situ hybridization was performed as described previously (35), confirming successful targeting in clones B9, C6 and D8. Mice from clones B9 and C6 were born in the expected Mendelian frequency and were healthy. Founder mice were crossed with C57BL/6 mice for 10 generations.
Mice
WT C57BL/6 mice were purchased from National Cancer Institute (Rockville, MD, USA). The OT-I and OT-II TCR transgenic mice in the C57BL/6 background are bred and maintained in our facility. Additionally, OT-I and OT-II mice in the C57BL/6 background were crossed with Tim3mut mice. CC10-OVA mice in the C57BL/6 background were generated as previously described (36), bred and maintained in our facility. Mice were used at 6–8 weeks of age and were sex matched for all experiments. All protocols were approved by the Massachusetts General Hospital Subcommittee on Research and Animal Care.
Flow Cytometry
Single cell suspensions of spleen, blood, lymph nodes, BAL fluid and lung were prepared and red blood cells were lysed. To extract leukocytes from lung tissue, lung lobes were removed, minced with scissors, and then digested for 45 minutes in RPMI with 0.28 Wunsh U/ml Liberase (Roche Applied Science, Indianapolis, IN) and DNase 30 U/ml (Sigma-Aldrich, St. Louis, MO) at 37°C. The digested tissues were then strained through a 70 µm filter prior to red blood cell lysis. Samples were blocked with purified CD16/CD32 monoclonal antibody (BD Biosciences, San Diego, CA) and then stained with fluorescently labeled antibodies to CD4, CD8, CD25, CD69 and Tim3 (BD Biosciences except for Tim3, which was obtained from R&D Systems, Minneapolis, MN). Some samples were also stained with fluorescently labeled tetramer to influenza nucleoprotein (366–374) (NIH Tetramer Facility, Atlanta, GA). For the apoptosis experiments, some cells were treated with 10 µM full-length recombinant gal-9 (Novus Biologicals, Littleton, CO) for 6 hours. Cells were then stained with Annexin V and propidium iodine (BD Biosciences). Flow cytometry was performed on an Accuri C6 analytical flow cytometer (Accuri® Cytometers, Ann Arbor, MI) and analyzed using FCS Express software (DeNovo™ Software, Los Angeles, CA).
OT-I and OT-II T Cell Preparation and Adoptive Transfer
Isolation and preparation of OT-I and OT-II T cells were performed as described previously (36, 37). Briefly, spleens and lymph nodes were harvested, single cell suspensions were prepared and CD8+ or CD4+ T cells were isolated using an antibody-mediated magnetic negative selection kit (EasySep® Mouse T Cell Enrichment Kit, StemCell Technologies, Vancouver, BC). Effector OT-I T cells were prepared by culturing purified CD8+ OT-I T cells with irradiated APCs from spleens of C57BL/6 or Tim3mut mice with 700 ng/ml SIINFEKL peptide, 2 µg/ml anti-CD28, 10 ng/ml recombinant IL-2, and 10 ng/ml recombinant IL-12. After 5 days of culture, CD8+ T cells were purified using gradient centrifugation. For the CC10-OVA adoptive transfer experiments, 2 × 105 effector CD8+ T cells were resuspended in sterile PBS and injected i.p. OT-II T cells were polarized to Th1 cells by co-culturing with irradiated APCs with 100 ng/ml OVA peptide, 1 µg/ml anti-CD28, 1 ng/ml IL-12 and 10 µg/ml anti-IL-4. In order to achieve maximal Tim3 expression, CD4+ T cells were re-stimulated with APCs and antigen on day 7 and purified on day 10.
Proliferation Assays
For in vitro proliferation studies, purified CD8+ and CD4+ T cells were labeled with CFSE (Invitrogen) and incubated for 48 to 72 hours. Proliferation was assessed by CFSE dilution on flow cytometry. In vivo proliferation studies were performed by injecting 2 mg BrdU (BD Pharmingen) i.p. into mice at day 6 after influenza infection. Tissues were collected twenty-four hours later and single cell suspensions were prepared. BrdU labeling was performed according to the manufacturer’s protocol and samples were analyzed by flow cytometry.
Cytotoxicity Assay
Purified effector OT-I T cells were co-incubated with antigen-loaded EL4 target cells. OT-I mediated cytotoxicity was measured using a commercially available bioluminescence kit (aCella Tox, Cell Technology, Mountain View, CA) according the manufacturer’s protocol.
Western Blot and Immunoprecipitation
Some cells were treated with 10 µM gal-9 for 2 or 5 minutes. Cells were lysed using ice cold NP40 lysis buffer (Invitrogen) containing a protease inhibitor cocktail (Complete Mini, Roche Applied Science) and a phosphatase inhibitor cocktail (Calbiochem, La Jolla, CA). Protein was quantified using a commercially available system (Pierce BCA Protein Assay Kit, Thermo Scientific, Rockford, IL) according to the manufacturer’s protocol. Equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane. The blot was incubated with an anti-phosphotyrosine antibody (clone 4G10, Millipore, Billerica, MA) and visualized by the ECL detection system (Amersham Pharmacia, GE Healthcare, Pittsburgh, PA). For immunoprecipitation (IP), cells were lysed, protein was quantified as above and equal amounts of protein were subjected to IP using an anti-CD3ζ antibody (Abcam, Cambridge, MA) and a commercially available kit (Pierce Co-Immunoprecipitation Kit, Thermo Scientific) according to the manufacturer’s protocol. Eluted protein was then subject to Western blotting as above using an anti-phosphotyrosine antibody.
Virus and Infections
Influenza A/Puerto Rico/8/34 (PR8) was obtained from the American Type Culture collection (ATCC; VR-1469, Manassas, VA, USA) and grown in Madin Darby canine kidney cells (MDCK) in our laboratory. Influenza A/Hong Kong/8/68- ×31 (×31) was provided by Dr. Troy Randall (Trudeau Institute) and grown in embryonated chicken eggs in our laboratory. The recombinant virus influenza A/WSN/33 OVAI (WSN-OVAI) was generated and kindly provided by Dr. David Topham (University of Rochester) (38). Viral pool titers were measured by MDCK plaque assay and appropriate infecting doses were determined by in vivo titration. Mice were infected with a dose of influenza PR8 that leads to about 80% mortality (lethal dose 80 or LD80; 103 PFU) at 6–8 weeks of age. For heterotypic memory experiments, mice were infected with a non-lethal dose of ×31 virus (5 × 102 PFU) and allowed to recover for 4 weeks. They were then infected with 3-fold the LD80 of PR8 (3 × 103 PFU) and followed for 28 days. For assessment of antigen-specific IFNγ production, mice were infected with a non-lethal dose of WSN-OVAI virus (200 PFU). Viruses were diluted to the appropriate dose using sterile PBS. Mice were anesthetized with ketamine (80 mg/kg)-xylazine (12 mg/kg) and 30 µl of virus was administered intranasally.
Histopathology
Mice were sacrificed at day 3 or 7 post-infection. Lung lobes were inflated with 10% buffered formalin and then placed in formalin. Paraffin-embedded 4 µm sections were prepared and stained with H&E. Histology was evaluated for severity by an investigator blinded to the genotype of the mice.
BAL Fluid Analysis
Mice were sacrificed at day 3 or 7 post-infection. BAL fluid was obtained by infusing six 0.5 ml washes of cold PBS with 0.12% 2mm EDTA intra-tracheally. Differential cell counts were obtained after spinning 1.5 × 105 cells onto slides and staining with Hema-3 (Fisher Scientific, Pittsburgh, PA). Differential counts were performed on at least 200 cells per slide. Cells were also analyzed by flow cytometry as above. Cytokine protein levels were analyzed using a commercially available bead-array (Procarta Cytokine Assay, Affymetrix, Santa Clara, CA) according to the manufacturer’s protocol.
Quantitative Real-Time PCR
RNA was purified using either chloroform extraction or a purification column (RNeasy; Qiagen, Valencia, CA). After a DNase step, 1 µg of RNA was converted to cDNA (Applied Biosystems, Carlsbad, CA). Specific primers for sequence detection of message for the Tim3 extracellular domain were forward primer 5’- CAGAAATCCAGCAGATACCAGCT-3’; reverse primer 5’-AGGTCCCATGGTCATCCAGA-3’; for the Tim3 cytoplasmic domain forward primer 5’-GAGGAAAATATCTACACCATCGAGGAGAAC-3’; reverse primer, 5’-CAGAAATGAAGGCGAGCCTTTAAAAGTG-3’. The primers for specific cytokines were selected from the Massachusetts General Hospital Primer Bank (http://pga.mgh.harvard.edu/primerbank/). Samples underwent amplification in the presence of SYBR Green (Applied Biosystems). The reaction was analyzed in real-time during amplification by the PCR machine (Mastercycler EP Realplex, Eppendorf, Hauppage, NY).
Intracellular Staining
Antigen-Specific
EL4 cells were pulsed with 700 ng/ml SIINFEKL peptide and then co-incubated for 24 hours with CD8+ T cells isolated from the lungs of mice 7 days after infection with WSN-OVA virus. The cells were then exposed to GolgiStop (BD Biosciences) to block cytokine secretion, fixed and permeabilized using a commercially available kit (BD Biosciences) and stained for IFNγ.
PMA / Ionomycin
CD8+ T cells were isolated from the lungs of mice 7 days after infection with PR8. They were stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml), exposed to GolgiStop, fixed, permeabilized and stained as above. Results were analyzed by flow cytometry.
Viral Titers
Infectious viral titers were determined using a modified MDCK cell assay as previously described (39, 40). Briefly, lungs were harvested on day 3 or 7 after influenza infection, homogenized, serially diluted and added to duplicate wells containing confluent monolayers of MDCK cells for 12 hours. The cells were then immunostained for surface expression of hemagglutinin and quantitatively imaged. In addition, the number of viral RNA copies per lung was determined by quantitative RT-PCR. RNA was prepared from whole lung homogenates at days 3, 7, 10 and 14 using TRIzol (Sigma-Aldrich), and 1 µg of RNA was converted to cDNA (Applied Biosystems). Quantitative PCR was performed as above to amplify the polymerase gene of the PR8 influenza virus using the following primers: forward primer 5’-CGGTCCAAATTCCTGCTGA-3’; reverse primer 5’-CATTGGGTTCCTTCCATCCA-3’.
RESULTS
Generation of a Tim3 mutant mouse
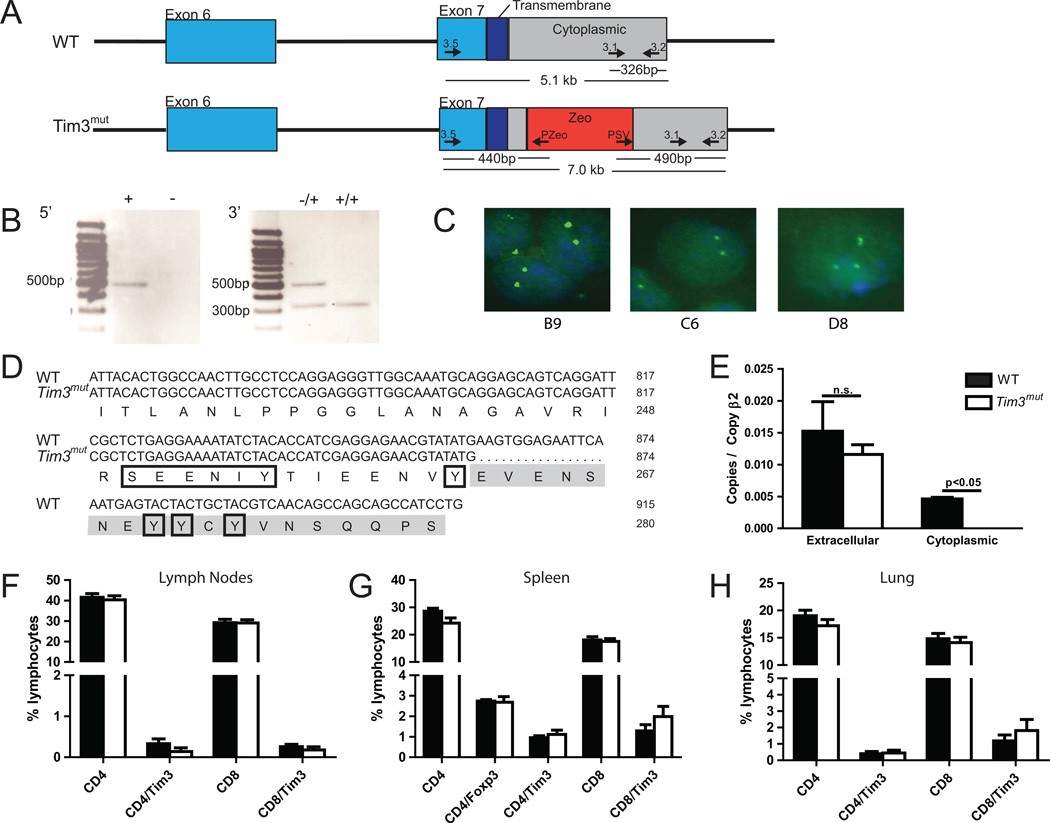
In order to investigate mechanisms of Tim3 signaling, we generated a BAC construct with disruption in the terminal portion of the cytoplasmic domain (exon 7) of the Tim3 gene by the zeomycin-resistance gene. The resulting mutant BAC is shown in Figure 1A. Recombinants were identified by PCR (Fig. 1B) and the construct was injected into embryonic stem cell clones. Targeting was confirmed by FISH analysis of several embryonic stem cell clones (Fig. 1C), which were used to generate Tim3mut mice. We performed sequence analysis of the cytoplasmic domain on genomic DNA. As shown in Fig. 1D, the mutation resulted in deletion of amino acids 263–280 while preserving two proximal tyrosine amino acids in the cytoplasmic tail. One of the tyrosines has previously been shown to be phosphorylated by the tyrosine kinase Itk after gal-9 binding (29), and both of the proximal tyrosines can be phosphorylated by the Src family tyrosine kinases Lck and Fyn (30). However, the signaling mechanisms of the deleted tyrosine residues are unknown. We further confirmed the deletion by RT-PCR analysis of splenocytes from Tim3mut mice using primers directed at either the extracellular domain or the deleted portion of the cytoplasmic domain. This demonstrated normal expression of RNA encoding the extracellular domain while we were unable to detect RNA encoding the targeted cytoplasmic domain (Fig. 1E).
Figure 1. Generation of a Tim3mut mouse.
A – Construct targeting the distal cytoplasmic tail of Tim3. Exon 7 of Tim3 is disrupted with the zeomycin (Zeo) gene. Primers used for checking the 5’ and 3’ integration sites are shown. B – PCR confirmation of Tim3 targeting. In the 5’ PCR reaction, correct integration results in a 440 bp fragment in the Tim3mut sequence. In the 3’ PCR reaction, the WT sequence results in a 326 bp fragment while correct integration results in a 490 bp fragment. C – FISH analysis of three embryonic stem cell clones confirms Tim3 targeting. D – Murine WT and Tim3mut sequence of exon 7 shown with the disrupted region highlighted in gray. Tyrosine residues predicted to be important in signaling are boxed. E – Tim3 RNA expression in splenocytes isolated from WT and Tim3mut mice. QPCR primers targeted the extracellular and disrupted cytoplasmic domains (n=2 mice per group; p<0.05 by Student’s t test; experiment was repeated with similar results). F – Lymph nodes, G – spleen and H – lungs were harvested from naïve 6–8 week old WT and Tim3mut mice and T cell populations were analyzed by flow cytometry (n=6 mice per group).
Tim3mut mice were viable, fertile and grossly normal in size and appearance. Analysis of lymph nodes, spleen and lung from 6–8 week old mice demonstrated normal percentages of CD4+, CD4+Foxp3+ and CD8+ T cells compared to WT mice. Additionally, there were no differences in Tim3 expression on naïve CD4+ or CD8+ T cells isolated from these mice (Fig. 1F–H).
Deletion of the terminal cytoplasmic domain potentiates the ability of Tim3 to down-regulate Tc1 cells
In order to study the functional effects of the Tim3 mutation in vitro, we crossed Tim3mut mice with OT-I and OT-II mice, which are transgenic for a TCR specific for an ovalbumin (OVA) peptide bound to class I and class II MHC, respectively. First, we assessed whether surface expression of the extracellular domain of Tim3 was normally up-regulated on CD8+ and CD4+ T cells from Tim3mut mice following activation. CD8+ and CD4+ T cells were isolated from WT and Tim3mut OT-I and OT-II mice and stimulated in vitro into Tc1 and Th1 cells using APCs and specific antigen. As expected, very few naïve T cells from WT and Tim3mut mice expressed Tim3. OT-II T cells required two rounds of in vitro polarization for maximal up-regulation of Tim3 expression. We found no significant differences in the surface expression of the extracellular domain of the mutant form of Tim3 in either CD8+ or CD4+ T cells compared to WT (Fig. 2A and 2B).
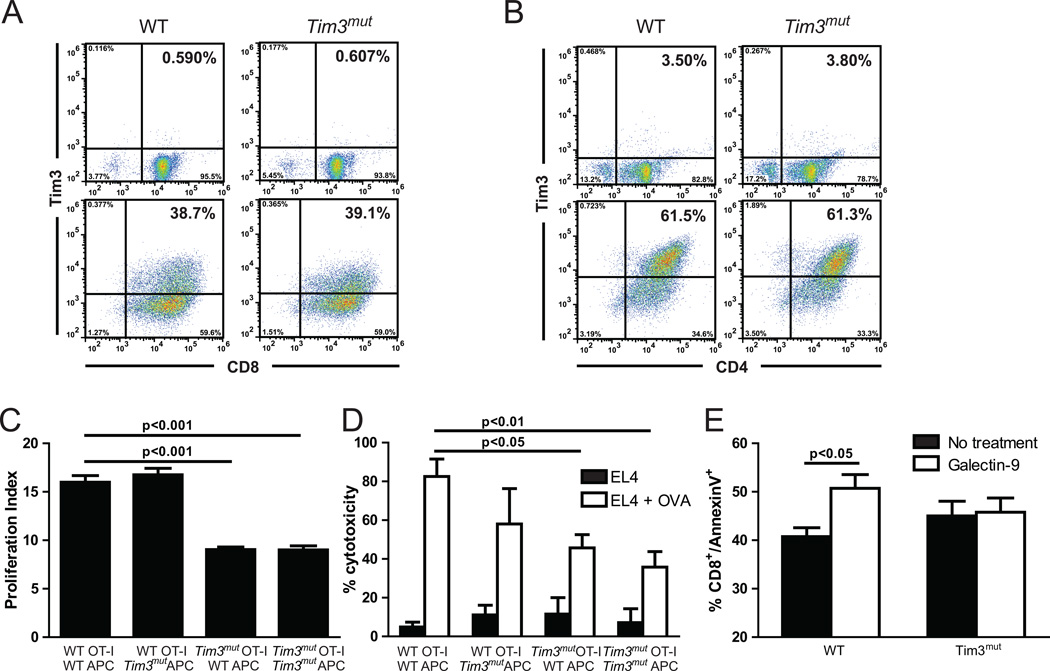
Figure 2. Deletion of the terminal cytoplasmic domain potentiates the ability of Tim3 to down-regulate Tc1 lymphocytes.
WT and Tim3mut OT-I and OT-II cells were stimulated in vitro and purified at day 5 or 10, respectively. Representative flow cytometry data of Tim3 surface expression on WT and Tim3mut A – OT-I CD8+ T cells at days 0 (upper plots) and 5 (lower plots) and B – OT-II CD4+ T cells at days 0 (upper plots) and 10 (lower plots). C – Activated effector CD8+ T cell proliferation 72 hours after purification assessed by CFSE dilution (2 independent experiments; p< 0.05 by 2way ANOVA). D – CD8+ T cell-mediated cytotoxicity against antigen-loaded target cells or unloaded cells as a control assessed using a bioluminescent assay (2 independent experiments; p<0.05 and p<0.01 by 2way ANOVA). E – Assessment of apoptosis by Annexin V+ staining before and after stimulation with 10 µM gal-9 for six hours (one experiment; p<0.05 by Student’s t test).
Activation of Tim3 on T cells has been shown to suppress effector T cell functions such as proliferation and cytotoxicity. To assess the functional effects of this Tim3 mutation, we first compared proliferation of Tim3mut OT-I to WT OT-I T cells by CFSE dilution. Because Tim3 is also expressed on APCs, we stimulated the T cells with either WT or Tim3mut APCs. WT and Tim3mut naïve CD8+ and CD4+ T cells, which express little to no Tim3, had normal proliferation and no differences were observed (data not shown). However, when we examined proliferation of activated effector CD8+ T cells at the time of peak Tim3 expression (day 5), there was a marked reduction in Tim3mut CD8+ T cell proliferation that was independent of Tim3 expression on APCs (Fig.2C). No differences were observed in CD4+ T cell proliferation in similar experiments (data not shown). We next assessed CD8+ T cell-mediated cytotoxicity by stimulating WT and Tim3mut OT-I T cells in vitro into effector T cells. CD8+ T cells were purified at day 5, co-incubated with OVA loaded EL4 target cells or unloaded cells, and cytotoxicity was assessed using a bioluminescent assay. This demonstrated a significant reduction in CD8+ T cell-mediated cytotoxicity by Tim3mut T cells when compared to WT T cells that was again independent of Tim3 expression on APCs (Fig. 2D). Taken together, these data suggest that deletion of the terminal cytoplasmic domain potentiates the ability of Tim3 to down-regulate proliferation and cytotoxicity of Tc1 cells. This activity is independent of Tim3 expression on APCs and is not replicated in Th1 cells.
Deletion of the terminal cytoplasmic domain eliminates gal-9 mediated CD8+ T cell apoptosis
The activation and clonal expansion of T cells is followed by a death phase, during which the majority of effector T cells are eliminated (41). In addition to its inhibitory effects on proliferation and cytotoxicity, Tim3 has been demonstrated to mediate T cell apoptosis (21). We therefore assessed whether there were differences in gal-9 mediated apoptosis by activating CD8+ T cells in vitro, incubating them alone or with 10 µM gal-9 for 6 hours, then staining with Annexin V and propidium iodide. There were no differences in apoptosis at baseline between WT and Tim3mut CD8+ T cells. However, while gal-9 was able to induce apoptosis on WT T cells, it had no effect on Tim3mut T cells (Fig. 2E). These studies indicate that deletion of the terminal cytoplasmic domain results in loss of gal-9 mediated apoptosis and that the negative regulatory and apoptotic functions of Tim3 may be differentially regulated.
Enhanced negative regulation of the Tc1 response is associated with decreased phosphorylation of the TCR-CD3ζ chain
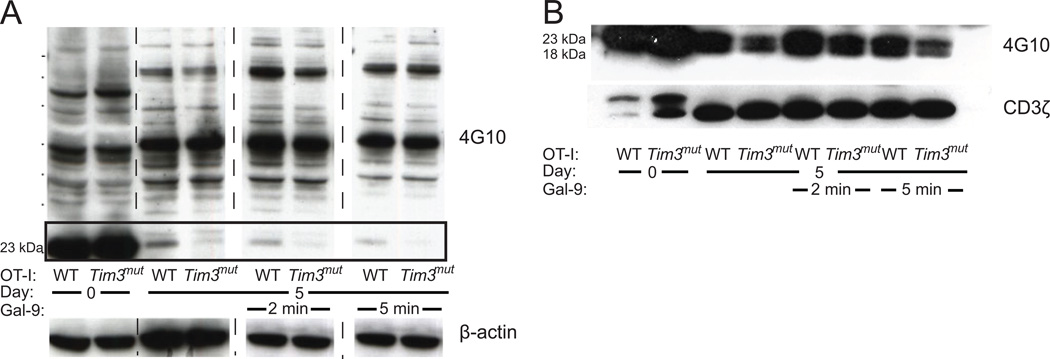
In order to confirm that the Tim3 mutation had altered signaling in CD8+ T cells, we assessed tyrosine phosphorylation by Western blot. WT and Tim3mut OT-I T cells were isolated, stimulated in vitro with WT APCs and purified on day 5. The CD8+ T cells were then incubated alone or with 10 µM gal-9 for 2 or 5 minutes. Protein lysates were blotted and probed with an anti-phosphotyrosine antibody (4G10). As expected there were no differences between naïve WT and Tim3mut CD8+ T cells at baseline, when Tim3 is expressed on very few T cells. However, at day 5 after activation there was appreciably decreased phosphorylation of a 23 kDa protein in Tim3mut T cells (Fig.3A). Phosphorylation of this protein further decreased after 2 and 5 minutes of gal-9 stimulation. Similar experiments in WT and Tim3mut CD4+ T cells failed to demonstrate any appreciable differences in phosphorylation (data not shown).
Figure 3. Mutation of Tim3 results in decreased phosphorylation of the TCR-CD3ζ chain.
WT and Tim3mut OT-I cells were stimulated in vitro into effector cells and purified at day 5. Some samples were treated with 10 µM gal-9 for 2 or 5 minutes and protein lysates were prepared. A - Anti-phosphotyrosine (4G10) Western blot. Phosphorylation differences can be seen in a 23 kDa protein. β-actin is shown as a loading control. (Representative blots from 2 independent experiments). B – Lysates were immunoprecipitated using an anti-CD3ζ antibody, blotted and probed with 4G10. Phosphorylation differences can be seen in the 18 and 23 kDa bands. Total CD3ζ is shown as a loading control. (Representative blots from two independent experiments).
Based on the 23 kDa size of the differentially phosphorylated protein, we hypothesized that this might represent the TCR-CD3ζ chain. Inhibition of CD3ζ phosphorylation has been shown to reduce cytokine production and proliferation in T cells (42). In order to test if the identified protein was CD3ζ, we repeated the experiment and then used an antibody to CD3ζ to immunoprecipitate (IP) the protein from cellular extracts. Precipitated protein was blotted and probed with 4G10. Phosphorylated CD3ζ typically results in the presence of two bands between 18 and 23 kDa. The IP increased the sensitivity of the detection, allowing both phosphorylated forms of CD3ζ to be visualized. Loading was assessed by re-probing the blot with anti-CD3ζ antibody. These results clearly demonstrate that there is less CD3ζ phosphorylation in the activated Tim3mut OT-I T cells compared to activated WT OT-I T cells both before and after gal-9 stimulation (Fig. 3B). These findings confirm that signaling differences are induced by the Tim3 mutation. In addition, given the functional role of CD3ζ phosphorylation for T cell effector functions, these data suggest a novel mechanism for the negative regulatory activity of Tim3 on CD8+ effector T cells.
Tim3 expression is up-regulated on T cells recruited to the lung after influenza infection
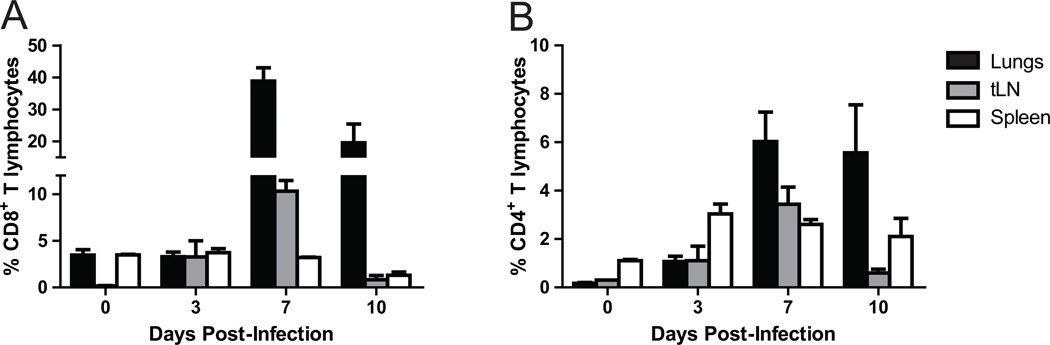
In order to explore whether Tim3 plays an important role in regulating the adaptive immune response during influenza infection, we infected WT mice with a dose of influenza A/Puerto Rico/8/34 (PR8) that is lethal in about 80% (LD80) and determined Tim3 expression on CD8+ and CD4+ T cells in the lung, draining thoracic lymph node (tLN) and spleen at various time points. During influenza infection, priming of naïve virus-specific T cells occurs within 72 hours. Priming is followed by sustained proliferation and accumulation of large numbers of virus-specific effector T cells that traffic to the airways and lung parenchyma at day 6–7 post-infection (43–45). These cells continue to accumulate from days 7 to 10 post-infection, resulting in rapid clearance of virus (46, 47). In naïve animals, very few T cells express Tim3. Up-regulation of Tim3 expression is seen on T cells in both the tLN and lung by day 3 post-infection and peaks at day 7. Levels of Tim3 expression are highest on CD8+ T cells in the lung, with nearly 40% of these cells expressing Tim3 at day 7 (Fig. 4 and Fig. S1). These results demonstrate significant up-regulation of Tim3 on CD8+ T cells at the site of inflammation, suggesting that Tim3 may play an important role in modulating the effector functions of these cells during influenza infection.
Figure 4. Tim3 expression is up-regulated on cells during influenza infection.
WT mice were infected with influenza PR8 and tissues collected at various time points. Percentages of Tim3 expressing A – CD8+ T cells and B – CD4+ T cells in the lungs, tLN and spleen at days 0, 3, 7 and 10 following influenza infection (n=3 mice per group per time point).
Tim3mut mice are protected from morbidity and mortality during influenza infection
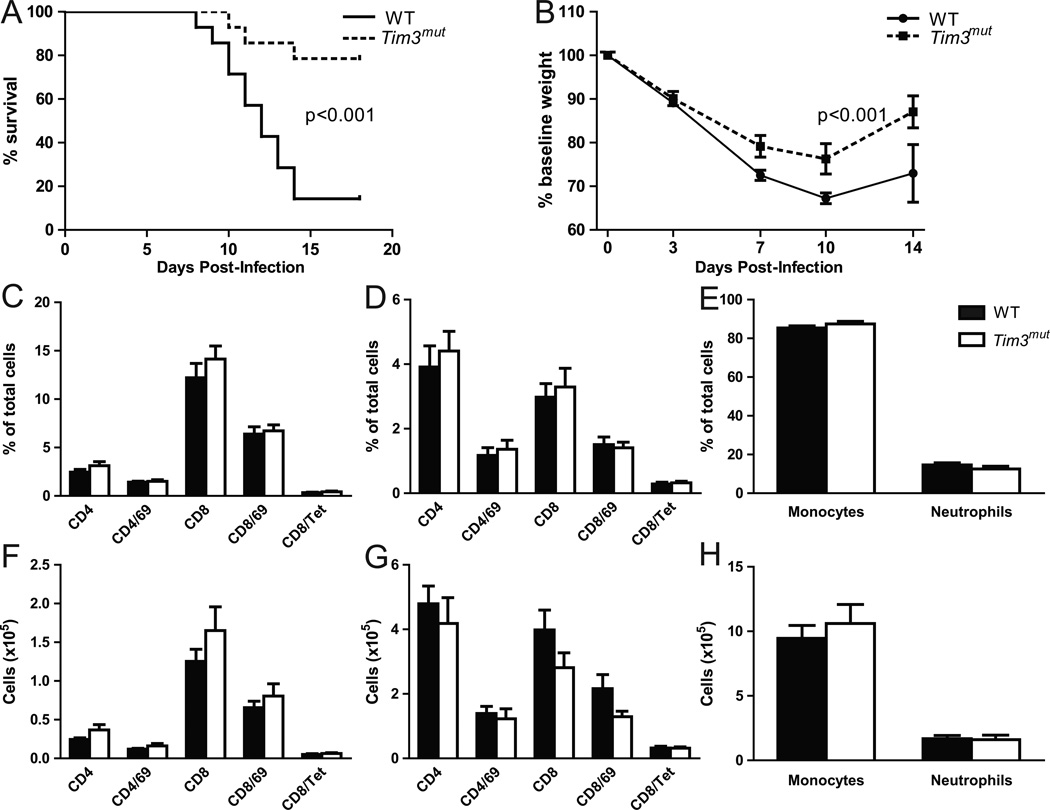
We next investigated the role of the Tim3 mutation in vivo by infecting WT and Tim3mut mice with LD80 influenza PR8. In this model, morbidity and mortality is dependent on immune mediated injury to the lung manifest as a severe pneumonitis, and prior work has suggested that CD8+ effector T cells are one of the critical mediators of this immunopathology (6, 48, 49). Mice were followed for weight loss and survival for 28 days. Mortality in Tim3mut mice was significantly reduced compared to WT mice (Fig. 5A). In addition, Tim3mut mice lost significantly less weight than their WT counterparts (Fig 5B). However, we found no significant differences in immunohistopathology on lung sections taken at day 3 or day 7 after PR8 infection (Fig. S2).
Figure 5. Tim3mut mice are protected from mortality and morbidity during influenza infection.
A – Survival of WT and Tim3mut mice following infection with LD80 PR8 (n=14 mice per group from 3 independent experiments; p<0.001 by log-rank test). B – Weight loss as a percentage of original weight during influenza infection (n=14 mice per group from 3 independent experiments; p< 0.001 by 2way ANOVA). Percentages and absolute numbers of cells in the BAL (C and F, respectively), lungs (D and G) and cytospin preparations (E and H) at day 7 following influenza infection (n=12 mice per group from 3 independent experiments). Tet is influenza nucleoprotein (366–374)-specific tetramer.
We therefore assessed the cellular infiltrate to the airways and lungs following PR8 infection. Analysis of BAL fluid and lung tissue at days 3 and 7 post-infection demonstrated no differences in mononuclear, neutrophilic or T cell infiltration of the respiratory system, including the number of antigen-specific CD8+ T cells as assessed by staining with class I tetramer for influenza nucleoprotein (366–374) (Figs. 5C–H, S3 and S4). We also did not detect differences in the number of NK cells in BAL fluid or lung tissue at day 3 following infection (data not shown).
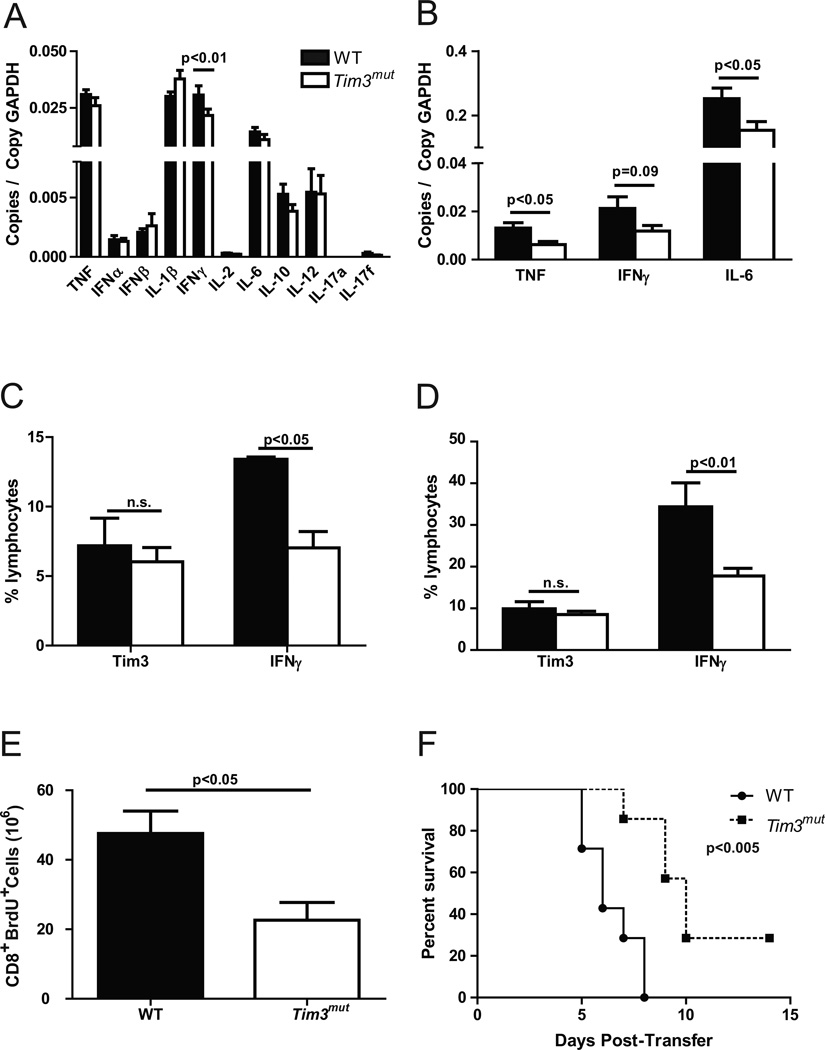
Both human and mouse studies have demonstrated that the magnitude of cytokines produced post-influenza infection may determine the extent of immunopathology, so we next assessed the levels and expression of inflammatory cytokines. Although we found no significant differences between WT and Tim3mut mice at day 3 after infection (Fig S3), at day 7 there was a trend toward lower expression of inflammatory cytokines in whole lung homogenates from Tim3mut mice, with significantly less IFNγ expression in Tim3mut mice after correction for multiple comparisons (p<0.01) (Fig. 6A). However, despite differences in cytokine RNA levels we were unable to detect differences in cytokine protein levels in BAL fluid from WT and Tim3mut mice at day 3 or day 7 after infection (data not shown). Discrepancies between RNA levels of the cytokines versus protein levels may relate to compartmentalization of protein secretion to different lung structures (i.e. airway versus interstitium), accumulation of protein in the lung, or the sensitivity of the protein assay. These data demonstrate that deletion of the terminal cytoplasmic domain of Tim3 is protective against morbidity and mortality during infection with influenza A and that this protection is associated with decreased IFNγ RNA expression.
Figure 6. Tim3mut CD8+ T cells express lower levels of inflammatory cytokines during influenza infection.
Cytokine expression by QPCR in A - whole lung homogenates (n=12 mice per group from 3 independent experiments; p<0.01 by 2way ANOVA after correction for multiple comparisons) and B – CD8+ T cells isolated from lungs (n=6 mice per group from one experiment; p<0.05, p=0.09 and p<0.05 by Student’s t test) of WT and Tim3mut mice at day 7 after influenza infection. Intracellular staining for IFNγ production by CD8+ T cells isolated from lungs at day 7 C - after infection WSN-OVAI and stimulation with OVA peptide (n=3 mice per group, p<0.05 by 2way ANOVA) and D - after infection with PR8 and stimulation with PMA and ionomycin (n=5 mice per group; p<0.01 by 2way ANOVA after correction for multiple comparisons; experiment was repeated with similar results). E – In vivo proliferation assessed at day 7. Numbers of CD8+Brdu+ T cells from whole lung homogenates are shown (two independent experiments, n=4 mice per group, p<0.05 by student’s t test). F – Survival of CC10-OVA mice after adoptive transfer of activated effector WT or Tim3mut OT-I cells (n=7 mice per group from two independent experiments, p<0.005 by log-rank test).
Tim3mut CD8+ T cells have impaired effector functions
Because Tim3 is expressed on multiple cells types that play an important role in the immune response to influenza, we wanted to further characterize the CD8+ T cell-specific effects. Since CD8+ and CD4+ T cells are the major sources of most of the key inflammatory cytokines, we isolated these cells from lung tissue at day 7 post-infection. CD8+ T cells from Tim3mut mice expressed lower RNA levels of TNF, IFNγ and IL-6 (Fig. 6B). However, no differences in cytokine expression were observed in CD4+ T cells (data not shown). We further investigated these findings by isolating CD8+ T cells from lung tissue at day 7 post-infection and performing intracellular staining for IFNγ. For these experiments, some mice were infected with a recombinant influenza A virus expressing OVA protein (WSN-OVAI), while others were infected with PR8. Using antigen-specific stimulation of CD8+ T cells isolated from the lungs of mice infected with WSN-OVAI and PMA and ionomycin stimulation of CD8+ T cells isolated from the lungs of mice infected with PR8, we were able to confirm lower levels of IFNγ production in Tim3mut mice (Fig. 6C and Fig. S4). Next, we examined in vivo CD8+ T cell proliferation in WT and Tim3mut mice. Mice were injected with BrdU at day 6 following influenza infection. Twenty-four hours later, lungs were collected, BrdU was labeled and proliferation was assessed by flow cytometry. Consistent with our in vitro data, we found fewer BrdU+ CD8+ T cells in the lungs of Tim3mut mice compared to WT (Fig. 6D and S4). It is important to note that since mice were injected with BrdU at day 6 post infection, these experiments provide us with only a snapshot of effector T cell proliferation at the time of maximal Tim3 expression. Therefore, despite differences in the number of proliferating cells, we may not see differences in total cell numbers. Finally, in order to confirm that Tim3mut CD8+ T cells have impaired ability to mediate pneumonitis, we utilized a CD8+ T cell specific model of severe lung injury. In this model, activated OT-I cells are adoptively transferred into a transgenic mouse that expresses membrane bound OVA in airway lining cells (CC10-OVA) (36, 50). Following transfer, OT-I cells are recruited into the lung where they proliferate and mediate lung injury leading to severe pneumonitis and death. OT-I cells were isolated from WT and Tim3mut mice, activated into effector cells in vitro, and adoptively transferred into CC10-OVA mice by i.p. injection. The adoptively transferred OT-I cells are the only cell type affected by the Tim3 mutation, allowing us to specifically assess the role of the Tim3 mutation in regulating CD8+ T cell effector functions. Effector activity of CD8+ T cells is correlated with mortality in this model (36, 50). As shown in Figure 6E, CC10-OVA mice that received Tim3mut OT-I cells had improved survival compared to mice that received WT OT-I cells, consistent with enhancement of the negative regulatory activity of Tim3 on these cells. Taken together, these data demonstrate that Tim3mut CD8+ T cells have decreased production of inflammatory cytokines, decreased proliferation and decreased effector function compared to WT cells.
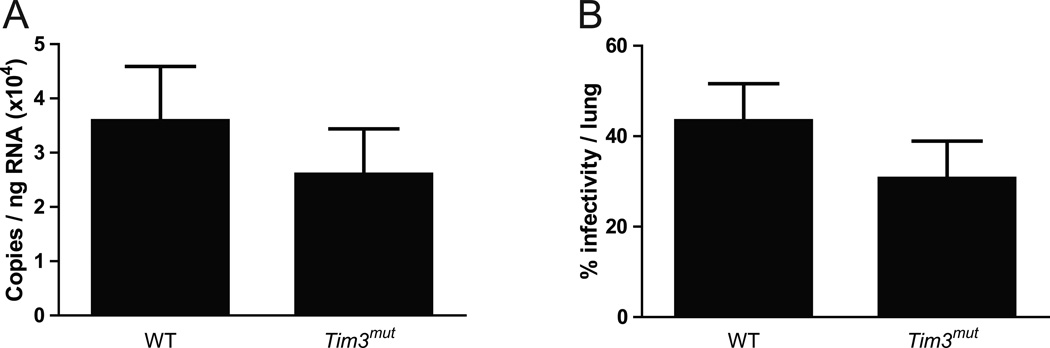
Tim3mut mice have normal viral clearance
We postulated that the reduced cytokine production and cytotoxicity of Tim3mut CD8+ T cells might lead to impaired viral clearance. In order to test this, lungs were collected from WT and Tim3mut mice following PR8 infection. Using QPCR to measure viral copies and a previously described MDCK infectivity assay to assess viral titer (39, 40), we found no differences in viral clearance between WT and Tim3mut mice (Fig. 7 and Fig. S3). These data demonstrate that despite the alteration of T cell effector function, there is no impairment in the ability of Tim3mut mice to clear influenza virus.
Figure 7. Tim3mut mice have normal viral clearance.
Viral titers in WT and Tim3mut mice at day 7 following influenza infection as assessed by A – QPCR (n=12 mice per group from 3 independent experiments) and B – MDCK infectivity assay (n=10 mice per group from 2 independent experiments).
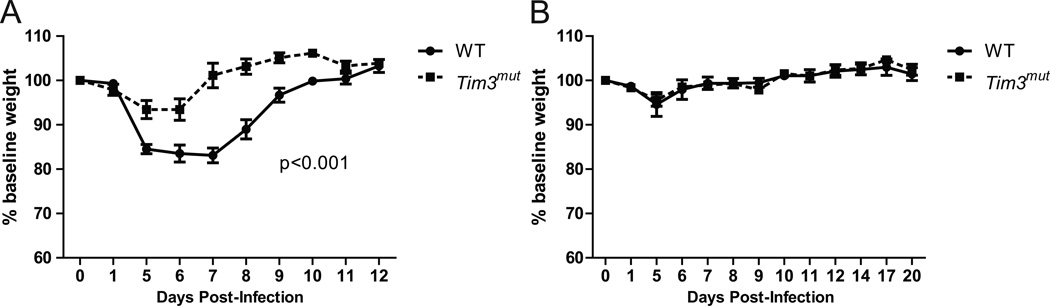
Tim3mut mice have intact functional heterotypic immunity
Recently published data suggest that Tim3-gal-9 interactions may limit the magnitude and efficiency of CD8+ T cell memory responses (21, 51). We therefore assessed if enhanced Tim3 activity would affect heterotypic memory. For these experiments, mice were infected with a non-lethal dose of influenza A / Hong Kong / x-31 (×31) virus, an H3N2 subtype. Mice recover for 4 weeks and are then challenged with an otherwise lethal dose of PR8 (3 × LD80), an H1N1 subtype. Effective immunity depends on CD8+ T cell memory in this model. Tim3mut mice lost less weight than WT controls during the ×31 infection, consistent with our findings in acute PR8 infection (Fig. 8A). Despite reduced CD8+ T cell effector function, Tim3mut mice had no weight loss and 100% survival similar to WT mice following secondary infection with PR8 (Fig. 8B). These data demonstrate that despite decreased effector functions of CD8+ T cells, functional heterotypic memory responses are intact in Tim3mut mice.
Figure 8. Tim3mut mice have preserved functional heterotypic immunity.
WT and Tim3mut mice were infected with influenza ×31, allowed to recover for four weeks, then re-infected with an otherwise lethal dose of influenza PR8. A – Weight loss as a percentage of original weight following infection with influenza ×31 (n=4 mice per group; p<0.001 by 2way ANOVA; experiment was repeated with similar results). B – Weight loss as a percentage of baseline following re-challenge with an otherwise lethal dose of influenza A/PR8 (n=4 mice per group; experiment was repeated with similar results).
DISCUSSION
These data clearly demonstrate that enhancement of the negative regulatory activity of Tim3 is protective against morbidity and mortality in a murine model of influenza infection. This protection is associated with decreased CD8+ T cell effector function including proliferation, cytotoxicity, and inflammatory cytokine expression. Experiments utilizing an adoptive transfer model of pneumonitis confirm that the effects of the Tim3 mutation specifically on CD8+ T cells can limit lung injury and mortality. Despite the decrease in CD8+ T cell effector function, viral clearance and functional heterotypic immunity remain intact. These findings suggest that enhancing the inhibitory activity of Tim3 may be beneficial in reducing immune mediated lung injury post-influenza infection.
Despite extensive characterization of its regulatory function, the signaling mechanisms of Tim3 remain largely undefined. Tim3 has a well-established role in inhibiting effector T cell function (52, 53). However, unlike other molecules that have been shown to inhibit effector T cells, there are no obvious inhibitory signaling motifs in the cytoplasmic tail. The cytoplasmic tail of Tim3 is considerably more complex than that of other Tim family members, containing five tyrosine residues that are good candidates for phosphorylation and several predicted serine/threonine kinase binding sites (54, 55). One of these tyrosine residues (human Y265; murine Y256) is phosphorylated following gal-9 binding by the tyrosine kinase Itk (29). This residue is intact and presumably available for signaling in our mutant construct. By deleting the terminal cytoplasmic domain of Tim3, we have demonstrated several novel aspects of Tim3 signaling. First, we have demonstrated that signaling through Tim3 involves not only the highly conserved Y256 but also occurs in the terminal cytoplasmic domain. Further, we have shown that deletion of the terminal cytoplasmic domain potentiates the ability of Tim3 to down-regulate Tc1 effector functions, suggesting that these residues are negative regulators of Tim3 function.
In a recent publication, a Tim3 truncation mutation similar to ours was generated to examine the role of cytoplasmic tyrosine residues in coupling T cell activation with downstream signaling events (30). Consistent with our data, this mutant construct demonstrated enhanced Tim3 signaling and activity relative to WT Tim3, confirming that the distal tyrosine residues of the cytoplasmic tail have negative regulatory activity on Tim3 signaling. Interestingly, their data also suggested that Tim3 might play a role in T cell activation. However, it should be noted that these experiments used an ectopic expression system whereby Tim3 was expressed at high-levels on naïve T cells prior to activation. This is not the case with primary naïve T cells or naïve T cells in vivo, which require several days of activation before there is significant up-regulation of Tim3. It is well documented that TCR signaling is different in naïve T cells compared to T cells after activation (56–58). Thus, we suspect that both mutants have enhanced Tim3 activity, but the effects of Tim3 are different in naïve T cells during activation versus effector T cells at the peak of the inflammatory response. Given the expression pattern of Tim3 in vivo, it seems likely that the predominant effect of enhanced Tim3 signaling will be on effector T cells, resulting in an anti-inflammatory role during viral infection.
The signaling differences we observed in effector T cells are associated with decreased phosphorylation of TCR-CD3ζ. Work by others has recently demonstrated that much like Tim1, Tim3 activity is coupled to TCR signaling pathways and that Tim3 interacts with multiple proteins involved in TCR signal transduction (30, 59). However, our studies are the first to suggest a potential interaction between Tim3 and signaling through CD3ζ. Interestingly, recent data suggest that Tim3 may be phosphorylated by the tyrosine kinase Lck which also phosphorylates CD3ζ (42). Phosphorylation of CD3ζ provides a critical link between antigen recognition by the TCR and diverse intracellular signal-transduction pathways. In addition, it has been suggested that CD3ζ plays a role in controlling T cell survival (60), and inhibition of CD3ζ phosphorylation has been shown to reduce cytokine production and proliferation in T cells (42). Furthermore, chronic immune dysfunction in diseases such as HIV, chronic HCV and certain malignancies has been associated with loss of expression of CD3ζ and with increased expression of Tim3 on T cells (20, 60–62). Thus, down-regulation of CD3ζ phosphorylation by Tim3 is one potential mechanism by which Tim3 may mediate its counter-regulatory effects on T cells.
Although this mutation enhanced the suppressive function of Tim3, it eliminated gal-9-mediated apoptosis, suggesting that the distal cytoplasmic portion of Tim3 plays an important role in this process. Apoptosis of T cells is a highly regulated and complicated process separate from activation (41). Although some studies have demonstrated that gal-9 can induce CD8+ T cell apoptosis (21), in disease processes such chronic viral infection antigen-specific CD8+ T cells that express high levels of Tim3 persist despite the presence of gal-9, suggesting that they are not susceptible to gal-9 mediated cell death (20, 63). These cells have been described as exhausted – a process by which highly activated effector cells are rendered dysfunctional through hierarchical loss of effector functions. Tim3 has been functionally linked to this process in studies that demonstrate that blockade of Tim3 results in restoration of effector functions (20, 61). Thus, it would appear that Tim3 is able to mediate its inhibitory effects separately from its effects on apoptosis. Our Tim3 mutation may provide important clues as to why exhausted T cells can persist.
Influenza primarily infects respiratory epithelial cells, leading to their necrosis (64). The resulting inflammation promotes maturation and trafficking of dendritic cells to the draining lymph nodes, where they present influenza antigen to T cells leading to their activation (65). Activated T cells then migrate to the lung (66), where CD8+ T cells constitute a major portion of this inflammatory cell infiltrate (47). It is well established that cytotoxic CD8+ T cells are critical for the clearance of influenza virus from the lung (46, 67, 68), and efficient viral clearance depends in part on inflammatory cytokine production by these cells (particularly IFNγ) as well as perforin and Fas-Fas ligand mediated cell death (47, 69–71). However, substantial evidence indicates that the mechanisms contributing to viral clearance can lead to severe pneumonitis and death, which is particularly apparent after infection with high dose infection, infection with virulent strains or when the host is unable to control viral loads (48, 49, 64). The ability to effectively utilize intrinsic negative regulatory molecules, such as Tim3, could offer a therapeutic means of preventing or treating severe pneumonitis following influenza infection.
Whether Tim3 plays an important role in regulating inflammation during acute viral infection and what effect manipulation of Tim3 activity would have in this setting has not been clearly established. Prior in vivo studies utilizing Tim3 deficient animals or antibodies that block Tim3 have demonstrated increased Th1 inflammation that results in greater immune-mediated tissue injury in models of transplant rejection and autoimmune disease (17, 19, 31). In a recent publication, gal-9 deficient mice infected with non-lethal influenza A produced more virus-specific CD8+ T cells, had improved viral clearance, and more robust CD8+ T cell recall responses compared to WT animals (51). These results are in agreement with an animal model of HSV ocular infection, in which blockade of Tim3-gal-9 interaction was associated with high-level cytokine production and an increase in the number of antigen-specific CD8+ T cells (21, 22). These results confirm our findings that Tim3 serves to down-regulate effector CD8+ T cell function during viral infection. While blockade of Tim3 signaling may be useful in generating more robust anti-viral response in non-lethal infection, our data clearly suggest that enhancement of Tim3 function is beneficial in severe pneumonitis and acute viral infection. Furthermore, these data provide strong evidence that increasing Tim3 activity could be used therapeutically to control inflammation in response to influenza infection without significant effects on viral clearance or the establishment of cellular memory.
Although our studies in the influenza model and CC10-OVA model clearly demonstrate that enhanced Tim3 activity on CD8+ T cells can limit immune-mediated lung injury and mortality, our data does not completely exclude that Tim3mut may also contribute to the observed phenotype in the influenza model via effects on CD4+ T cells, NK cells or APCs. However, our Tim3 mutation did not result in any appreciable phenotypic or signaling differences in CD4+ T cells. Furthermore, our in vitro studies did not show an effect of our Tim3 mutation on APCs, and we did not detect any differences in cellular infiltrate or IFNγ expression in our in vivo influenza model at day 1 (data not shown) or day 3, during which time inflammation is primarily driven by the innate immune system.
In summary, we have shown that Tim3 is an important regulator of the adaptive immune response to influenza, and that enhanced Tim3 activity can protect against morbidity and mortality following infection. Experiments performed both in vitro and in vivo demonstrate a significant defect in effector functions of Tim3mut CD8+ T cells, suggesting that enhancing Tim3 activity may reduce immune mediated lung injury post-influenza infection due in part to inhibition of CD8+ T cell effector function. Finally, our experiments suggest a potentially novel mechanism for Tim3-mediated down-regulation of T cell effector function via alteration of CD3ζ phosphorylation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Khristianna Jones for technical support with this research.
Abbreviations used
- BAL
bronchoalveolar lavage
- gal-9
galectin-9
- IP
immunoprecipitation
- LD80
lethal dose 80
- PR8
influenza A/Puerto Rico/8/34
- Tet
influenza nucleoprotein (366–374) tetramer
- Tim3
cell immunoglobulin and mucin domain 3
- tLN
thoracic lymph node
- ×31
influenza A/Hong Kong/8/68-×31
Footnotes
Grant Support: This work was supported by grants from the National Institutes of Health F32 HL099070 (to J.L. Cho), T32 HL07874 (to B.D. Medoff), Roche Organ Transplant Research Foundation Award to BDM, and R01 HL088297 (to B.D. Medoff and R.J. Xavier).
Nonstandard abbreviations used: Tim3, T cell immunoglobulin and mucin domain 3; BAL, bronchoalveolar lavage; gal-9, galectin-9; tLN, thoracic lymph node.
Conflict of interest: The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Murphy BR, W R. Orthomyxoviruses. In: BN F, editor. Fields Virology. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 1397–1445. [Google Scholar]
- 2.Couch RB. Influenza: prospects for control. Annals of internal medicine. 2000;133:992–998. doi: 10.7326/0003-4819-133-12-200012190-00015. [DOI] [PubMed] [Google Scholar]
- 3.Simonsen L, Fukuda K, Schonberger LB, Cox NJ. The impact of influenza epidemics on hospitalizations. The Journal of infectious diseases. 2000;181:831–837. doi: 10.1086/315320. [DOI] [PubMed] [Google Scholar]
- 4.Heltzer ML, Coffin SE, Maurer K, Bagashev A, Zhang Z, Orange JS, Sullivan KE. Immune dysregulation in severe influenza. Journal of leukocyte biology. 2009;85:1036–1043. doi: 10.1189/jlb.1108710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nature medicine. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kash JC, Tumpey TM, Proll SC, Carter V, Perwitasari O, Thomas MJ, Basler CF, Palese P, Taubenberger JK, Garcia-Sastre A, Swayne DE, Katze MG. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan KM, Monto AS, Longini IM., Jr Estimates of the US health impact of influenza. American journal of public health. 1993;83:1712–1716. doi: 10.2105/ajph.83.12.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermejo-Martin JF, Martin-Loeches I, Rello J, Anton A, Almansa R, Xu L, Lopez-Campos G, Pumarola T, Ran L, Ramirez P, Banner D, Cheuk Ng D, Socias L, Loza A, Andaluz D, Maravi E, Gomez-Sanchez MJ, Gordon M, Gallegos MC, Fernandez V, Aldunate S, Leon C, Merino P, Blanco J, Martin-Sanchez F, Rico L, Varillas D, Iglesias V, Marcos MA, Gandia F, Bobillo F, Nogueira B, Rojo S, Resino S, Castro C, Ortiz de Lejarazu R, Kelvin D. Host adaptive immunity deficiency in severe pandemic influenza. Critical care (London, England) 2010;14:R167. doi: 10.1186/cc9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagau N, Slavcovici A, Gonganau DN, Oltean S, Dirzu DS, Brezoszki ES, Maxim M, Ciuce C, Mlesnite M, Gavrus RL, Laslo C, Hagau R, Petrescu M, Studnicska DM. Clinical aspects and cytokine response in severe H1N1 influenza A virus infection. Critical care (London, England) 2010;14:R203. doi: 10.1186/cc9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauad T, Hajjar LA, Callegari GD, da Silva LF, Schout D, Galas FR, Alves VA, Malheiros DM, Auler JO, Jr, Ferreira AF, Borsato MR, Bezerra SM, Gutierrez PS, Caldini ET, Pasqualucci CA, Dolhnikoff M, Saldiva PH. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med. 2009;181:72–79. doi: 10.1164/rccm.200909-1420OC. [DOI] [PubMed] [Google Scholar]
- 11.Nin N, Soto L, Hurtado J, Lorente JA, Buroni M, Arancibia F, Ugarte S, Bagnulo H, Cardinal P, Bugedo G, Echevarria E, Deicas A, Ortega C, Frutos-Vivar F, Esteban A. Clinical characteristics and outcomes of patients with 2009 influenza A(H1N1) virus infection with respiratory failure requiring mechanical ventilation. J Crit Care. 2011;26:186–192. doi: 10.1016/j.jcrc.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Salomon R, Hoffmann E, Webster RG. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12479–12481. doi: 10.1073/pnas.0705289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomon R, Webster RG. The influenza virus enigma. Cell. 2009;136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol. 2007;81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK, Mytle N, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Rowe T, Kelvin DJ. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008;82:11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchroo VK, Dardalhon V, Xiao S, Anderson AC. New roles for TIM family members in immune regulation. Nat Rev Immunol. 2008;8:577–580. doi: 10.1038/nri2366. [DOI] [PubMed] [Google Scholar]
- 17.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA, Freeman GJ, Kuchroo VK. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536–541. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 18.Oikawa T, Kamimura Y, Akiba H, Yagita H, Okumura K, Takahashi H, Zeniya M, Tajiri H, Azuma M. Preferential involvement of Tim-3 in the regulation of hepatic CD8+ T cells in murine acute graft-versus-host disease. J Immunol. 2006;177:4281–4287. doi: 10.4049/jimmunol.177.7.4281. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T, Kuchroo VK, Gutierrez-Ramos JC, Coyle AJ, Strom TB. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nature immunology. 2003;4:1093–1101. doi: 10.1038/ni987. [DOI] [PubMed] [Google Scholar]
- 20.Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, Wong JC, Satkunarajah M, Schweneker M, Chapman JM, Gyenes G, Vali B, Hyrcza MD, Yue FY, Kovacs C, Sassi A, Loutfy M, Halpenny R, Persad D, Spotts G, Hecht FM, Chun TW, McCune JM, Kaul R, Rini JM, Nixon DF, Ostrowski MA. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. The Journal of experimental medicine. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS pathogens. 2010;6 doi: 10.1371/journal.ppat.1000882. e1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sehrawat S, Suryawanshi A, Hirashima M, Rouse BT. Role of Tim-3/galectin-9 inhibitory interaction in viral-induced immunopathology: shifting the balance toward regulators. J Immunol. 2009;182:3191–3201. doi: 10.4049/jimmunol.0803673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nature immunology. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 24.Hastings WD, Anderson DE, Kassam N, Koguchi K, Greenfield EA, Kent SC, Zheng XX, Strom TB, Hafler DA, Kuchroo VK. TIM-3 is expressed on activated human CD4+ T cells and regulates Th1 and Th17 cytokines. Eur J Immunol. 2009;39:2492–2501. doi: 10.1002/eji.200939274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, Bruce JN, Kane LP, Kuchroo VK, Hafler DA. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science (New York, N.Y. 2007;318:1141–1143. doi: 10.1126/science.1148536. [DOI] [PubMed] [Google Scholar]
- 26.Hirashima M, Kashio Y, Nishi N, Yamauchi A, Imaizumi TA, Kageshita T, Saita N, Nakamura T. Galectin-9 in physiological and pathological conditions. Glycoconjugate journal. 2004;19:593–600. doi: 10.1023/B:GLYC.0000014090.63206.2f. [DOI] [PubMed] [Google Scholar]
- 27.Dai SY, Nakagawa R, Itoh A, Murakami H, Kashio Y, Abe H, Katoh S, Kontani K, Kihara M, Zhang SL, Hata T, Nakamura T, Yamauchi A, Hirashima M. Galectin-9 induces maturation of human monocyte-derived dendritic cells. J Immunol. 2005;175:2974–2981. doi: 10.4049/jimmunol.175.5.2974. [DOI] [PubMed] [Google Scholar]
- 28.Imaizumi T, Kumagai M, Sasaki N, Kurotaki H, Mori F, Seki M, Nishi N, Fujimoto K, Tanji K, Shibata T, Tamo W, Matsumiya T, Yoshida H, Cui XF, Takanashi S, Hanada K, Okumura K, Yagihashi S, Wakabayashi K, Nakamura T, Hirashima M, Satoh K. Interferon-gamma stimulates the expression of galectin-9 in cultured human endothelial cells. Journal of leukocyte biology. 2002;72:486–491. [PubMed] [Google Scholar]
- 29.van de Weyer PS, Muehlfeit M, Klose C, Bonventre JV, Walz G, Kuehn EW. A highly conserved tyrosine of Tim-3 is phosphorylated upon stimulation by its ligand galectin-9. Biochemical and biophysical research communications. 2006;351:571–576. doi: 10.1016/j.bbrc.2006.10.079. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Su EW, Zhu C, Hainline S, Phuah J, Moroco JA, Smithgall TE, Kuchroo VK, Kane LP. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol Cell Biol. 2011;31:3963–3974. doi: 10.1128/MCB.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatos CA, Chakravarti S, Cha E, Schubart A, Sanchez-Fueyo A, Zheng XX, Coyle AJ, Strom TB, Freeman GJ, Kuchroo VK. Interaction of Tim-3 and Tim-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nature immunology. 2003;4:1102–1110. doi: 10.1038/ni988. [DOI] [PubMed] [Google Scholar]
- 32.Seki M, Oomizu S, Sakata KM, Sakata A, Arikawa T, Watanabe K, Ito K, Takeshita K, Niki T, Saita N, Nishi N, Yamauchi A, Katoh S, Matsukawa A, Kuchroo V, Hirashima M. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clinical immunology (Orlando, Fla. 2008;127:78–88. doi: 10.1016/j.clim.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Wang F, He W, Yuan J, Wu K, Zhou H, Zhang W, Chen ZK. Activation of Tim-3-Galectin-9 pathway improves survival of fully allogeneic skin grafts. Transpl Immunol. 2008;19:12–19. doi: 10.1016/j.trim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, He W, Zhou H, Yuan J, Wu K, Xu L, Chen ZK. The Tim-3 ligand galectin-9 negatively regulates CD8+ alloreactive T cell and prolongs survival of skin graft. Cell Immunol. 2007;250:68–74. doi: 10.1016/j.cellimm.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Medoff BD, Seed B, Jackobek R, Zora J, Yang Y, Luster AD, Xavier R. CARMA1 is critical for the development of allergic airway inflammation in a murine model of asthma. J Immunol. 2006;176:7272–7277. doi: 10.4049/jimmunol.176.12.7272. [DOI] [PubMed] [Google Scholar]
- 36.Medoff BD, Seung E, Wain JC, Means TK, Campanella GS, Islam SA, Thomas SY, Ginns LC, Grabie N, Lichtman AH, Tager AM, Luster AD. BLT1-mediated T cell trafficking is critical for rejection and obliterative bronchiolitis after lung transplantation. The Journal of experimental medicine. 2005;202:97–110. doi: 10.1084/jem.20042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikhak Z, Fukui M, Farsidjani A, Medoff BD, Tager AM, Luster AD. Contribution of CCR4 and CCR8 to antigen-specific T(H)2 cell trafficking in allergic pulmonary inflammation. J Allergy Clin Immunol. 2009;123:67–73. e63. doi: 10.1016/j.jaci.2008.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Topham DJ, Castrucci MR, Wingo FS, Belz P, Doherty C. The role of antigen in the localization of naïve, acutely activated, and memory CD8(+) T cells to the lung during influenza pneumonia. J Immunol. 2001;167:6983–6990. doi: 10.4049/jimmunol.167.12.6983. [DOI] [PubMed] [Google Scholar]
- 39.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M. Influenza A virus neuraminidase limits viral superinfection. J Virol. 2008;82:4834–4843. doi: 10.1128/JVI.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nature immunology. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 42.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence CW, Braciale TJ. Activation, differentiation, and migration of naive virus-specific CD8+ T cells during pulmonary influenza virus infection. J Immunol. 2004;173:1209–1218. doi: 10.4049/jimmunol.173.2.1209. [DOI] [PubMed] [Google Scholar]
- 44.Roman E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G, Swain SL. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. The Journal of experimental medicine. 2002;196:957–968. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohlmeier JE, Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 46.Eichelberger M, Allan W, Zijlstra M, Jaenisch R, Doherty PC. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. The Journal of experimental medicine. 1991;174:875–880. doi: 10.1084/jem.174.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 48.Xu L, Yoon H, Zhao MQ, Liu J, Ramana CV, Enelow RI. Cutting edge: pulmonary immunopathology mediated by antigen-specific expression of TNF-alpha by antiviral CD8+ T cells. J Immunol. 2004;173:721–725. doi: 10.4049/jimmunol.173.2.721. [DOI] [PubMed] [Google Scholar]
- 49.Moskophidis D, Kioussis D. Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. The Journal of experimental medicine. 1998;188:223–232. doi: 10.1084/jem.188.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seung E, Cho JL, Sparwasser T, Medoff BD, Luster AD. Inhibiting CXCR3-dependent CD8+ T cell trafficking enhances tolerance induction in a mouse model of lung rejection. J Immunol. 2011;186:6830–6838. doi: 10.4049/jimmunol.1001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma S, Sundararajan A, Suryawanshi A, Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY, Rouse BT. T cell immunoglobulin and mucin protein-3 (Tim-3)/Galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proceedings of the National Academy of Sciences of the United States of America. 2012;108:19001–19006. doi: 10.1073/pnas.1107087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuchroo VK, Meyers JH, Umetsu DT, DeKruyff RH. TIM family of genes in immunity and tolerance. Adv Immunol. 2006;91:227–249. doi: 10.1016/S0065-2776(06)91006-2. [DOI] [PubMed] [Google Scholar]
- 53.Su EW, Lin JY, Kane LP. TIM-1 and TIM-3 proteins in immune regulation. Cytokine. 2008;44:9–13. doi: 10.1016/j.cyto.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kane LP. T cell Ig and mucin domain proteins and immunity. J Immunol. 2010;184:2743–2749. doi: 10.4049/jimmunol.0902937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obenauer JC, L C, MB Yaffe. 2010 Oct 4; http://scansite.mit.edu/index.html. [Google Scholar]
- 56.Chandok MR, Farber DL. Signaling control of memory T cell generation and function. Semin Immunol. 2004;16:285–293. doi: 10.1016/j.smim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 57.Kimachi K, Sugie K, Grey HM. Effector T cells have a lower ligand affinity threshold for activation than naive T cells. Int Immunol. 2003;15:885–892. doi: 10.1093/intimm/dxg087. [DOI] [PubMed] [Google Scholar]
- 58.Teixeiro E, Daniels MA, Hamilton SE, Schrum AG, Bragado R, Jameson SC, Palmer E. Different T cell receptor signals determine CD8+ memory versus effector development. Science (New York, N.Y. 2009;323:502–505. doi: 10.1126/science.1163612. [DOI] [PubMed] [Google Scholar]
- 59.Binne LL, Scott ML, Rennert PD. Human TIM-1 associates with the TCR complex and up-regulates T cell activation signals. J Immunol. 2007;178:4342–4350. doi: 10.4049/jimmunol.178.7.4342. [DOI] [PubMed] [Google Scholar]
- 60.Baniyash M. TCR zeta-chain downregulation: curtailing an excessive inflammatory immune response. Nat Rev Immunol. 2004;4:675–687. doi: 10.1038/nri1434. [DOI] [PubMed] [Google Scholar]
- 61.Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. The Journal of experimental medicine. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golden-Mason L, Palmer BE, Kassam N, Townshend-Bulson L, Livingston S, McMahon BJ, Castelblanco N, Kuchroo V, Gretch DR, Rosen HR. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol. 2009;83:9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMahan RH, Golden-Mason L, Nishimura MI, McMahon BJ, Kemper M, Allen TM, Gretch DR, Rosen HR. Tim-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J Clin Invest. 120:4546–4557. doi: 10.1172/JCI43127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bender BS, Small PA., Jr Influenza: pathogenesis and host defense. Seminars in respiratory infections. 1992;7:38–45. [PubMed] [Google Scholar]
- 65.Harmsen AG, Muggenburg BA, Snipes MB, Bice DE. The role of macrophages in particle translocation from lungs to lymph nodes. Science (New York, N.Y. 1985;230:1277–1280. doi: 10.1126/science.4071052. [DOI] [PubMed] [Google Scholar]
- 66.Cerwenka A, Morgan TM, Dutton RW. Naive, effector, and memory CD8 T cells in protection against pulmonary influenza virus infection: homing properties rather than initial frequencies are crucial. J Immunol. 1999;163:5535–5543. [PubMed] [Google Scholar]
- 67.Epstein SL, Lo CY, Misplon JA, Bennink JR. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol. 1998;160:322–327. [PubMed] [Google Scholar]
- 68.Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. The Journal of experimental medicine. 1997;186:2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allan W, Tabi Z, Cleary A, Doherty PC. Cellular events in the lymph node and lung of mice with influenza. Consequences of depleting CD4+ T cells. J Immunol. 1990;144:3980–3986. [PubMed] [Google Scholar]
- 70.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159:5197–5200. [PubMed] [Google Scholar]
- 71.Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunological reviews. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.