Abstract
HIV-1 replicates preferentially in IL-4 producing CD4 T cells for unclear reasons. We show increased HIV-1 expression is irrespective of viral tropism for chemokine receptors as previously suggested, but rather transcription of the HIV-1 long terminal repeat (LTR) is increased in IL-4 producing CD4 T cells. Increased expression of HIV-1 message is also confirmed in IL-4 producing CD4 T cells from HIV-1 infected individuals ex vivo. In exploring a transcriptional mechanism, we identify a novel c-maf (required for IL-4 expression) transcription factor binding site just upstream of the dual NFκB/NFAT binding sites in the proximal HIV-1 LTR. We demonstrate c-maf binds this site in vivo and synergistically augments HIV-1 transcription in cooperation with NFAT2 and NFκB p65, but not NFAT1 nor NFκB p50. Conversely, siRNA inhibition of c-maf reduces HIV-1 transcription in IL-4 producing T cells. Thus, c-maf increases HIV-1 expression in IL-4 producing CD4 T cells by binding the proximal HIV-1 LTR and augmenting HIV-1 transcription in partnership with NFAT2 and NFκB p65, specifically. This has important implications for selective targeting of transcription factors during HIV-1 infection since, over the course of HIV-1 progression/AIDS, IL-4 producing T cells frequently predominate and substantially contribute to disease pathology.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection is characterized by viremia, progressive depletion of CD4 T lymphocytes, and the development of acquired immune deficiency syndrome (AIDS). Although the CD4 T cell count provides an indication of the intactness of the CD4 T cell compartment and its ability to prevent opportunistic infections, the peripheral blood viral load provides an indication of how rapidly the CD4 T cell compartment will be depleted as a result of pathological processes driven by viral replication (1). The profound contribution of highly active antiretroviral therapy (HAART) is to interrupt HIV replication with a resultant decrease in viral load and an increase in CD4 T cell count and function (2). Therefore, the viral load assay is one of the most sensitive and reliable parameters to assess HAART effectiveness, despite the fact that use of immunological criteria are suggested for making treatment decisions about changing from a first to a second line regimen under the current World Health Organization HIV-1 treatment guidelines in resource-limited settings (3–5). Similarly, failure of intermittent antiretroviral therapy is marked by a strong viral rebound followed by more severe and longer depletion of CD4 T cells and disease progression (6, 7). Notably, viral replication substantially contributes to the viral load in an infected individual and thus plays a critical role in HIV-1/AIDS pathogenesis.
HIV-1 largely infects and replicates in activated human CD4 T cells. In particular, HIV-1 shows a preferential replication in Th2 and Th0 subsets of activated effector T cells without clearly defined reasons. Early studies found that HIV-1 infection might induce a shift in the relative proportion of different CD4 T helper cell subsets, and the virus preferentially replicated in CD4 T cells that produced Th2-type cytokines (8, 9). This preferential replication phenomenon by HIV-1 was later confirmed in several studies by using primary HIV-1 isolates capable of binding either the CCR5 or the CXCR4 chemokine receptors required as co-receptors for viral entry into CD4 T cells (10–14). However, these studies were conducted in vitro using conditionally polarized Th0, Th1, and Th2 clones derived from HIV-1 patients or healthy donors, and thus not necessarily reflecting infection of CD4 T cell subsets in vivo. Therefore, analysis of HIV-1 expression levels ex vivo in different CD4 T cell populations from infected individuals will be much more relevant to understanding HIV-1/AIDS progression (15).
Because of the significance of viral replication in HIV-1/AIDS pathogenesis, it is important to understand the mechanism underlying HIV-1 preferential replication in Th2 and/or Th0 effector CD4 T cells. Initially, one of the possible explanations put forward was related to a difference on T cell subsets in expression of chemokine receptors required for HIV-1 infection. Namely, Th1 cells express more CCR5 (16) and consequently are more easily infected by R5-tropic HIV-1; conversely, Th2 cells express a high density of CXCR4 (17) and are more susceptible to X4-tropic HIV-1 infection. However, over the time course of infection, R5 viral replication surprisingly increased in Th2 cells and drastically decreased in Th1 cells. By comparison, X4 viruses consistently maintained a high level of replication in Th2 cells and a low level in Th1 cells (10, 11). In addition, a proposed blockade of CCR5 by associative ligands, such as RANTES, MIP-1α, and MIP-1β, does not provide a satisfactory explanation because they seem to be generated and secreted constitutively, and a variety of chemokines might play different roles during HIV-1 replication (10, 18, 19). Thus, differential chemokine receptor expression does not appear to explain increased HIV-1 expression in IL-4 producing CD4 T cells.
An alternative hypothesis is that HIV-1 gene transcription influences differential viral replication in host CD4 T cell subsets (20). Along this line, we herein investigate whether a Th2 specific transcriptional mechanism affects HIV-1 replication. Specifically, we focus on a Th2-restricted factor, c-maf, exploring a potential mechanism that augments HIV-1 gene expression in IL-4 producing CD4 T cells. The proto-oncogene, c-maf, is the cellular counterpart of oncogenic v-maf (21). The c-maf protein belongs to a large family of basic region leucine zipper domain transcription factors, and exerts its transcriptional role through binding to a Maf recognition element (MARE) (22). c-maf is expressed in Th2 but not Th1 clones, and plays a critical and selective role in transcriptional activation of IL-4 (in conjunction with NFAT), IL-10, and IL-21 cytokine gene transcription (23–26). NFAT transcription factors, especially NFAT2, are also involved in the positive regulation of IL-4 gene expression (27). Moreover, NFAT2 is capable of inducing a highly permissive state for HIV-1 replication in primary CD4 T cells (28). NFAT proteins, as well as their relatively homologous Rel family members, NFκB RelA (p65) and p50, have been clearly shown to directly bind to and augment transcription of the HIV-1 LTR via the dual proximal NFκB/NFAT binding sites (28, 29). Moreover, they have even been reported to be essential for enhancing HIV-1 transcription (30–32) and likely play a role in governing latency maintenance (33). Herein, we show the transcription factor, c-maf, contributes to increased HIV-1 transcription in IL-4 producing CD4 T cells via binding to the LTR in cooperation with NFAT2 and NFκB p65, specifically. Our studies provide new insight into the preferential replication of HIV-1 in IL-4 producing CD4 T cells, and further suggest means to disrupt this increased HIV-1 expression or, alternatively, augment transcription of latently infected cells.
Materials and Methods
Plasmids and reagents
The original expression vectors for c-maf from Dr. Xiaojing Ma (34) and for NFAT2 (29) have been described. Luciferase reporter plasmids pLTR-Luc and pIL4-Luc were previously described (35). The pLTR-GFP, pIFN-γ-GFP, and pIL-4-GFP constructs were generated by sub-cloning the 3′ HIV-1 LTR segment (598 bp), human IFN-γ promoter segment (646 bp), and human IL-4 promoter segment (805 bp), respectively, into pEGFP-N1 to substitute the CMV promoter (Clontech). Similarly, the pc-maf-GFP plasmid was generated with the minimum c-maf-binding promoter, in which two tandem MARE consensus sequences (5′TGCTGACTCAGCA) were inserted just upstream of the minimal c-maf-binding fragment (position −206 to +30) of the human IL-10 promoter (34). rIL-2 was supplied by the NIH AIDS Research and Reference Reagent Program (Germantown, MD). rIL-4 was obtained from R&D Systems. Fluorescence-conjugated anti-IL-4, anti-IFN-γ, anti-CXCR4, and anti-CCR5 antibodies were purchased from BD Bioscience, as was 7AAD. Fluorescence-conjugated anti-HIV-1 core antigen (p24) antibody was bought from Beckman Coulter.
Cell isolation and expansion of IL-4 producing CD4 T cells
Institutional Review Board approval for protection of human subjects was obtained from the University of Pennsylvania and the University of Alabama at Birmingham. Primary human peripheral blood CD4 T cells from healthy donors were isolated by negative selection using a proprietary antibody mix (StemCell Technologies) as described (36). To polarize IL-4 producing CD4 T cells, the primary T cells were activated with PHA (200 ng/ml) for 3 days in the presence of IL-4 (10 ng/ml), and irradiated (3,000 rads) syngeneic PBMC (37). The activated cells were cultured for another 8–12 days in the presence of rIL-2 (30 IU/ml) and rIL-4 (10 ng/ml) with fresh culture media replacement every 2–3 days.
Transient transfection and reporter gene assays
Freshly isolated human CD4 T cells were transiently transfected with expression vector(s), alone or in combination with GFP or luciferase reporter plasmids, using Human T cell Nucleofector kits (Lonza Inc.), as described elsewhere (38). In primary infection experiments, the transfected cells were activated with PHA (1.5 μg/ml) and rIL-2 (20 U/ml) for 3 days followed by HIV-1 infection. In c-maf-knockdown experiments, the polarized CD4 T cells were transiently transfected with a siRNA expression vector or its control as previously detailed (39). The cells were then cultured in the presence of rIL-2 (30 U/ml) prior to analysis. All GFP reporter expression was detected by flow cytometry.
Electricity Mobility Shift Assay (EMSA) and in vitro footprinting
Nuclear extracts were prepared from polarized human CD4 T cells. Oligonucleotide probes were labeled with γ-32P-ATP (Table 1). EMSA was performed as previously described (36). Baculovirus generated recombinant mafK and NFAT2 were prepared and studied for binding to the proximal HIV-1 LTR by in vitro footprinting as previously described (29).
Table 1.
Listing of oligonucleotides used in this study*
| EMSA probes | ||
| NFAT binding sequence (mouse IL-2 promoter) | TCGAGCCCAAAGAGGAAAATTTGTTTCATG | |
| NFκB binding sequence (human Igκ promoter) | ATGTGAGGGGACTTTCCCAGGC | |
| c-maf binding sequence (human IL-4 promoter) | AATTGCTGACTCAGCATTACT | |
| HIV-1 LTR-NFAT/NFkB dual site sequence | CAAGGGACTTTCCGCTGGGGACTTTCCAGGG | |
| HIV-1 LTR-MARE | AACTGCTGACATCGAGCTTGTTACAAGGGACTTTCCGCT | |
| Real-Time PCR for HIV-1 gag, 18S, c-maf, and IL-4 | ||
| HIV-1 semi-nested, forward primer | CTTAGGCATCTCCTATGGCAGGAA | |
| HIV-1 semi-nested, reverse primer | GGATCTGTCTCTGTCTCTCTCTCCACC | |
| HIV-1 forward primer | ACAGTCAGACTCATCAAGTTTCTCTATCAAAGCA | |
| HIV-1 reverse primer | The same as semi-nested reverse primer | |
| HIV-1 probe | FAM-TTCCTTCGGGCCTGTCGGGTCCC-BHQ2 | |
| 18SRNA forward primer | TTTCGATGGTAGTCGCCG | |
| 18SRNA reverse primer | TGGATGTGGTAGCCGTTT C | |
| 18SRNA probe | FAM-CCACGGGTGACGGGGAATCAGG-BHQ2 | |
| c-maf forward primer | GTCAGCAAGGAGGAGGTGAT | |
| c-maf reverse primer | TTCTTCTCCGACTCCAGGAC | |
| c-maf probe | FAM-ATGCCCAGTCCTGCCGCTTC-BHQ2 | |
| IL-4 forward primer | CCA CGG ACA CAA GTG CGA TA | |
| IL-4 reverse primer | CCC TGC AGA AGG TTT CCT TCT | |
| IL-4 probe | FAM- TCT GTG CAC CGA GTT GAC CGT AAC AGAC-BHQ | |
| Real-Time PCR for ChIP | ||
| HIV-1 LTR forward primer | TTGACAGCCGCCTAGCATT | |
| HIV-1 LTR reverse primer | CACGCCTCCCTGGAAAGTC | |
| HIV-1 LTR probe | FAM-CATCACGTGGCCCGAGAGCTGC-BHQ | |
| siRNA and scrambled control | ||
| c-maf specific siRNA | CGGCGAAGCTTTTTCCAAAAAAGAGACCGACCGC ATCATCACTACACAAATGATGATGCGGTCGGTCT CCGGTGTTTCGTCCTTTCCACAAG (AF055376, start position 918) (core oligo. 19 bp) | |
| Scrambled sequence control | CGGCGAAGCTTTTTCCAAAAAAGCACAACTAACG TCGCACGCTACACAAACGTGCGACGTTAGTTGTG CCGGTGTTTCGTCCTTTCCACAAG (core oligo 19 bp) | |
Underlined bases are the core oligonucleotide binding sequences for corresponding transcription factors, or for c-maf siRNA and its scrambled control.
Chromatin immunoprecipitation (ChIP) and real-time PCR
Cytokine polarized human CD4 T cells were specifically infected with the HIV-1 NL4-3-GFP strain, which was kindly provided by Dr. David N. Levy (New York University). Prior to the ChIP assay, HIV-1 infected cells (GFP+) were live sorted by flow cytometry following activation with PMA and ionomycin for 3 hrs. The ChIP assay was performed on the sorted GFP+ cells according to the product guidelines (Upstate Biotechnology). Purified DNA precipitated by anti-NFAT1, -NFAT2, -NFκB p65, -NFκB p50, -c-maf, -Oct1 antibodies, or IgG (Santa Cruz Biotechnologies) was subjected to real-time PCR. The amplicon was designed to overlap with the dual NFκB/NFAT sites and the nearby upstream MARE site within the proximal HIV-1 LTR (Table 1). The reaction was performed with the Taqman Universal PCR Master Mix (Applied Biosystems). Fold transcription factor binding relative to negative control antibody precipitation was calculated by the formula, fold binding=2(ΔCt), where ΔCt is calculated as Ct of antibody control minus Ct of the transcription factor antibody of interest.
HIV-1 infection and p24/gag detection
The HIV-1 virus strains, NL4-3, 89.6, Bal-1, were supplied by the Center for AIDS Research of the University of Pennsylvania. The NA420/BB3 HIV-1 strain was kindly provided by Dr. Phillip Smith (University of Alabama at Birmingham, AL). The activated (or polarized) CD4 T cells were infected with viral stock (p24, 50–100 ng/106 cells) in the presence of DEAE-Dextran (4μg/ml) for 2 hrs or overnight at 37°C. The cells were then cultured in the presence of rIL-2 for an additional 3–5 days prior to analysis. Intracellular p24/gag staining and analysis was performed as previously described (40).
Intracellular cytokine analysis and cell proliferation assays
Cytokine-polarized CD4 T cells were HIV-1 (or mock) infected and then stimulated with PMA (25 ng/ml) and ionomycin (1.5 μM) for 4–6 h at 37°C in the presence of brefeldin A (10 μg/ml) followed by intracellular staining according to the manufacturer’s suggestions (BD Pharmingen). For proliferation assays during HIV-1 infection, cytokine-polarized cells were first labeled by using cardoxyfluorescein diacetate succinimidyl ester (CFSE) prior to viral infection. Two days later, the CFSE-labeled HIV-1 infected cells were stimulated with anti-CD3 (5 μg/ml) and anti-CD28 (5 μg/ml) antibodies in the presence of irradiated syngeneic PBMC for another 3 days before analyzing intracellular cytokine production, CFSE dilution, and HIV-1 p24 expression. Flow cytometry data were collected using a BD Calibur or LSRII flow cytometer, and analyzed using Flowjo software (TreeStar Inc.).
Isolation of IL-4(+) cells and detection of HIV-1 mRNA from HIV-1 infected individuals
Sixty ml of peripheral blood was collected from HIV-1 infected individuals with high viral titers (viral loads were between 65,000 – 950,000 copies/ml; CD4 counts ranged between 127–350/μl). CD4 positive cells were then purified by negative selection using a proprietary antibody mix as previously described (35), followed by stimulation of PMA plus ionomycin overnight to allow for IL-4 expression. IL-4 positive and negative cells were then separated by using IL-4 cell enrichment and detection kits (Miltenyi Biotech) according to the product guidelines. RNA was isolated from each population of cells and converted to cDNA with SuperScript III (Invitrogen) reverse transcriptase. Two rounds of PCR amplification were carried out to determine the HIV-1 mRNA abundance as described (41). Briefly, the first round of PCR was performed on a conventional PCR machine with the setting as follows: 94°C for 3 min, followed by 21 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. These products were subsequently used as templates for the second, semi-nested, real-time, PCR, performed on the iQ5 multicolor real-time detection machine (Bio-Rad). The TaqMan detection chemistry was used and real-time PCR settings were as follows: 50°C for 2 min, then 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Real-time RT-PCR analyses were also performed for 18S RNA, IL-4, and c-maf mRNA levels. Primers and probes used for detection are listed in Table 1. Differences in message levels between IL-4(+) and IL-4(−) cells in individual patients were calculated using the Wilcoxon matched paired test (GraphPad Prism 5), and statistical significance was set at p<0.05.
Results
HIV-1 preferentially replicates in IL-4 producing CD4 T cells
The majority of previous studies examining HIV-1 infectivity and replication were carried out using in vitro separately derived Th1 and Th2 cell lines from HIV-1 infected individuals, healthy donors, or umbilical cord blood samples. HIV-1 infection and replication efficiency was then compared by monitoring p24/gag levels in culture supernatants (8, 9). There are, however, major shortcomings of these types of studies: 1) the infection conditions differed for the different environments used to support the Th1 and Th2 cell lines, and 2) assaying the p24 level in the bulk culture supernatant does not provide data at an individual cell level within a bulk population.
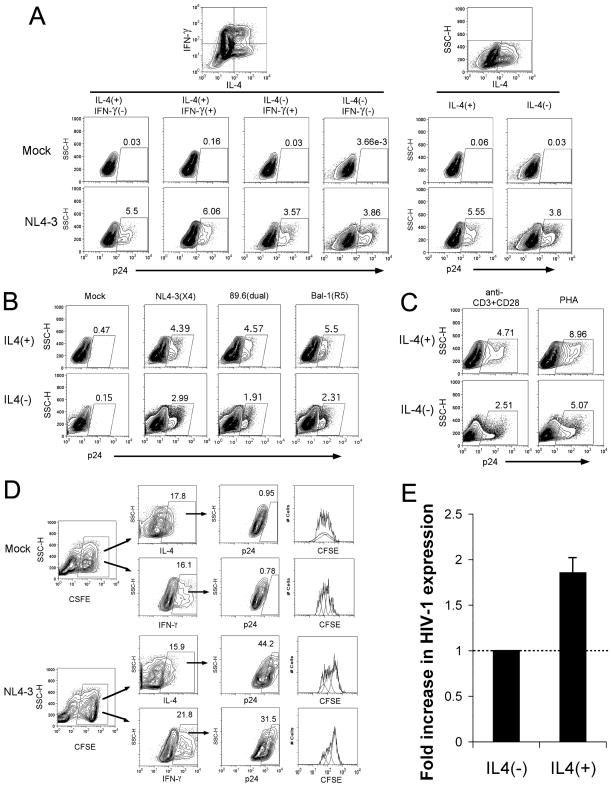
To study the fidelity of HIV-1 preferential replication in Th2 cells and in order to overcome the two aforementioned problems, we generated in vitro, using a combination of IL-2 and IL-4, an admixture of Th1-like (IFN-γ+, IL-4−), Th2-like (IFN-γ-, IL-4+), and Th0-like (IFN-γ+, IL-4+) CD4 T cells in one bulk population. After 8–10 days of differentiation in vitro, the cells were infected with HIV-1 for 3–5 days in the presence of IL-2 and IL-4. The production of cytokines and the expression of virus protein, p24, were measured by intracellular staining. Notably, IL-4 single positive (Th2) or IL-4 plus IFN-γ double positive (Th0) cells generate a higher percentage of p24 than IFN-γ single positive (Th1) or IFN-γ and IL-4 double negative cells (Fig. 1A). This increased expression in IL-4+ cells is not due to a relative increased rate of apoptosis in IFN-γ+ cells following HIV-1 infection, as HIV-1 infection increased the rate of apoptosis (as detected by 7AAD) virtually identically (~1.5-fold) in both IFN-γ+ and IL-4+ CD4 T cells (Supplemental Fig. 1). Therefore, IL-4 producing CD4 T cells preferentially support NL4-3 replication.
Figure 1. HIV-1 preferentially replicates in IL-4 producing CD4 T cells.
IL-4 polarized human CD4 T cells were infected with HIV-1 and tested for cytokine production and viral core protein, p24, expression by intracellular staining and flow cytometric analysis. (A) Mimic and HIV-1 NL4-3 infected cells were analyzed for IL-4 and IFN-γ along with p24 expression by 3-color flow cytometry. HIV-1 p24 expression is depicted for the different subsets of cytokine expression patterns. (B) HIV-1 p24 expression was compared between IL-4(+) and IL-4(−) subsets following infection with 3 different HIV-1 strains (NL4-3, 89.6, Bal-1) with different tropisms for chemokine receptor expression. (C) Primary human CD4 T cells were stimulated by anti-CD3 and anti-CD28 antibodies plus IL-4, or by PHA plus IL-4, for 3 days. Both were then cultured in the presences of IL-2 and IL-4 to allow the development of IL-4 producing CD4 T cells, and infected with the HIV-1 NL4-3 strain. HIV-1 p24 expression was compared between IL-4(+) and IL-4(−) subsets in each of polarized cells with initially different stimulations. (D) IL-4 polarized CD4 T cells were labeled with CFSE, followed by mock or HIV-1 NL4-3 strain infection. The cells were further stimulated with anti-CD3 and anti-CD28 antibodies in the presence of irradiated syngeneic PBMC. Cytokine production, cell proliferation (i.g. CFSE dilution), and p24 expression were detected by flow cytometry. (E) The average (from 12 separate infections) fold increase (1.86±0.189) in HIV-1 expression in IL-4(+) cells versus IL-4(−) cells is shown.
To further exclude the potential influences of viral tropisms on viral replication described elsewhere (42), an X4-tropic strain, NL4-3, a R5-tropic strain, Bal-1, and a X4R5 dual tropic strain, 89.6, were compared for their abilities to infect an admixture of polarized Th0, Th1, and Th2 CD4 T cells. To our surprise, IL-4 producing cells consistently and preferentially support HIV-1 viral expression irrespective of the viral tropism (Fig. 1B). Furthermore, this preference is consistent using different initial stimuli (anti-CD3 plus anti-CD28 mAb versus PHA) to polarize the host cells (Fig. 1C). As a complimentary alternative approach, we also show that surface CXCR4 expression is similar between IFN-γ+ and IL-4+ CD4 T cells, whereas CCR5 expression is higher on IFN-γ+ cells following T cell activation (Supplemental Fig. 2A). Nevertheless, both X4-and R5-tropic HIV-1 strains are expressed at similar levels in CXCR4+ and CCR5+ CD4 T cells (Supplemental Fig. 2B) despite increased expression in IL-4+ relative to IFN-γ+ CD4 T cells (Supplemental Fig. 2C). Thus, differences in chemokine receptor expression or in HIV-1 viral tropism do not explain increased HIV-1 expression in IL-4+ CD4 T cells.
In order to exclude the possibility that high level expression of HIV-1 in IL-4 producing T cells is a result of a relative rapid growth during HIV-1 infection, cytokine-polarized CD4 T cells were labeled by using CFSE to allow for tracking of cell proliferation during HIV-1 infection. As shown in Fig. 1D, HIV-1 infection does not result in increased proliferation of IL-4 producing CD4 T cells. Thus, our data demonstrate HIV-1 strains of any tropism are capable of replicating in either Th1 or Th2 cells, but IL-4-producing cells preferentially support replication of all varieties of HIV-1 tropism. Averaging the results of several experiments (n=12) reveals that IL-4 producing cells support HIV-1 replication approximately twice as well as IL-4 negative cells (Fig. 1E). This increased expression of HIV-1 did not appear to be related to either increased proliferation nor decreased HIV-1 driven apoptosis of IL-4+ relative to IFN-γ+ CD4 T cells.
Higher levels of HIV-1 gag mRNA are detected in IL-4 producing CD4 T cells from HIV-1 infected individuals
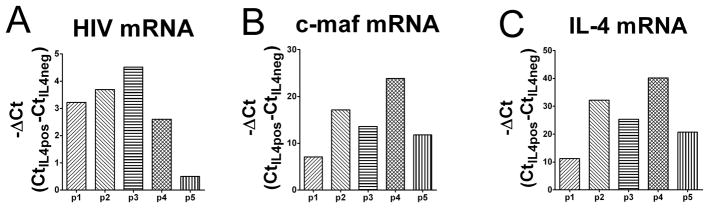
To substantiate the increased difference in p24 production between IL-4 producing and non-producing CD4 T cells seen in vitro, HIV-1 levels were assessed in peripheral blood CD4 T cells from HIV-1 infected individuals ex vivo. CD4 T cells from peripheral blood in HIV-1 infected individuals were stimulated ex vivo with PMA and ionomycin to allow for production of cytokines. IL-4(+) and IL-4(−) cells were then sorted by an IL-4 detection and enrichment method and used for RNA purification. HIV-1 gag mRNA was detected by real-time RT-PCR. Similarly, IL-4 mRNA and c-maf mRNA were also quantified. In these assays, 18S RNA levels served as an internal control and all other cycle threshold (Ct) values were normalized to it. Analyses were performed on peripheral blood samples from 5 different HIV-1 infected individuals with notably elevated HIV-1 levels in their blood. There was a remarkable and statistically significant (p=0.01) increase in HIV-1 message in IL-4(+) cells as assessed by Ct values (mean ± SEM is 26.89±1.29 for IL-4(+) cells, and 30.33±1.24 for IL-4(−) cells; ΔCt=3.44 or ~10.8 fold increase in HIV-1 mRNA between IL-4-producing versus non-producing cells) for the group as a whole (Fig. 2A). As predicted, based on the importance of c-maf for IL-4 expression (24), c-maf mRNA levels were only detected to any significant level in the IL-4(+) cells (Fig. 2B). Likewise, IL-4 mRNA was essentially limited to IL-4 producing cells (Fig. 2C). Thus, HIV-1 mRNA levels are about 10-fold higher on average in c-maf expressing, IL-4 producing CD4 T cells from peripheral blood of HIV-1 infected individuals. Since the cells were sorted based on IL-4 expression, the relative levels of IL-4 and associated c-maf are markedly higher in the IL-4+ cells compared to the IL-4(−) cells (e.g. 210–40) as expected. However, since the difference in HIV-1 was only expected to be near 2-fold (based on the in vitro data, Fig. 1) between IL-4+ and IL-4(−) cells, it is difficult to demonstrate a linear correlation among the 3 genes. Nevertheless, using a Wilcoxon matched paired test, with an n=5, IL-4(+) cells demonstrate higher HIV-1 viral RNA levels than IL-4(−) cells for each patient studied with a p value of 0.0313.
Figure 2. Higher levels of HIV-1 mRNA are detected in IL-4(+) CD4 T cells from HIV-1-infected individuals.
(A) The mRNA levels of HIV-1 gag in IL-4(+) and IL-4(−) CD4 T cells from HIV-1-infected patients’ peripheral blood were determined by real-time RT-PCR. Before real-time RT-PCR, HIV-1 specific semi-nested primer PCR was carried out after reverse transcription of total RNA extracts. 18S RNA acted as the internal control. The Ct (threshold cycle) values of HIV-1 gag mRNA in IL-4(−) CD4 T cells were normalized to that in IL-4(+) cells by correcting them according to the 18S Ct values. The p value is 0.01 when comparing with IL-4(+) cells. The -ΔCt calculated [-(Ct of IL-4(+) minus Ct of IL-4(−) cells)] values are shown for each individual patient (Δmean=3.44, where p1=patient 1, p2=patient 2, and so on; n=5). Similarly, the Ct values of c-maf mRNA (B) and IL-4 mRNA (C) were calculated, and the p values are 0.001 and 0.0002, respectively, when comparing between IL-4(+) and IL-4(−) cells. The -ΔCt calculated [-(Ct of IL-4(+) minus Ct of IL-4(−) cells)] values of c-maf mRNA and IL-4 mRNA are shown for each individual patient (Δmean=15.51 for c-maf mRNA, =25.89 for IL-4 mRNA, where p1=patient 1, p2=patient 2, and so on; n=5).
Increased HIV-1 LTR transcriptional activity is detected in IL-4 producing CD4 T cells
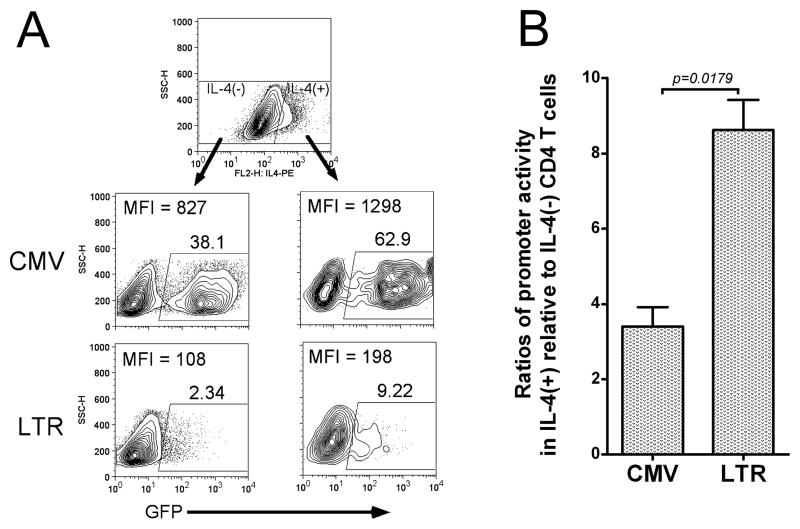
In order to explore a role for differential HIV-1 LTR transcriptional regulation in IL-4(+) versus IL-4(−) CD4 T cells, IL-4 polarized human CD4 T cells were infected with a GFP-expressing recombinant lentivirus driven by the HIV-1 LTR. A CMV-driven GFP lentiviral infection served as a relative control. As shown in Fig. 3A, both CMV and LTR transcriptional activities are higher in IL-4 producing T cells than in non-IL-4 producing cells. Nevertheless, the promoter activity ratio for IL-4(+) relative to IL-4(−) CD4 T cells driven by the HIV-1 LTR (8.62±1.40) is significantly (p=0.0179) higher than that from the CMV immediate early promoter (3.4±0.90) (Fig. 3B). Interestingly, the LTR activity ratio calculated in these in vitro experiments [8.6-fold higher in IL-4(+) cells] is very close to the difference in HIV-1 mRNA levels detected in the in vivo patient samples (Fig. 2A, ΔCt=3.44, mRNA level in IL-4 producing cells is 23.44=10.8 times higher than in non-IL-4 producing cells). Therefore, HIV-1 LTR-directed transcription is substantially higher in IL-4(+) than IL-4(−) cells and correlates with increased HIV-1 mRNA levels.
Figure 3. HIV-1 LTR transcriptional activity is increased in IL-4(+) CD4 T Cells.
(A) IL-4 polarized human CD4 T cells were infected by a recombinant GFP-expressing lentiviruses driven by either the immediate-early CMV promoter or HIV-1 LTR. GFP expression and IL-4 production were detected by flow cytometry. Results from one representative experiment out of 5 are depicted demonstrating that both CMV and LTR promoters show higher transcriptional activity in IL-4(+) CD4 T cells than in IL-4(−) cells, but HIV-1 LTR transcriptional activity was notably higher than CMV promoter activity in IL-4(+) cells relative to IL-4(−) cells. (B) The mean ratios of transcriptional promoter activities of IL-4(+) cells relative to IL-4(−) cells were calculated [percent positive multiplied by MFI in IL-4(+) cells divided by percent positive times MFI in IL-4(−) cells)] and presented as bar graphs. The HIV-1 LTR shows a higher relative ratio [IL-4(+)/IL-4(−)] of transcriptional activity than CMV [mean±SEM of ratio for CMV promoter: 3.4±0.90; for HIV-1 LTR: 8.62±1.40; p = 0.0179 using a student t-test (n=5)].
c-maf, NFAT2, and NFκB p65 preferentially bind the proximal HIV-1 LTR
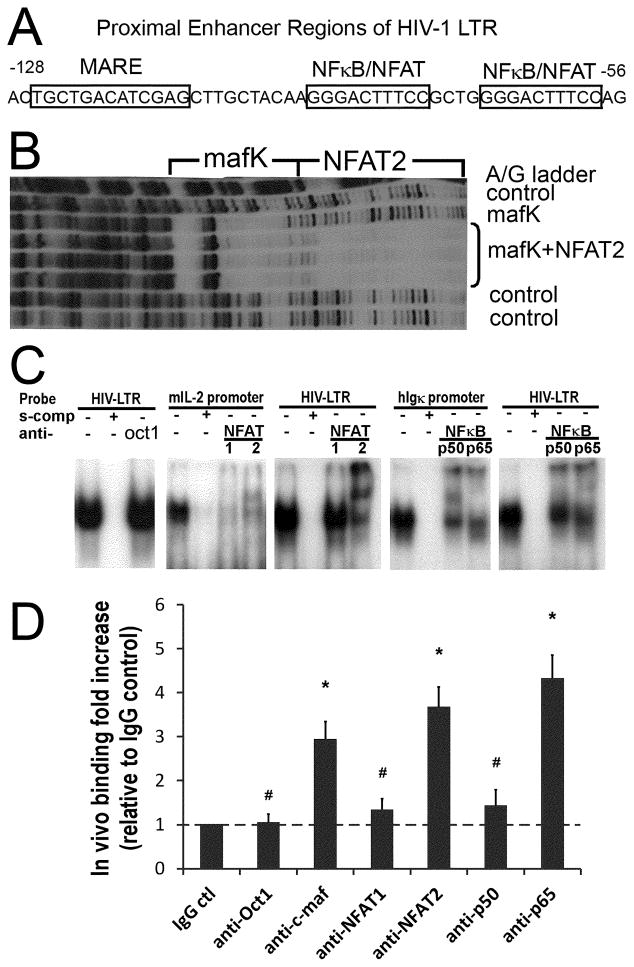
Since increased HIV-1 expression in IL-4(+) cells correlated with increased HIV-1 mRNA levels and transcription rates, we next focused on factors critical to IL-4 transcription. In particular, the Th2 restricted transcription factor, c-maf, binds to a MARE in the proximal IL-4 promoter and cooperates with a neighboring NFAT binding site to specifically enhance transcription of IL-4 (24, 35). In comparison with a c-maf consensus-binding sequence (43), 5′TGCTGACTCAGCA, a novel homologous sequence, 5′TGCTGACATCGAG, was identified and located just upstream (5′) of the dual NFκB/NFAT sites in the HIV-1 proximal LTR (Fig. 4A). We next explored the ability of c-maf to bind the HIV-1 LTR.
Figure 4. c-maf, NFAT2, and NFκB p65 preferentially bind the proximal HIV-1 LTR.
(A) The dual NFκB/NFAT binding sites and the predicted c-maf binding site (MARE) in the proximal HIV-1 LTR are depicted. (B) The in vitro footprint of the HIV-1 LTR DNase I digestion shows a mafK induced footprint of the LTR just upstream and adjacent to the NFAT2 footprint. (C) Nuclear extracts prepared from activated IL-4 primed human CD4 T cells bind the HIV-1 LTR NFκB/NFAT dual site probe (panel 1, 3, 5), the mouse proximal IL-2 promoter NFAT site (panel 2), and the human Igκ promoter NFκB site (panel 4) in gel shift assays (see Materials and Methods for details). NFAT1 and NFAT2 bind the IL-2 promoter similarly (panel 2), whereas, NFAT2 preferentially binds over NFAT1 to the HIV-1 proximal LTR (panel 3). Likewise, NFκB p65 and p50 bind the Igκ promoter NFκB site similarly (panel 4), but NFκB p65 binds preferentially, relative to NFκB p50, to the proximal HIV-1 LTR (panel 5). Oct1 protein acts as a control that does not bind to the proximal HIV-1 LTR (panel 1). Results are representative of 3 independent experiments. (D) Real-Time PCR was performed to quantify transcription factors bound to the integrated HIV-1 LTR in vivo as detected by ChIP. The results reveal c-maf, and NFAT2 over NFAT1, and NFκB p65 over p50, preferentially bind to the HIV-1 LTR in vivo. Rabbit IgG and anti-Oct1 antibodies are negative controls. The relative fold increases of transcription factor binding are calculated in comparison with the rabbit IgG control. # p>0.05 when comparing with IgG control; * p<0.05 when comparing with IgG control; n=5.
c-maf belongs to a large family of maf proteins, all of which share a very homologous DNA-binding domain (44). To facilitate preparation of recombinant protein, we tested the ability of mafK, a small maf family member with a virtually identical DNA binding domain to c-maf, to bind to the proximal HIV-1 LTR by in vitro footprinting. mafK clearly protected DNA residues from DNase I digestion (i.e., bound the LTR in vitro) corresponding to the predicted MARE (Fig. 4B). Moreover, recombinant NFAT2 in conjunction with mafK extended the protected residues through the NFκB/NFAT region (Fig. 4B). Interestingly, even though there are 10 residues between the predicted MARE and the NFκB/NFAT region, the site protected by mafK is adjacent to the NFAT2 binding site. This suggests the 2 factors may interact and potentially cooperate when binding to the proximal HIV-1 LTR.
The ability of NFAT and NFκB proteins present in polarized human CD4 T cells to bind in conjunction with c-maf on the proximal HIV-1 LTR was then explored. Prior work, including our own, has shown that recombinant NFκB family members, p65 and p50, and NFAT family members, NFAT1 and NFAT2, are all capable of binding to the HIV-1 LTR NFκB/NFAT sites in vitro (29, 31). In the present studies, the LTR NFκB/NFAT dual site probe was incubated with activated IL-4 polarized CD4 T cell nuclear extracts in the presence of specific anti-transcription factor antibodies during the gel shift assays. Interestingly, NFκB p65 is preferentially present in the shifted complexes relative to NFκB p50, and NFAT2 preferentially binds the LTR over NFAT1 (Fig. 4C). This preferential binding of NFκB p65 and NFAT2 is unique to the HIV-1 LTR and not a function of the particular supershifting antibodies, as NFκB p50, NFκB p65, NFAT1, and NFAT2 contained in the identical nuclear extracts were all relatively equally supershifted by the same antibodies when tested for binding to NFκB and NFAT sites derived from the Igκ and IL-2 promoters, respectively (Fig. 4C). Thus, the preferential binding of NFκB p65 and NFAT2 to the HIV-1 LTR may reflect subtle differences in the HIV-1 LTR DNA binding or flanking sequences, and/or cooperative protein-protein interactions that can only be detected in physiologically relevant CD4 T cell nuclear extracts.
Binding of c-maf to the HIV-1 LTR in vitro was also explored by gel shift assay. A gel retarded band, from nuclear extracts from IL-4 primed CD4 T cells, migrated identically comparing a consensus MARE probe and a probe containing the putative HIV-1 LTR MARE (Supplemental Fig. 3). These bands were both fully competed with excess unlabeled self-probe, and they were both partially diminished in intensity (decreased by 48% for the consensus MARE probe, and decreased by 40% for the HIV-1 MARE probe, by densitometry) using a blocking anti-c-maf antibody (Supplemental Fig. 3). This suggests that c-maf present in IL-4 primed CD4 T cell nuclear extracts is capable of binding the HIV-LTR MARE in vitro just upstream of the dual NFκB/NFAT sites.
To confirm the binding of c-maf and the individual NFκB and NFAT family members to the HIV-1 LTR in vivo, IL-4 primed human CD4 T cells were infected with the NL4.3-GFP strain (45). ChIP assays were then performed using the purified GFP+ (infected) cells with anti-c-maf, -NFκB, and -NFAT antibodies, and immunoprecipitated DNA was quantified by real-time PCR. We previously demonstrated that octamer transcription factors do not bind the HIV-1 LTR in vitro (36), so anti-Oct1 was chosen as a negative transcription factor antibody control, in addition to the isotype control antibody. As shown in Fig. 4D, the fold-binding to the HIV-1 LTR for each transcription factor was calculated according to the respective Ct value of the specific transcription factor antibody relative to the IgG control antibody. Similar to in vitro binding results (Figs. 4B and 4C, and Supplemental Fig. 3), there was clear evidence for binding to the HIV-1 LTR in vivo for c-maf, NFκB p65, and NFAT2, but not for Oct1, NFκB p50, and NFAT1 (Fig. 4D). Not only did this imply a role for c-maf in HIV-1 regulation, but it confirmed the preferential binding of unique NFκB (p65 over p50) and NFAT (NFAT2 over NFAT1) transcription factor family members to the HIV-1 LTR.
c-maf cooperates with NFAT2 and NFκB p65 to augment HIV-1 LTR transcription
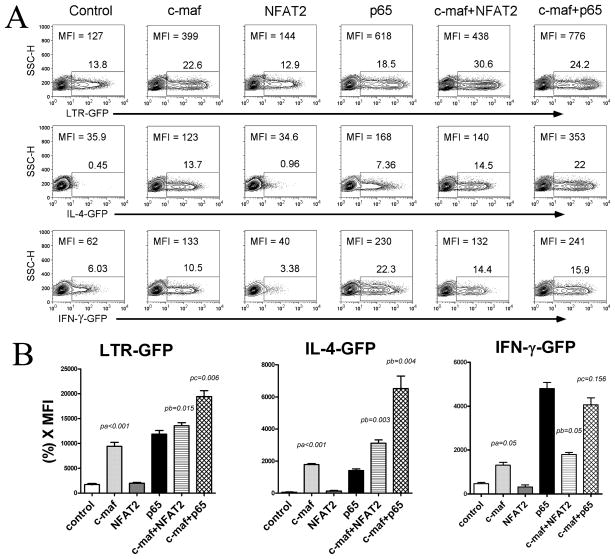
The functional significance of c-maf alone, c-maf plus NFAT2, or c-maf plus NFκB p65 to HIV-1 expression was next explored. First, primary human CD4 T cells were transiently transfected with an HIV-1 LTR-driven GFP reporter plasmid and co-tranfected with c-maf, NFAT2, NFκB p65, c-maf plus NFAT2, or c-maf plus NFκB p65 expression vectors. It is evident that c-maf alone increases HIV-1 LTR activity (MFI: 127 to 399) without increasing cell survival (less than 10% difference in cell number following transfection). The increased HIV-1 LTR transcriptional activity is further and significantly enhanced when co-expressing NFAT2 (MFI: 438) or NFκB p65 (MFI: 776) (Fig. 5A, top panel), suggesting a cooperative interaction between c-maf and NFAT2 and NFκB p65.
Figure 5. c-maf synergizes with NFAT2 and NFκB p65 to augment HIV-1 and hIL-4 promoter transcription.
(A) Primary human CD4 T cells were transfected with single or dual expression vector(s) together with HIV-1 LTR, hIL-4, or hIFN-γ promoter GFP reporter plasmids. GFP expression was detected by flow cytometry after 24 hours. Expression of c-maf alone is capable of increasing HIV-1 LTR and hIL-4 promoter activity, but not hIFN-γ promoter activity. This effect is further and significantly enhanced when co-transfecting c-maf and NFAT2, or c-maf and NFκB p65. A typical result out of 5 independent experiments is shown. (B) Promoter reporter activity was calculated as GFP percentage multiplied by MFI (%*MFI) and are expressed as means±SEM from 5 independent experiments. c-maf alone, and in cooperation with NFAT2 or NFκB p65, statistically significantly increase HIV-1 LTR and hIL-4 promoter activity (pa=p value, c-maf transfected cells in comparison with control; pb=p value, c-maf and NFAT2 dual transfected cells in comparison with c-maf alone; pc=p value, c-maf and NFκB p65 dual transfected cells in comparison with NFκB p65 alone; n=5).
Was this cooperative transcriptional activation by these factors relatively unique to the HIV-1 LTR? This possibility was explored by comparing hIL-4 promoter-, hIFN-γ promoter-, and HIV-1 LTR-driven GFP reporter gene activity in primary human CD4 T cells. As expected, c-maf notably increases HIV-1 LTR and hIL-4 promoter (MFI: 35.9 to 123) transcriptional activity, but only modestly augments hIFN-γ promoter activity (MFI: 62 to 133). Increased HIV-1 LTR and IL-4 promoter transcription can be augmented in the presence of either NFAT2 or NFκB p65 (Fig. 5A, top and middle panels). The activity of each promoter was calculated [%(+) cells × MFI] and averaged from 5 independent experiments and is shown in Fig. 5B. NFκB p65, but not NFAT2, increased individual promoter transcription. However, both significantly cooperate with c-maf to increase LTR and IL-4, but not IFN-γ promoter transcription. These results demonstrate that c-maf in cooperation with NFAT2 or NFκB p65 notably augments HIV-1 LTR and hIL-4 promoter, but not hIFN-γ promoter, transcription.
Inhibiting c-maf expression dramatically decreases HIV-1 LTR activity
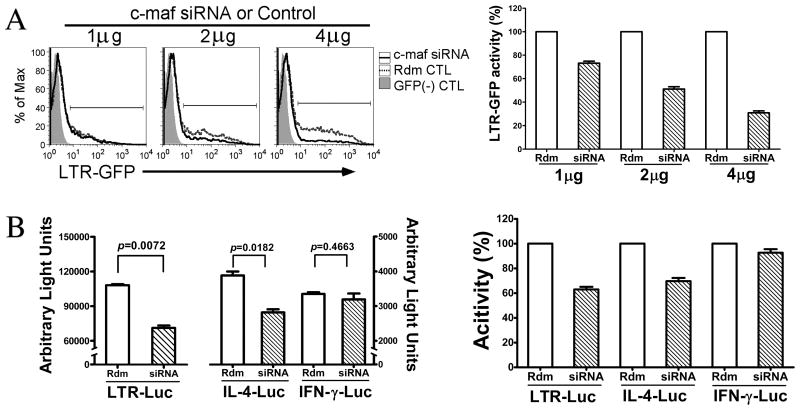
The role of endogenous c-maf during HIV-1 transcription was tested by introduction of c-maf-specific siRNA into IL-4 polarized CD4 T cells using our own published protocol (39). We first determined the efficiency and specificity of c-maf specific siRNA designed by ourselves by verifying decreased expression of endogenous c-maf in IL-4 polarized CD4 T cells at both the mRNA and protein levels, and a decreased activity of a c-maf responsive reporter plasmid, when transfecting c-maf siRNA into those cells (Supplemental Fig. 4A, B, and C, respectively). Therefore, the c-maf specific siRNA can be used to assess the role of endogenous c-maf during HIV-1 infection.
To first test HIV-1 LTR transcriptional activity, IL-4 producing CD4 T cells were co-transfected with an HIV-1 LTR-GFP reporter plasmid plus the c-maf-specific siRNA expression vector or scrambled control. Compared with control siRNA, HIV-1 transcriptional activity is notably diminished in the presence of c-maf siRNA. Moreover, increasing doses of the c-maf specific siRNA demonstrate dose-dependent inhibition of HIV-1 transcription (Fig. 6A, both panels). Similarly, c-maf specific siRNA inhibits hIL-4 promoter- but not hIFN-γ promoter-driven transcription in IL-4 producing CD4 T cells (Fig. 6B, left panel). When visualized as percentage inhibition of transcription averaged over several experiments, it is clear that c-maf specific siRNA inhibits both HIV-1 LTR- and hIL-4 promoter-driven transcription, but not transcription of the hIFN-γ promoter (Fig. 6B, right panel). Thus, the c-maf specific siRNA is specific to its target genes, and the results with its use suggest that endogenous c-maf present IL-4-producing CD4 T cells is not only important for optimal IL-4 gene transcription but for that of HIV-1 transcription as well.
Figure 6. Inhibition of endogenous c-maf expression decreases HIV-1 LTR and IL-4 promoter transcription.
(A) IL-4 polarized CD4 T cells were co-transfected with c-maf specific siRNA plus a HIV-1 LTR GFP reporter. GFP expression was detected 24 hours later by flow cytometry. GFP expression in c-maf siRNA transfected cells is decreased compared with the scrambled sequence control siRNA, and the inhibition is in a dose-dependent manner relative to the amount of transfected siRNA. Representative flow cytometry histograms of GFP expression at the different siRNA (solid line – c-maf; dashed line – scrambled control) concentrations are shown on the left. The percent LTR transcriptional activity, relative to the scrambled control, was calculated using the formula, [(MFI*GFP% in the c-maf siRNA sample)/(MFI*GFP% in scrambled control sample)], and a summary graph of the means±SEM LTR activities in the presence of escalating amounts of c-maf specific siRNA is depicted on the right (n=3). (B) IL-4 polarized CD4 T cells were co-transfected with scrambled control or c-maf specific siRNA plus various luciferase reporter plasmids as depicted. Transcriptional activities (relative light units) are shown as means±SEM of 3 similar experiments on the left. Percent transcriptional activities in the presence of c-maf siRNA for the LTR and IL-4 and IFN-γ promoters, relative to scrambled control, are shown on the right.
c-maf alone, and c-maf in cooperation with NFAT2 or NFκB p65, augment HIV-1 expression
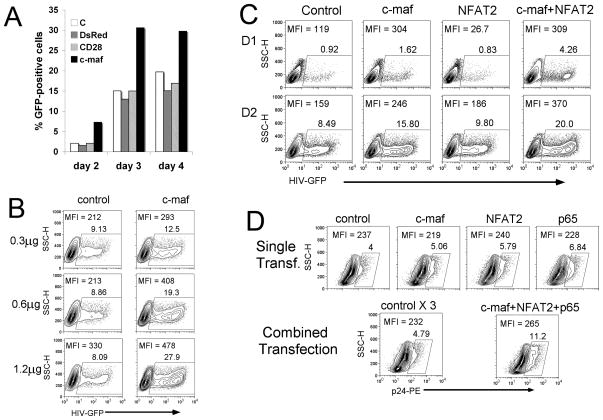
Based on c-maf binding to the proximal HIV-1 LTR and its ability to increase HIV-1 LTR transcription, we next examined the effect of c-maf on HIV-1 replication. To generate stable c-maf expression, a c-maf expressing retrovirus was used to infect Jurkat T cells prior to the infection of HIV-1 NL4-3-GFP. HIV-1 expression, as detected by GFP, was analyzed 2–4 days following HIV-1 infection. In comparison to 3 different control expression retroviruses (empty, DsRed, CD28), c-maf clearly led to increased HIV-1 infection at all 3 time points (Fig. 7A). Using another model system, HEK-293T cells were transfected with increasing amounts of a c-maf expression plasmid followed by infection with HIV-1 NLENG-vsvg-GFP. In comparison with the control vector, c-maf was capable of increasing HIV-1 expression in a dose dependent manner (Fig. 7B). Thus, enhanced c-maf expression led to increased HIV-1 expression in 2 different cell lines.
Figure 7. c-maf alone, or in cooperation with NFAT2 and NFkB p65, increases HIV-1 expression.
(A) Jurkat T cells were infected with recombinant retroviruses that express nothing, deRed2, CD28, or c-maf prior to infection of a HIV-1-GFP reporter virus. GFP expression was analyzed at 2, 3, and 4 days post-infection. c-maf significantly increases HIV-1 replication compared with the controls. One representative result out of 3 independent experiments is shown. (B) HEK-293T cells were transfected with increasing amounts of c-maf expression or control plasmid 12–16 hours prior to HIV-1-GFP virus infection. GFP expression was detected by flow cytometry 24 hours following infection. c-maf increases HIV-1 infection in a dose dependent fashion. Results from 1 of 3 representative experiments are shown. (C). HEK-293T cells were transfected with control, c-maf, NFAT2, or c-maf plus NFAT2, expression plasmids, 12–16 hours prior to HIV-1-GFP virus infection. GFP expression was detected by flow cytometry at days 1 and 2 after infection. c-maf notably increases HIV-1 expression and this is augmented in the presence of NFAT2 co-transfection. One typical result out of 3 independent experiments is shown. (D) Freshly isolated human peripheral blood CD4 T cells were transfected with c-maf, NFAT2, or NFκB p65 alone, or the combination of all 3 factors. The control plasmid was also transfected in 1x or 3x quantities of DNA to serve as a negative control. Cells were subsequently infected with the HIV-1 NL4-3 virus, and HIV-1 expression was detected by flow cytometry (intracellular p24 staining) at day 3 post-infection. HIV-1 p24 expression is modestly enhanced when individual transcription factors are expressed; however, HIV-1 expression is notably augmented when all 3 factors are co-expressed, compared with the 3x control. Results are representative of 1 of 3 similar experiments.
Since c-maf is known to cooperate with NFAT in driving IL-4 transcription (46), we explored the ability of c-maf to cooperate with NFAT2 in driving HIV-1 expression. HEK-293T cells were transfected with c-maf alone, NFAT2 alone, or co-transfected with c-maf and NFAT2 expression vectors. While NFAT2 did not increase HIV-1 expression alone, NFAT2 clearly augments c-maf enhanced HIV-1 expression, particularly at day 2 post infection (Fig. 7C). Most importantly, we wished to confirm this effect in primary human CD4 T cells. CD4 T cells were first transfected with control, c-maf, NFAT2, or NFκB p65 expression vectors prior to HIV-1 NL4-3 infection. HIV-1 expression was analyzed at 3–5 days post-infection. While none of the transcription factors in isolation dramatically augments HIV-1 expression in primary CD4 T cells (Fig. 7D, top row), when all 3 expression vectors (c-maf, NFAT2, and NFκB p65) were co-transfected, the combined over-expression of these 3 factors notably augments HIV-1 viral expression (Fig. 7D, bottom row). Thus, there does appear to be a cooperative/synergistic effect of c-maf, NFAT2, and NFκB p65 on increasing HIV-1 expression in primary human CD4 T cells.
Discussion
In exploring a previously noted shift from a Th1- to Th2/Th0 phenotype occurring during HIV-1 infection and progression to AIDS, investigators found a bias of HIV-1 replication in different CD4 T cell subsets or clones (8, 9). Focusing on HIV-1 infection and expression in individual cells from a bulk culture, we confirm that HIV-1 preferentially replicates in either Th2 or Th0, namely IL-4 producing, cells. Attempting to explain this phenomenon, others have previously reported that cloned Th1 cells are more susceptible to HIV-1 induced apoptosis (47). However, using cytokine primed bulk CD4 T cells, we demonstrate that preferential HIV-1 expression is not secondary to a rapid growth of IL-4 producing cells (Fig. 1D), nor related to an increased rate of HIV-1 triggered apoptosis of non-IL-4 producing cells (Supplemental Fig. 1). Moreover, we show that increased HIV-1 expression in IL-4(+) cells is independent of viral tropism (Fig. 1B) or the level of chemokine receptor expression (Supplemental Fig. 2). These results are consistent with the majority of previous reports (10–14, 18), but different from others where the HIV-1 R5 strain alone, or both R5 and X4 strains, were found to replicate (equally) in Th1 and Th2 cells (48–50). Although our in vitro infection is not identical to the infection occurring in vivo, by using primary human CD4 T cells in bulk culture it more closely resembles the natural infection process than cell lines or clones. Therefore, we believe our results are pathophysiologically relevant.
The findings of high levels of the ex vivo HIV-1 mRNA in IL-4 producing CD4 T cells from infected individuals, and increased HIV-1 LTR activity in IL-4-polarized human CD4 T cells in vitro, stimulated exploration of whether an IL-4-restricted transcriptional mechanism was involved in optimal HIV-1 gene expression. Accordingly, we identified a functional binding site for the Th2-restricted transcription factor, c-maf, in the proximal region of the HIV-1 LTR adjacent to the dual NFκB/NFAT sites. In addition, NFκB factors, p65 over p50, and NFAT factors, NFAT2 over NFAT1, were shown to preferentially bind to the LTR dual NFκB/NFAT sites. It has reported that both p65 and NFAT2 are increased in human IL-4 expression (51, 52). In this regard, a synergistic effect between c-maf and NFAT2, or between c-maf and p65, specifically increased HIV-1 LTR and hIL-4 promoter, but not hIFN-γ promoter, transcriptional activity. Taken together, c-maf contributes to increased HIV-1 transcription in IL-4 producing T cells via binding to the proximal LTR in cooperation with specific NFAT (NFAT2) and NFκB (p65) family members.
Based on our findings, as well as a recent report describing a physical interaction specifically between NFAT2 and NFκB p65 which synergistically promotes cardiac hypertrophy and ventricular remodeling (53), we postulate that a similar cooperative interaction among c-maf, NFAT2, and NFκB p65 contributes to increased HIV-1 transcription in IL-4(+) CD4 T cells. Moreover, this synergism is likely an important mechanism contributing to the frequently observed preferential replication of HIV-1 in IL-4 producing CD4 T cells. Nevertheless, HIV-1 may lower c-maf expression, as recently suggested by Ahmad and colleagues (54), thus placing more importance on a cooperative interaction of transcription factors needed to promote HIV-1 transcription.
HIV-1 gene transcription and expression regulate viral replication, and consequently influence AIDS progression. Along these lines, it has been reported that HIV-1 LTR transcriptional activity is increased 3-fold and 10-fold in neonatal cord blood T lymphocytes and macrophages, respectively, in comparison to adult blood cells (55). The authors conclude that it is increased viral gene expression derived from enhanced LTR transcription, rather than differences in cell proliferative capacities, cells surface receptor expression, viral RNA genome reverse transcriptional activity, or translocation of the pre-integration complex into the nucleus, that directly results in a higher level of viremia and faster disease progression in neonates than adults (55). They further identify a complex transcriptional mechanism regulating control of HIV-1 gene transcription and expression in neonatal mononuclear cells (20). In the present study, we find an 8.6-fold higher HIV-1 LTR transcriptional activity and almost 2-fold higher HIV-1 p24 expression in IL-4 producing CD4 T cells compared to non-IL-4 producing T cells. Therefore, one may postulate that viral production in Th2 and Th0 cells substantially contributes to viral load and viremia during HIV-1 infection and AIDS progression. Over a decade or more of infection, even 2-fold differences in viral production will give rise to substantial differences in total viral burden. Considering the hypothesis that a Th1 to Th2 (or Th0) shift occurs under certain circumstances during HIV-1 infection (8, 9, 56), it seems reasonable that relatively high levels of IL-4 will help maintain the stability of the Th2 compartment and concomitantly inhibit the generation of the Th1 compartment. This scenario predicts facilitation of HIV-1 proliferation via the virus taking advantage of Th2 cell-specific transcriptional machinery to optimize its replication while avoiding Th1-mediated anti-viral immune responses. Moreover, individuals who resist HIV-1 infection by secreting an IL-4 variant with a deletion of exon 2, a natural antagonist of IL-4, further support pivotal roles of IL-4 and Th2 cells during HIV-1 infection (57, 58). Taking together these previous reports and our present study, we propose that the preferential support of HIV-1 replication by IL-4 producing T cells, along with the relative cell stability in the presence of IL-4, are critical pathophysiologic consequences of HIV-1 infection and AIDS progression. Our findings are of significant importance in helping to increase our current understanding of HIV-1 biology and AIDS pathogenesis.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Dr. David B. Lewis (Stanford University, Palo Alto, CA, USA), in whose laboratory this work was begun, Dr. Michael Brunner (Humigen, Hamilton, NJ, USA) for technical assistance, and Dr. Timothy Beukelman (University of Alabama at Birmingham, AL, USA) for statistical support. E.C. was a Pediatric AIDS Foundation scholar.
This was work was supported by a grant from the Elizabeth Glaser Pediatric AIDS Foundation and by NIH R21 AI-68574, both to R.Q.C. R.M.L. was supported by the Creative and Novel Ideas in HIV Research program. This program is funded through a supplement to the University of Alabama at Birmingham (UAB) Center for AIDS Research (CFAR), grant number P30 AI027767, and this funding is made possible by NIAID and the Office of AIDS Research.
Footnotes
Contribution: Conceived and designed experiments: M.Z., A.C., S.S., F.H.W., P.A.G., O.K., R.M.L., R.Q.C. Performed experiments: M.Z., A.C., E.C., L.J., G.W., T.R., S.S., F.H.W., R.Q.C. Analyzed the data: M.Z., A.C., S.S., F.H.W., P.A.G., O.K., R.M.L., R.Q.C. Contributed reagents/materials/analysis tools: M.Z., A.C., J.Y., F.H.W., P.A.G., O.K., R.Q.C. Wrote the paper: M.Z., R.Q.C.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 2.Hart JE, Jeon CY, Ivers LC, Behforouz HL, Caldas A, Drobac PC, Shin SS. Effect of directly observed therapy for highly active antiretroviral therapy on virologic, immunologic, and adherence outcomes: a meta-analysis and systematic review. J Acquir Immune Defic Syndr. 2010;54:167–179. doi: 10.1097/QAI.0b013e3181d9a330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bongaarts J, Over M. Public health. Global HIV/AIDS policy in transition. Science. 328:1359–1360. doi: 10.1126/science.1191804. [DOI] [PubMed] [Google Scholar]
- 4.Bunupuradah T, Puthanakit T, Kosalaraksa P, Kerr S, Boonrak P, Prasitsuebsai W, Lumbiganon P, Mengthaisong T, Phasomsap C, Pancharoen C, Ruxrungtham K, Ananworanich J. Immunologic and virologic failure after first-line NNRTI-based antiretroviral therapy in Thai HIV-infected children. AIDS Res Ther. 2011;8:40. doi: 10.1186/1742-6405-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantor R, Diero L, Delong A, Kamle L, Muyonga S, Mambo F, Walumbe E, Emonyi W, Chan P, Carter EJ, Hogan J, Buziba N. Misclassification of first-line antiretroviral treatment failure based on immunological monitoring of HIV infection in resource-limited settings. Clin Infect Dis. 2009;49:454–462. doi: 10.1086/600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ledergerber B, Lundgren JD, Walker AS, Sabin C, Justice A, Reiss P, Mussini C, Wit F, d’Arminio Monforte A, Weber R, Fusco G, Staszewski S, Law M, Hogg R, Lampe F, Gill MJ, Castelli F, Phillips AN. Predictors of trend in CD4-positive T-cell count and mortality among HIV-1-infected individuals with virological failure to all three antiretroviral-drug classes. Lancet. 2004;364:51–62. doi: 10.1016/S0140-6736(04)16589-6. [DOI] [PubMed] [Google Scholar]
- 7.Trotta MP, Cozzi-Lepri A, Ammassari A, Vecchiet J, Cassola G, Caramello P, Vullo V, Soscia F, Chiodera A, Ladisa N, Abeli C, Cauda R, Buonuomi AR, Antinori A, d’Arminio Monforte A. Rate of CD4+ cell count increase over periods of viral load suppression: relationship with the number of previous virological failures. Clin Infect Dis. 2011;51:456–464. doi: 10.1086/655151. [DOI] [PubMed] [Google Scholar]
- 8.Maggi E, Mazzetti M, Ravina A, Annunziato F, de Carli M, Piccinni MP, Manetti R, Carbonari M, Pesce AM, del Prete G, et al. Ability of HIV to promote a TH1 to TH0 shift and to replicate preferentially in TH2 and TH0 cells. Science. 1994;265:244–248. doi: 10.1126/science.8023142. [DOI] [PubMed] [Google Scholar]
- 9.Romagnani S, Maggi E, Del Prete G. HIV can induce a TH1 to TH0 shift, and preferentially replicates in CD4+ T-cell clones producing TH2-type cytokines. Res Immunol. 1994;145:611–617. doi: 10.1016/s0923-2494(05)80042-2. discussion 617–618. [DOI] [PubMed] [Google Scholar]
- 10.Moonis M, Lee B, Bailer RT, Luo Q, Montaner LJ. CCR5 and CXCR4 expression correlated with X4 and R5 HIV-1 infection yet not sustained replication in Th1 and Th2 cells. Aids. 2001;15:1941–1949. doi: 10.1097/00002030-200110190-00005. [DOI] [PubMed] [Google Scholar]
- 11.Ofori H, Jagodzinski PP. Increased in vitro replication of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates in Th2 lymphocytes may correlate with AIDS progression. Scand J Infect Dis. 2004;36:46–51. doi: 10.1080/00365540310017087. [DOI] [PubMed] [Google Scholar]
- 12.Ofori H, Prokop J, Jagodzinski PP. Replication at the level of reverse transcription of HIV-1SF2 strain in Th1 and Th2 cells. Biomed Pharmacother. 2003;57:15–19. doi: 10.1016/s0753-3322(02)00330-x. [DOI] [PubMed] [Google Scholar]
- 13.Vicenzi E, Panina-Bodignon P, Vallanti G, Di Lucia P, Poli G. Restricted replication of primary HIV-1 isolates using both CCR5 and CXCR4 in Th2 but not in Th1 CD4(+) T cells. J Leukoc Biol. 2002;72:913–920. [PubMed] [Google Scholar]
- 14.Vyakarnam A, Matear PM, Martin SJ, Wagstaff M. Th1 cells specific for HIV-1 gag p24 are less efficient than Th0 cells in supporting HIV replication, and inhibit virus replication in Th0 cells. Immunology. 1995;86:85–96. [PMC free article] [PubMed] [Google Scholar]
- 15.Rubbo PA, Tuaillon E, Bollore K, Foulongne V, Bourdin A, Nagot N, Van de Perre P, Desgranges C, Israel-Biet D, Vendrell JP. The potential impact of CD4+ T cell activation and enhanced Th1/Th2 cytokine ratio on HIV-1 secretion in the lungs of individuals with advanced AIDS and active pulmonary infection. Clin Immunol. 2011;139:142–154. doi: 10.1016/j.clim.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer JM. CCR5 is characteristic of Th1 lymphocytes. Nature. 1998;391:344–345. doi: 10.1038/34814. [DOI] [PubMed] [Google Scholar]
- 17.Jagodzinski PP, Trzeciak WH. Differential expression of chemokine receptor CXCR4 in Th1 and Th2 subtypes of CD4+ lymphocytes. Folia Histochem Cytobiol. 2000;38:21–23. [PubMed] [Google Scholar]
- 18.Annunziato F, Galli G, Nappi F, Cosmi L, Manetti R, Maggi E, Ensoli B, Romagnani S. Limited expression of R5-tropic HIV-1 in CCR5-positive type 1-polarized T cells explained by their ability to produce RANTES, MIP-1alpha, and MIP-1beta. Blood. 2000;95:1167–1174. [PubMed] [Google Scholar]
- 19.Ansari AW, Heiken H, Meyer-Olson D, Schmidt RE. CCL2: a potential prognostic marker and target of anti-inflammatory strategy in HIV/AIDS pathogenesis. Eur J Immunol. 2011;41:3412–3418. doi: 10.1002/eji.201141676. [DOI] [PubMed] [Google Scholar]
- 20.Sundaravaradan V, Mehta R, Harris DT, Zack JA, Ahmad N. Differential expression and interaction of host factors augment HIV-1 gene expression in neonatal mononuclear cells. Virology. 2010;400:32–43. doi: 10.1016/j.virol.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci U S A. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 23.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 25.Kim JI, I, Ho C, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–751. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, Ochando JC, Bromberg JS, Ding Y. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida H, Nishina H, Takimoto H, Marengere LE, Wakeham AC, Bouchard D, Kong YY, Ohteki T, Shahinian A, Bachmann M, Ohashi PS, Penninger JM, Crabtree GR, Mak TW. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity. 1998;8:115–124. doi: 10.1016/s1074-7613(00)80464-1. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 29.Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin Immunol. 2000;94:179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- 30.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 31.Jacque JM, Fernandez B, Arenzana-Seisdedos F, Thomas D, Baleux F, Virelizier JL, Bachelerie F. Permanent occupancy of the human immunodeficiency virus type 1 enhancer by NF-kappa B is needed for persistent viral replication in monocytes. J Virol. 1996;70:2930–2938. doi: 10.1128/jvi.70.5.2930-2938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chene L, Nugeyre MT, Barre-Sinoussi F, Israel N. High-level replication of human immunodeficiency virus in thymocytes requires NF-kappaB activation through interaction with thymic epithelial cells. J Virol. 1999;73:2064–2073. doi: 10.1128/jvi.73.3.2064-2073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O. Determinants of the establishment of human immunodeficiency virus type 1 latency. J Virol. 2009;83:3078–3093. doi: 10.1128/JVI.02058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–3492. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cron RQ, Bort SJ, Wang Y, Brunvand MW, Lewis DB. T cell priming enhances IL-4 gene expression by increasing nuclear factor of activated T cells. J Immunol. 1999;162:860–870. [PubMed] [Google Scholar]
- 36.Zhang M, Genin A, Cron RQ. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology. 2004;321:323–331. doi: 10.1016/j.virol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Calabresi PA, Allie R, Mullen KM, Yun SH, Georgantas RW, 3rd, Whartenby KA. Kinetics of CCR7 expression differ between primary activation and effector memory states of T(H)1 and T(H)2 cells. J Neuroimmunol. 2003;139:58–65. doi: 10.1016/s0165-5728(03)00127-9. [DOI] [PubMed] [Google Scholar]
- 38.Cron RQ. CD154 transcriptional regulation in primary human CD4 T cells. Immunol Res. 2003;27:185–202. doi: 10.1385/IR:27:2-3:185. [DOI] [PubMed] [Google Scholar]
- 39.Yin J, Ma Z, Selliah N, Shivers DK, Cron RQ, Finkel TH. Effective gene suppression using small interfering RNA in hard-to-transfect human T cells. J Immunol Methods. 2006;312:1–11. doi: 10.1016/j.jim.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 40.Selliah N, Finkel TH. HIV-1 NL4–3, but not IIIB, inhibits JAK3/STAT5 activation in CD4(+) T cells. Virology. 2001;286:412–421. doi: 10.1006/viro.2001.0994. [DOI] [PubMed] [Google Scholar]
- 41.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, Lukashov VV. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki Y, Koyanagi Y, Tanaka Y, Murakami T, Misawa N, Maeda N, Kimura T, Shida H, Hoxie JA, O’Brien WA, Yamamoto N. Determinant in human immunodeficiency virus type 1 for efficient replication under cytokine-induced CD4(+) T-helper 1 (Th1)- and Th2-type conditions. J Virol. 1999;73:316–324. doi: 10.1128/jvi.73.1.316-324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutsch O, Benveniste EN, Shaw GM, Levy DN. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol. 2002;76:8776–8786. doi: 10.1128/JVI.76.17.8776-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodge MR, Chun HJ, Rengarajan J, Alt A, Lieberson R, Glimcher LH. NF-AT-Driven interleukin-4 transcription potentiated by NIP45. Science. 1996;274:1903–1905. doi: 10.1126/science.274.5294.1903. [DOI] [PubMed] [Google Scholar]
- 47.Accornero P, Radrizzani M, Delia D, Gerosa F, Kurrle R, Colombo MP. Differential susceptibility to HIV-GP120-sensitized apoptosis in CD4+ T-cell clones with different T-helper phenotypes: role of CD95/CD95L interactions. Blood. 1997;89:558–569. [PubMed] [Google Scholar]
- 48.Mikovits JA, Taub DD, Turcovski-Corrales SM, Ruscetti FW. Similar levels of human immunodeficiency virus type 1 replication in human TH1 and TH2 clones. J Virol. 1998;72:5231–5238. doi: 10.1128/jvi.72.6.5231-5238.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salgame P, Guan MX, Agahtehrani A, Henderson EE. Infection of T cell subsets by HIV-1 and the effects of interleukin-12. J Interferon Cytokine Res. 1998;18:521–528. doi: 10.1089/jir.1998.18.521. [DOI] [PubMed] [Google Scholar]
- 50.Vicenzi E, Bordignon PP, Biswas P, Brambilla A, Bovolenta C, Cota M, Sinigaglia F, Poli G. Envelope-dependent restriction of human immunodeficiency virus type 1 spreading in CD4(+) T lymphocytes: R5 but not X4 viruses replicate in the absence of T-cell receptor restimulation. J Virol. 1999;73:7515–7523. doi: 10.1128/jvi.73.9.7515-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranger AM, Hodge MR, Gravallese EM, Oukka M, Davidson L, Alt FW, de la Brousse FC, Hoey T, Grusby M, Glimcher LH. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity. 1998;8:125–134. doi: 10.1016/s1074-7613(00)80465-3. [DOI] [PubMed] [Google Scholar]
- 52.Li-Weber M, Salgame P, Hu C, Davydov IV, Laur O, Klevenz S, Krammer PH. Th2-specific protein/DNA interactions at the proximal nuclear factor-AT site contribute to the functional activity of the human IL-4 promoter. J Immunol. 1998;161:1380–1389. [PubMed] [Google Scholar]
- 53.Liu Q, Chen Y, Auger-Messier M, Molkentin JD. Interaction between NFkappaB and NFAT coordinates cardiac hypertrophy and pathological remodeling. Circ Res. 2012;110:1077–1086. doi: 10.1161/CIRCRESAHA.111.260729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iannello A, Boulassel MR, Samarani S, Debbeche O, Tremblay C, Toma E, Routy JP, Ahmad A. Dynamics and consequences of IL-21 production in HIV-infected individuals: a longitudinal and cross-sectional study. J Immunol. 2010;184:114–126. doi: 10.4049/jimmunol.0901967. [DOI] [PubMed] [Google Scholar]
- 55.Sundaravaradan V, Saxena SK, Ramakrishnan R, Yedavalli VR, Harris DT, Ahmad N. Differential HIV-1 replication in neonatal and adult blood mononuclear cells is influenced at the level of HIV-1 gene expression. Proc Natl Acad Sci U S A. 2006;103:11701–11706. doi: 10.1073/pnas.0602185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clerici M, Shearer GM. A TH1-->TH2 switch is a critical step in the etiology of HIV infection. Immunol Today. 1993;14:107–111. doi: 10.1016/0167-5699(93)90208-3. [DOI] [PubMed] [Google Scholar]
- 57.Becker Y. The molecular mechanism of human resistance to HIV-1 infection in persistently infected individuals--a review, hypothesis and implications. Virus genes. 2005;31:113–119. doi: 10.1007/s11262-005-2503-5. [DOI] [PubMed] [Google Scholar]
- 58.Becker Y. The spreading of HIV-1 infection in the human organism is caused by fractalkine trafficking of the infected lymphocytes--a review, hypothesis and implications for treatment. Virus genes. 2007;34:93–109. doi: 10.1007/s11262-006-0056-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.