Abstract
Kinesin motor proteins transport a wide variety of molecular cargoes in a spatially and temporally regulated manner. Kinesin motor domains, which hydrolyze ATP to produce a directed mechanical force along a microtubule, are well conserved throughout the entire superfamily. Outside of the motor domains, kinesin sequences diverge along with their transport functions. The non-motor regions, particularly the tails, respond to a wide variety of structural and molecular cues that enable kinesins to carry specific cargoes in response to particular cellular signals. Here, we demonstrate that intrinsic disorder is a common structural feature of kinesins. A bioinformatics survey of the full-length sequences of all 43 human kinesins predicts that significant regions of intrinsically disordered residues are present in all kinesins. These regions are concentrated in the non-motor domains, particularly in the tails and near sites for ligand binding or post-translational modifications. In order to experimentally verify these predictions, we expressed and purified the tail domains of kinesins representing three different families (Kif5B, Kif10, and KifC3). Circular dichroism (CD) and NMR spectroscopy experiments demonstrate that the isolated tails are disordered in vitro, yet they retain their functional microtubule-binding activity. Based on these results, we propose that intrinsic disorder is a common structural feature that confers functional specificity to kinesins.
Keywords: kinesin, tail domain, structure prediction, intrinsic disorder, circular dichroism, NMR spectroscopy
Introduction
Kinesin motor proteins are involved in a myriad of cellular processes, from the transport of neurotransmitters, to mitosis, to the distribution of mRNA during development1-3. All kinesins have a common motor domain that, in most instances, hydrolyzes ATP and reversibly binds to microtubules (MTs). In addition to the motor domain, most kinesins contain coiled-coil stalk domains of up to several hundred residues that allow the kinesins to form multimers, and a tail domain adjacent to the end of the stalks opposite the motor (Figure S1). There are 43 known members of the human kinesin superfamily, and each one appears to be tailored to its specific cellular functions via its non-motor domains (reviewed by Hirokawa et al, 20094). It follows that, unlike the superfamily-conserved motor domains, kinesin non-motor domains are structurally diverse.
The literature contains detailed and sophisticated structural and functional studies of kinesin motor domains from several families. In contrast, there are relatively few investigations of the stalk or tail domains. Even for the most well-studied flagship kinesin motor, Kif5B, the mechanisms of cargo selection and regulation are still being discovered5-7. C-terminal to the motor, Kif5B (conventional kinesin) contains a mostly coiled-coil stalk domain of roughly 550 residues followed by a 50 residue tail region of unknown structure. The Kif5B tail is auto-regulatory, binding directly to the motor domains to inhibit their ATPase activity5,8. However, the same region of the tail binds specifically to several cargo proteins, including microtubules (MTs)9, Fez110,11, and RanBP212. The Kif5B tail is therefore a critical node for regulation and cargo selection. Frustratingly, the Kif5B tail and most other kinesin tails do not contain any known conserved protein-protein interaction sequences. Therefore, we sought to determine the general structural features of the kinesin non-motor domains, particularly the tails, in order to better understand how cargo proteins are selected and bound.
In this work, we have performed a bioinformatics survey to identify intrinsically disordered (ID) regions within the sequences of all 43 human kinesin motors. The resulting predictions indicate that significant ID regions are found throughout the kinesin superfamily, particularly in the non-motor domains. An ID protein or domain is a polypeptide chain that lacks a single, folded, “native” state, but rather exists as an ensemble of locally interconverting structural states. The structural details and biological functions of ID proteins have been reviewed in detail elsewhere13-15. Intrinsic disorder is common; it is estimated that 35-50% of all eukaryotic proteins contain at least one region of forty or more consecutive ID residues16. The predicted ID regions we identified in this work have different distributions of length and net charge depending on whether they are located in the motor, stalk, or tail domains, suggesting that kinesin ID regions have domain-specific properties or functions.
We tested the predictions resulting from our bioinformatics survey by performing CD and NMR spectroscopies on the tail regions of a selection of three kinesins: Kif5B, Kif10, and KifC3. Our results indicate that these tails do not contain any stable secondary structural elements, validating the ID prediction results. The tails retain their abilities to bind to a common known ligand (microtubules), showing that these intrinsically disordered tails are also functional9,17,18. ID domains are particularly useful in situations where structural flexibility, regulation by post-translational modifications, and / or high-affinity high-specificity ligand binding are desirable (reviewed by Dyson, 201119). The tails of the three kinesins featured in this work, like those of many other kinesin families, are hubs for cargo binding and regulatory interactions with other proteins. A lack of stable secondary structure may therefore be a functionally useful property of ID regions in kinesin motors.
Methods
Bioinformatics methods
The canonical sequences for each of the 43 human kinesin superfamily members for which complete protein sequences are available, as listed in the UniProt database (http://www.uniprot.org/), made up the primary data set for this work. Sequences were aligned, and sequence identity and similarity were calculated using ClustalW20,21. GlobPlot was used to determine the boundaries of known globular domains22. Isoelectric points were calculated using the “calculate pI/MW” tool on the ExPASy server23 (http://web.expasy.org/compute_pi/). Coiled-coil domains were predicted with the program COILS (http://www.ch.embnet.org/software/COILS_form.html)24.
For intrinsic disorder prediction, the combined outputs of a set of algorithms can theoretically yield a consensus prediction that is considered more accurate than the results of a single algorithm25. In the present work, the outputs of seven intrinsic disorder prediction algorithms that utilize different datasets, protein or sequence characteristics indicative of ID, or data analysis methods were combined to achieve a consensus as to which residues are most likely to be disordered for each human kinesin isoform. The seven ID prediction algorithms used were GlobPlot (http://globplot.embl.de/)22, DisEMBL (http://dis.embl.de/)26, DISOPRED2 (http://bioinf.cs.ucl.ac.uk/disopred/)27, IUPred (http://iupred.enzim.hu/index.html)28, DISpro (http://scratch.proteomics.ics.uci.edu/)29, OnD-CRF (http://babel.ucmp.umu.se/ond-crf)30, and DRIP-PRED (http://www.sbc.su.se/~maccallr/disorder/).
The results of each algorithm were converted to a simple binary output of ordered or disordered based on the default cut-off values for disorder prediction for each algorithm, and the number of algorithms predicting that a residue was intrinsically disordered (0-7) were tallied and plotted on a per residue basis for each kinesin sequence. We found that elements known to be disordered in the motor and stalk domains were best identified as such by a consensus between at least 3 of the 7 predictors, and therefore, in our final analysis, we classified a residue as disordered if at least 3 of 7 predictors identified it as disordered (further details are described in the Results and Discussion section below).
Protein Expression and Purification
The following human protein constructs were used in this work: Kif5B 822-963, Kif5B 860-963, Kif5B 905-963, Kif10 2600-2701, and KifC3 1-99. All constructs contained a C-terminal histidine hexamer tag for purification purposes. Proteins for CD and microtubule pull-down assays were expressed in BL21 DE3 RP competent bacteria (Aligent Technologies, Santa Clara CA) grown in 2.0 L TPM media (20 g/L tryptone, 15 g/L bactoyeast extract, 150 mM NaCl, 15 mM Na2HPO4, 7 mM KH2PO4) supplemented with 50 mg/L kanamycin and 34 mg/L chloramphenicol. Uniformly 15N-labeled proteins for NMR experiments were grown in 2.0 L M9 media (50 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2) supplemented with 50 mg/L kanamycin, 34 mg/L chloramphenicol, and 1 g/L 15N-ammonium sulfate (Cambridge Isotope Laboratories Inc., Andover MA). Cells were pelleted at 7000xg, lysed by sonication, and pelleted again at 70,000xg. The soluble fraction was bound to Ni-NTA resin(Qiagen, Valencia, CA), washed with several column volumes of wash buffer (50 mM phosphate buffer pH 7.0, 200 mM NaCl, 75 mM imidazole), eluted with elution buffer (50 mM phosphate buffer pH 7.0, 200 mM NaCl, 300 mM imidazole), and then further purified with a 5 mL HiTrap SP HP column (GE Healthcare, Piscataway, NJ). Fractions containing the purified protein were identified by gel analysis, pooled, concentrated, and buffer exchanged into CD buffer (10 mM phosphate buffer pH 7.0, 10 mM NaCl) or NMR buffer (50 mM phosphate buffer pH 7.0, 100 mM NaCl, 10% D2O v/v). Protein concentrations were determined by measuring the intensities of protein gel bands relative to known lysozyme standards with ImageJ (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD). Protein identities were confirmed by mass spectrometry analysis. Porcine microtubules were prepared as previously described9.
CD Spectroscopy
Circular dichroism measurements were performed on samples containing 0.05 to 0.12 mg/mL protein in a quartz cuvette with a 2.0 mm path length. Measurements were made on a Jasco J-815 CD spectrometer (Jasco, Eason, MD) at 20°C. Spectra between 195 and 260 nm were collected in triplicate and automatically averaged, baseline measurements for buffer alone were automatically subtracted from the spectra, and the final spectra were the average of measurements from three separate protein samples. Data between 200 to 241 nm were analyzed for secondary structure content with the K2D program on the DichroWeb server31.
NMR Spectroscopy
NMR measurements were performed on samples containing 0.1 to 0.5 mM protein. 1H 15N HSQC (heteronuclear single quantum coherence) spectra were collected on a Varian Inova 600 MHz spectrometer at 25°C.
Microtubule Pull-Down Assay
1.0 μM kinesin tail proteins with and without 10 μM polymerized paclitaxel-stabilized microtubules were incubated at room temperature for 15 minutes in 30 μL total volume of binding buffer (50 mM HEPES pH 7.0, 50 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 1% v/v paclitaxel). The samples were centrifuged at 50,000×g for 15 minutes at 25°C, the supernatants were removed, the pellets were resuspended in an equal volume of binding buffer, and 50% of each supernatant and pellet sample were run on a gel and stained with Coomassie Blue.
Results and Discussion
ID is a structural element predicted to be found throughout the kinesin superfamily
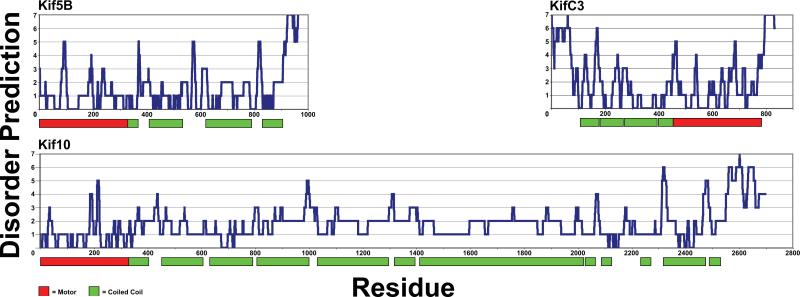
In order to investigate regions of kinesin molecules whose structures are unknown, the sequences of all 43 human kinesins were analyzed by a set of 7 predictors of intrinsic disorder. The plots in Figure 1 and Figure S1 show the number of predictors (0-7) that classify a residue as ID verses the residue number. An inspection of these plots indicates that there is at least one prominent region in each kinesin isoform in which the combined outputs of the predictors suggest that it is likely to be disordered. These particular seven predictors were selected because they all employ significantly different learning datasets, protein or sequence characteristics indicative of ID, or data analysis methods (summarized in Dosztanyi, et al32). Similar to other published combinations of algorithms assembled into “meta-predictors”, our set of algorithms is not entirely orthogonal33. Methods for combining ID prediction algorithms into meta-predictors are being refined constantly, and combining them using a non-weighted “majority vote” manner similar to the method here has been shown to yield a more accurate prediction by overriding the least accurate predictors for a given input34. Despite these considerations, some regions may still be erroneously classified as ID by our set of algorithms, and therefore we took advantage of the structural information that is available for the motor and stalk domains in order to establish a threshold level of consensus amongst the ID prediction algorithms that must be met for a residue to be classified as disordered.
Figure 1. Kinesin molecules contain significant discrete regions of intrinsic disorder.
The disorder predictions for three representative kinesin molecules are shown. The x-axis corresponds to the residue number, and the y-axis is the number of disorder predictors indicating that a particular residue is disordered (0-7). The colored bars below the graphs represent the boundaries of the predicted motor and coiled-coil domains.
An examination of the structures of the Kif5B motor domain that are available in the Protein Data Bank (PDB) showed that approximately 30% of the motor domain is made up of surface-accessible loops that may or may not have stable secondary structures in solution. In addition, approximately 70% of the Kif5B stalk is predicted to be coiled-coil (Table S2), implying that 30% of the stalk has an unknown structure. These two estimates are therefore upper bounds for the percentage of residues that may be disordered in the respective motor and stalk domains of Kif5B. In order to filter out some of the noise associated with the disorder predictors, and thus decrease the likelihood of falsely predicting that a residue is disordered, we required that there was a consensus of three or more predictors in order to consider a prediction of disorder to be valid. Using a consensus of 3 out of 7 algorithms as a lower threshold for ID prediction, the average kinesin motor domain is predicted to be 18.2% disordered, and the average stalk domain is predicted to be 29.5% disordered, while the average ID content for an entire kinesin molecule is predicted to be 34.8% (Table I). These ID predictions are in reasonably good agreement with the above ID estimates for the motor and stalk domains based on experimentally determined structures of the motors and coiled-coil predictions for the stalks. Consequently, we believe that our ID prediction approach and constraints are appropriate and sufficient to produce generally accurate structural information about the non-globular regions of kinesins.
Table I.
Sequence conservation and predicted intrinsic disorder are inversely related.
| Domain | Average Sequence Identity | Average Sequence Similarity | Average % Disorder |
|---|---|---|---|
| Motor | 40.1 ± 19.1 | 69.3 ± 15.5 | 18.2 ± 6.6 |
| Stalk | 13.3 ± 17.3 | 32.3 ± 25.5 | 29.5 ± 19.5 |
| Tail | 5.4 ± 6.9 | 19.3 ± 12.7 | 71.8 ± 23.9 |
| Total Protein | 20.5 ± 18.7 | 40.77 ± 23.4 | 34.8 ± 13.3 |
Surprisingly, the average tail domain was predicted to be 71.8% disordered, suggesting that ID may be the dominant structural feature of many kinesin tails. This finding is at odds with the widely held, but poorly substantiated, belief in the kinesin field that kinesin tails are for the most part globular. In addition, an analysis of the sequence similarity and identity amongst the various kinesin subfamilies showed that outside of the motor domains, kinesin sequences are generally not well conserved (Table S1). Interestingly, the percentage of predicted ID content per domain is inversely related to the amount of sequence identity or similarity for that domain (Table I), suggesting that ID regions are more likely to be found in the unique portions of kinesin molecules (non-motor) than the conserved regions (motor).
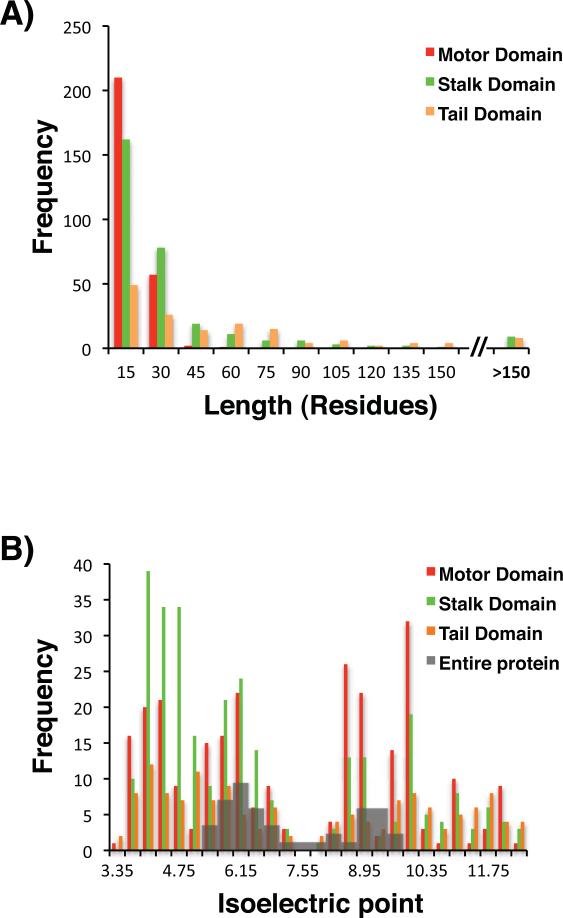
The predicted ID regions have a wide range of lengths (Figure 2A), and these lengths are also domain-dependent. Within the motor domain the median length of an ID region is 9 residues, 14 residues in the stalk, and 31 residues in the tail (Table II). ID regions in the motor domain correspond to various short flexible loops that have been observed in many of the atomic resolution kinesin motor structures (Table S3), while ID regions in non-motor domains, particularly the tails, are significantly longer, suggesting that ID regions may contribute to different structural or functional roles in the motor and non-motor domains.
Figure 2. Lengths and isoelectric points of predicted kinesin ID regions are domain-specific.
A) The lengths of the predicted kinesin ID regions found in specific kinesin domains are indicated on the x-axis, and the numbers of predicted ID regions that fall within each bin are indicated on the y-axis. B) The isoelectric points of the predicted kinesin ID regions are indicated on the x-axis, and the numbers of predicted ID regions that fall within each bin are indicated on the y-axis. The motor, stalk, and tail domains are color-coded as indicated. The shaded gray bars show the distribution of the isoelectric points of the entire kinesin proteins.
Table II.
Lengths and isoelectric points of predicted kinesin ID regions are domain-specific.
| Domain | n | Median Length of ID Regions | % Regions or Proteins with pI ≤ 7.0 | Average pI (pI ≤ 7.0 range) | % Regions or Proteins with pI > 7.0 | Average pI (pI > 7.0 range) |
|---|---|---|---|---|---|---|
| Motor ID | 270 | 9 | 51.9 | 5.0 ± 1.0 | 48.1 | 9.6 ± 1.1 |
| Stalk ID | 299 | 14 | 70.6 | 4.9 ± 0.9 | 29.4 | 9.9 ± 1.2 |
| Tail ID | 151 | 31 | 52.3 | 4.9 ± 1.0 | 47.7 | 10.2 ± 1.4 |
| Total Protein | 43 | - | 60.5 | 6.0 ± 0.5 | 39.5 | 8.7 ± 0.6 |
ID domains generally have a greater propensity for charged amino acids than globular domains, so we examined the isoelectric points (pIs) of the predicted ID sequences within kinesins (Table S II). The distribution of the isoelectric points (pI) of predicted ID sequences (Figure 2B) is bimodal with two peaks at pI values of approximately 5.0 and 10.0. The disordered regions in the motor and tail domains are evenly divided between positive and negative pIs, while the percentages of positively charged and negatively charged ID regions are 70.6% and 29.4%, respectively, in the stalk domain (Table II). The distribution of pIs for the entire sequences of all 43 kinesins is also bimodal (Figure 2B), with peak pI values of approximately 6.0 and 9.0 (Table II). This is reminiscent of distributions that have been constructed for entire eukaryotic proteomes, which also generally have bimodal distributions of pIs, with the acidic peak centered at a pI value of 5.5 and the basic peak at 9.035. The average acidic pI of the predicted ID regions is more acidic than the average acidic pI of the whole kinesin proteins at a statistically significant level as determined by a student's t-test (p < 0.01), and similarly, the average basic pI of the predicted ID regions is more basic than the average basic pI of the whole kinesin proteins at a statistically significant level as determined by a student's t-test (p < 0.01) (Table II). The more acidic and alkaline average pIs observed for the ID regions relative to the whole kinesin proteins or whole eukaryotic proteomes likely reflect the higher propensity of disordered regions to contain charged amino acids relative to structured regions, and may have functional implications for these ID regions.
Predicted ID segments correspond to functional loops in kinesin motor domains
The motor domain is the common structural link amongst the various kinesin proteins, and even where the primary sequences are not well conserved, the secondary and tertiary structures are. The ID prediction plots indicate that there are discrete regions of disorder in each of the motors (Figure 1, Figure S1). A comparison of the ID predictions to the known structural topology of the motor domain assigns these ID regions to known loop structures (see Table S3 for a summary of these predictions). The motor structures that are most consistently predicted to contain ID residues include Loop 1, Loop 2, P-loop, Loop 7, Switch I, Loop 10, Switch II, Loop 12, and the neck-linker. Many of these loops are known to undergo conformational changes in response to the nucleotide state and / or microtubule binding and release36. Several kinesin motors also contain inserts of ID sequence in various loops, relative to the canonical motor Kif5B, which contribute to the different motile or functional properties of these motors (see Table S3 for a description of the most significant differences). For example, all Kinesin-13 motors contain an unstructured insert in Loop 2 that is important for the microtubule depolymerase activity of these motors37. Therefore, disordered structural elements are important components of the communication link between the ATP and microtubule binding-sites and the neck-linker, and of loops that impart unique functions to a given motor. In addition, the fact that our bioinformatics approach was able to identify known disordered or structurally flexible regions in the motor domains, gave us confidence in the reliability of the ID predictions for the non-motor domains of the molecules.
Predicted ID segments correspond to non-globular regions of the stalk and tail domains
The non-motor domains of kinesin molecules facilitate the formation of multimers and contain the majority of the known sites of post-translational modifications and ligand binding. While it has been assumed that kinesin stalks fold into roughly continuous coiled-coils and the tails are globular structures, the ID predictions presented in Figure 1 and Figure S1 show that this is not generally the case. For example, the stalks of the Kinesin-11 proteins are over 1000 residues long and are predicted to be almost completely disordered, while the stalk of Kif10 is over 2000 residues long and is predicted to be nearly all coiled-coil with several unstructured breaks of approximately 10-100 residues each. A possible purpose of ID residues in the midst of coiled-coils in the stalk domain has been established in Kif5B, in which it has been experimentally determined that the stalk contains several short flexible hinge regions that enable the molecule to fold into a compact conformation under certain conditions38, and these hinge regions correspond to sequences that have a high ID probability (Figure 1). Whether disordered regions in the stalks of other kinesins serve a similar purpose remains to be experimentally determined, but clearly the structures of kinesin stalks are more complex than an uninterrupted coiled-coil.
ID predictions in regions of predicted coiled-coil should be viewed with caution since coiled-coils are often cross-predicted to be ID39. These cross-predictions were minimized in our analysis by requiring a consensus of 3 of 7 predictors in order for a residue to be considered disordered. Using this cutoff, 14.2% of the stalk residues that are predicted to be coiled-coil are also predicted to be ID. Most of these cross-predicted residues are within the first or last 10 residues of the predicted coiled-coil region (Table S2), suggesting that much of the error associated with these cross-predictions is due to an imprecise definition of the boundary between the predicted ID and coiled-coil regions. Thus, while the general trend of coiled-coil regions broken by ID regions seen in our data is probably correct in most instances, the precise boundaries between ID and coiled-coil regions may differ from our analysis.
The stalk and tail domains of some Kinesin-3, -4, -10, and -13 families contain small globular domains including FHA (forkhead-associated), PH (pleckstrin homology), PX (phox), CAP-Gly (cytoskeleton-associated protein Gly-rich), WD40 (Trp-Asp 40), HhH (helix-hairpin-helix), and SAM (sterile alpha motif) domains (Figure S1). Most of these small globular domains are known to facilitate protein-ligand interactions, and while the functions of many of these domains in the context of the whole kinesin molecules are unknown, a few have been shown to bind to various ligands40-44. Since only a minority of kinesin isoforms contain these small globular domains, the majority of the other kinesin isoforms must interact with their ligands via another structural mechanism. Also, these globular domains were clearly predicted to be structured by the ID algorithms, providing further support for the accuracy of the ID predictions (Figure S1).
The tail domains are defined here as the region of the kinesin molecule that extends from the end of the stalk coiled-coil to the C-terminus of the protein (N-terminus for some kinesins including members of the Kinesin-14 family, whose motor domains are located at the C-terminus). The results of the prediction algorithms for these tail domains were striking. For 10 out of 14 kinesin families whose tails do not contain any known globular domains, a strong consensus of algorithms predicted that the tails are disordered (Figure 1, Figure S1), and the tail domains that contain ordered residues also contain various small globular domains (Kinesin-3, 4, 10, and 13 isoforms). The ID tail regions within most kinesin families vary in length from tens to hundreds of residues, and are involved in the auto-regulation of the motor domain, are substrates for a variety of post-translational modifications, and contain numerous ligand binding-sites (reviewed by Hirokawa et al, 20094 and Gindhart, 200645). Thus, our bioinformatics predictions indicate that intrinsic disorder is a conserved structural element utilized extensively for a variety of purposes by the kinesin non-motor domains, challenging the notion that the kinesin stalk and tail domains are predominantly structured.
Experimental evidence for intrinsic disorder: structural analysis of tail domains from three kinesin families
We next sought to experimentally verify the findings of our in silico predictions, particularly the striking finding of ID in large portions of the kinesin tail domains. We examined several tail domains that were predicted to contain ID residues by circular dichroism (CD) and NMR spectroscopies. Three constructs of 50-100 residues from the human Kif5B C-terminus were examined (Kif5B 822-963 dimer, Kif5B 860-963 dimer, Kif5B 905-963 monomer), along with the Kif10 C-terminal tail (Kif10 2600-2701 monomer), and the KifC3 N-terminal tail (KifC3 1-99 monomer). The motor domains of Kif5B and Kif10, and most kinesins, are located at the N-terminal ends of the molecules, whereas in KifC3, the motor domain is located at the C-terminal end of the molecule. This alternative arrangement of domains results in the KifC3 motor moving towards the minus end of the MT and the Kif5B and Kif10 motors moving towards the plus end of the MT. Therefore, ID in the kinesin tails is not contingent on whether the tails are located at the C-terminal or N-terminal ends of the molecule (Figure 1).
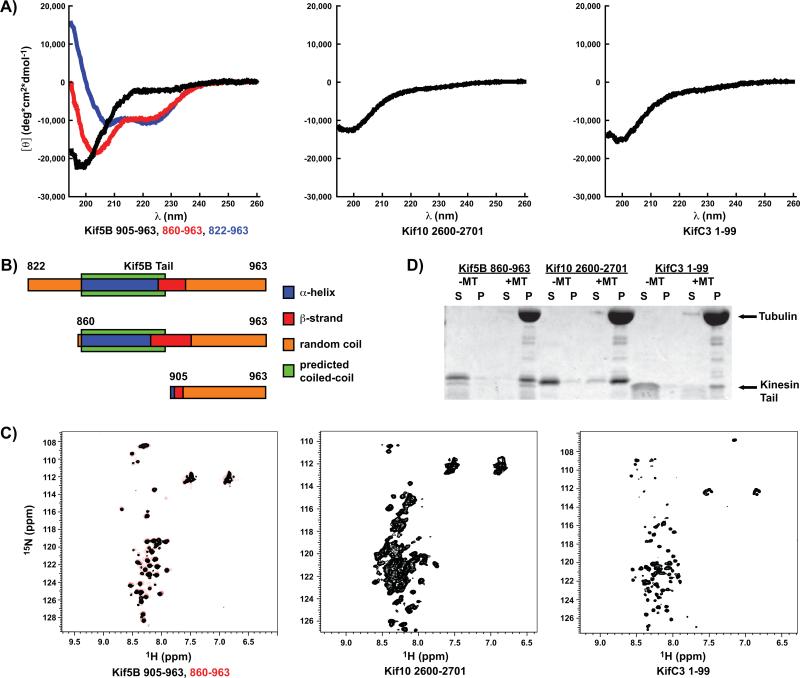
CD spectroscopy is a standard method for determining the secondary structural content of proteins, and the CD spectra of disordered proteins show a characteristic minimum at approximately 200 nm46. The CD spectra of Kif5B 905-963, Kif10 2600-2701 and KifC3 1-99 are typical for disordered polypeptides in that there is a single minimum at 200 nm and no other significant features (Figure 3A), indicating that these tails contain little to no stable secondary structure. The dimeric Kif5B 822-963 and 860-963 constructs both contain the final predicted 49-residue coiled-coil region of the stalk domain, and their CD spectra show a strong α-helical component in addition to the disordered component observed in the monomeric 905-963 construct. Estimates of the secondary structure content of the Kif5B tail constructs obtained from fits of the CD data are remarkably consistent with the structure of the Kif5B C-terminus as predicted by the ID algorithms and the coiled-coil predictor COILS (Table S4). Our reconstruction of the Kif5B C-terminus based on the CD data and coiled-coil predictions shows that the tail domain is in fact disordered (Figure 3B). Importantly, both monomeric and dimeric tail constructs contain ID segments of approximately the same length. This argues that the lack of stable secondary structure within the Kif5B tail constructs is not due to their being expressed as truncated recombinant proteins, but rather reflects the actual structures of these tail domains.
Figure 3. Kinesin tails isolated from three distinct families are intrinsically disordered and functional.
A) CD spectra of three Kif5B tail constructs of varying lengths (left), as well as Kif10 (center) and KifC3 (right) tail constructs. B) A reconstruction of the secondary structure of the three Kif5B tail constructs based on a best-fit of the CD spectroscopy data (Table S4) and the predicted coiled-coil boundaries (Table S2). C) 1H15N-HSQC spectra of two different Kif5B tail constructs (left), and Kif10 (center) and KifC3 (right) tail constructs. D) Protein gel of a microtubule pull-down assay showing that the Kif5B, Kif10, and KifC3 unstructured tail constructs sediment with 10 μM microtubules. MT = microtubule, S = supernatant, P = pellet.
We also examined the Kif5B 860-963, Kif5B 905-963, Kif10 2600-2701, and KifC3 1-99 tail constructs by NMR spectroscopy. The 1H 15N HSQC spectra of disordered proteins have a characteristically narrow range of spectral dispersions, particularly in the 1H axis46. All of these constructs had very narrow spectral dispersions in both axis (1H < 1 ppm and 15N < 20 ppm) (Figure 3C), indicating that the tail domains of these kinesins do not contain any stable secondary structure that is visible on the timescale of the HSQC experiment. Some ID proteins have been shown to contain regions of preformed or transient structure54, and although we did not observe any such structures in our NMR experiments, we cannot exclude the possibility that some disordered kinesin tails may contain preformed or transient structures. Interestingly, we were only able to observe peaks for residues 905-963 in the Kif5B 860-963 construct, as indicated by the nearly identical 1H 15N HSQC spectra of the Kif5B 860-963 and 905-963 constructs (Figure 3C). The peaks from residues 860-905, which are predicted to form a coiled-coil, may not be visible as a result of decreased tumbling in solution due to the rod-like shape of the coiled-coil, while the disordered tail residues are visible due to their less restricted range of motion. The fact that the NMR spectra of Kif5B 860-963 and 905-963 can be superimposed so well indicates that the tail of Kif5B remains disordered in the context of the coiled-coil dimeric stalk domain, and therefore the two tails of a Kif5B dimer are structurally distinct entities. This conclusion is consistent with the CD data described above. Both the Kif10 and KifC3 tail constructs were designed such that they would not include any of the predicted coiled-coil stalk domains, and therefore their spectra appear very similar to the Kif5B tail spectra, in that none of these tails appear to contain any stable secondary structure.
The kinesin tail domains are the substrates for a variety of post-translational modifications and the binding-sites for various ligands. We tested the disordered Kif5B, 10, and C3 tail constructs used in the above assays to determine if they retained their ligand-binding functionality. The Kif5B, Kif10, and, by sequence similarity to its Drosophila homolog Ncd, KifC3 tails have all been shown to bind to microtubules9,17,18. A microtubule pull-down assay showed that all three of these disordered tail constructs retain their abilities to bind to microtubules (Figure 3D). Thus, our experimental results together confirm the ID predictions by showing that the Kif5B, Kif10, and KifC3 tail domains are both disordered and functional.
Conclusions
In this work, we show that regions of ID having a wide range of lengths and charges are predicted for every member of the human kinesin superfamily. ID in the motor domain is localized to surface loops, most of which are involved in communicating structural changes between the nucleotide and microtubule binding-sites and the neck-linker. ID residues are also found throughout the non-motor domains. The stalks are a mixture of coiled-coil regions and unstructured regions, both of varying lengths. There was a striking consensus of the seven algorithms used, predicting that kinesin tail domains contain significant unstructured regions. We verified our conclusions experimentally by examining the tail domains of Kif5B, Kif10, and KifC3 by CD and NMR spectroscopies and a microtubule pull-down assay. These assays together demonstrated that these three tail domains are both disordered and functional.
The question remains as to why intrinsic disorder is such a prominent structural component in kinesin proteins. One can imagine that three properties of ID proteins may be particularly useful to kinesins: structural flexibility, accommodation of post-translational modifications, and high-affinity high-specificity ligand binding14. Structural flexibility might allow the efficient transmission of conformational changes between the nucleotide binding pocket and microtubule binding interface and the neck-linker in the motor domain. Structural flexibility in the stalk might allow various kinesins to adopt compact auto-inhibited conformations that have been observed in several distinct kinesin isoforms, including Kif5B38, Kif1747, Kif1A48, Kif13B49, and Kif1050. In the stalk and tail regions, ID sequences may also provide structurally compact substrates for high-affinity and high-specificity interactions with kinesin cargoes. Binding interactions of this nature have been observed in a variety of situations including the interactions between the dynein light chain protein LC8 and its various ligands. LC8 has been shown to bind to over 22 different proteins, and these various binding interactions are all accommodated while maintaining binding specificity through specific interactions between disordered residues on both LC8 and its ligands51. Post-translational modifications have also been shown to regulate kinesin-ligand interactions52, and therefore a post-translational modification of an ID ligand binding-site may alter the structure and consequently the affinity of that binding-site for ligands in a regulated manner.
The present work provides a structural insight into how intrinsic disorder may enable kinesins to accomplish their specific tasks in vivo. Further experimentation will be necessary to test whether these ideas are applicable to kinesin proteins. In particular, regions of ID often contain ligand binding-sites53, therefore it will be intriguing to identify the specific tail ID residues responsible for kinesin-binding partner interactions, and to determine if and how kinesin tails become ordered upon binding to their ligands, as has been observed for other ID proteins54.
Supplementary Material
Figure S1. All kinesins contain significant, discrete regions of intrinsic disorder. The disorder predictions for all 43 human kinesin molecules are shown. The x-axis corresponds to the residue number, and the y-axis corresponds to the number of intrinsic disorder prediction algorithms (0-7) indicating that a region is likely to be disordered. The colored bars below the graphs represent the boundaries of the predicted motor domains, coiled-coils, and known globular domains.
Acknowledgments
This work is supported by the National Institutes of Health (R01GM072656).
Footnotes
All work was completed at Northwestern University
Contributor Information
Mark A. Seeger, Northwestern University
Yongbo Zhang, Northwestern University.
Sarah E. Rice, Northwestern University
References
- 1.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68(4):610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Ferenz NP, Gable A, Wadsworth P. Mitotic functions of kinesin-5. Semin Cell Dev Biol. 2010;21(3):255–259. doi: 10.1016/j.semcdb.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock SL. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans. 2011;39(5):1161–1165. doi: 10.1042/BST0391161. [DOI] [PubMed] [Google Scholar]
- 4.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10(10):682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 5.Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333(6044):883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Schwarz TL. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136(1):163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moua P, Fullerton D, Serbus LR, Warrior R, Saxton WM. Kinesin-1 tail autoregulation and microtubule-binding regions function in saltatory transport but not ooplasmic streaming. Development. 2011;138(6):1087–1092. doi: 10.1242/dev.048645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2(5):257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 9.Seeger MA, Rice SE. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem. 2010;285(11):8155–8162. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176(1):11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gindhart JG, Chen J, Faulkner M, Gandhi R, Doerner K, Wisniewski T, Nandlestadt A. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol Biol Cell. 2003;14(8):3356–3365. doi: 10.1091/mbc.E02-12-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho KI, Cai Y, Yi H, Yeh A, Aslanukov A, Ferreira PA. Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic. 2007;8(12):1722–1735. doi: 10.1111/j.1600-0854.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 13.Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 16.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 17.Karabay A, Walker RA. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38(6):1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- 18.Liao H, Li G, Yen TJ. Mitotic regulation of microtubule cross-linking activity of CENP-E kinetochore protein. Science. 1994;265(5170):394–398. doi: 10.1126/science.8023161. [DOI] [PubMed] [Google Scholar]
- 19.Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44(4):467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 21.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–699. doi: 10.1093/nar/gkq313. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31(13):3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5009):1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Peng Z, Kurgan L. On the complementarity of the consensus-based disorder prediction. Pac Symp Biocomput. 2012:176–187. [PubMed] [Google Scholar]
- 26.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11(11):1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Sweredoski M, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Mining and Knowledge Discovery. 2005;11(3):213–222. [Google Scholar]
- 30.Wang L, Sauer UH. OnD-CRF: predicting order and disorder in proteins using [corrected] conditional random fields. Bioinformatics. 2008;24(11):1401–1402. doi: 10.1093/bioinformatics/btn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dosztanyi Z, Meszaros B, Simon I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief Bioinform. 2010;11(2):225–243. doi: 10.1093/bib/bbp061. [DOI] [PubMed] [Google Scholar]
- 33.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Obradovic Z. Unsupervised Integration of Multiple Protein Disorder Predictors: The Method and Evaluation on CASP7, CASP8 and CASP9 Data. Proteome Sci. 2011;9(Suppl 1):S12. doi: 10.1186/1477-5956-9-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiller GF, Caraux G, Sylvester N. The modal distribution of protein isoelectric points reflects amino acid properties rather than sequence evolution. Proteomics. 2004;4(4):943–949. doi: 10.1002/pmic.200200648. [DOI] [PubMed] [Google Scholar]
- 36.Sindelar CV. A seesaw model for intermolecular gating in the kinesin motor protein. Biophys Rev. 2011;3(2):85–100. doi: 10.1007/s12551-011-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116(4):591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 38.Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem. 1999;274(21):14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 39.Szappanos B, Suveges D, Nyitray L, Perczel A, Gaspari Z. Folded-unfolded cross-predictions and protein evolution: the case study of coiled-coils. FEBS Lett. 2010;584(8):1623–1627. doi: 10.1016/j.febslet.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109(3):347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sagona AP, Nezis IP, Pedersen NM, Liestol K, Poulton J, Rusten TE, Skotheim RI, Raiborg C, Stenmark H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12(4):362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 42.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174(3):425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueno H, Huang X, Tanaka Y, Hirokawa N. KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell. 2011;20(1):60–71. doi: 10.1016/j.devcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Shen X, Meza-Carmen V, Puxeddu E, Wang G, Moss J, Vaughan M. Interaction of brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 and kinesin motor protein KIF21A. Proc Natl Acad Sci U S A. 2008;105(48):18788–18793. doi: 10.1073/pnas.0810104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gindhart JG. Towards an understanding of kinesin-1 dependent transport pathways through the study of protein-protein interactions. Brief Funct Genomic Proteomic. 2006;5(1):74–86. doi: 10.1093/bfgp/ell002. [DOI] [PubMed] [Google Scholar]
- 46.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189(6):1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7(3):e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46(35):10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espeut J, Gaussen A, Bieling P, Morin V, Prieto S, Fesquet D, Surrey T, Abrieu A. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol Cell. 2008;29(5):637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Nyarko A, Hall J, Hall A, Hare M, Kremerskothen J, Barbar E. Conformational dynamics promote binding diversity of dynein light chain LC8. Biophys Chem. 2011;159(1):41–47. doi: 10.1016/j.bpc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Guillaud L, Wong R, Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat Cell Biol. 2008;10(1):19–29. doi: 10.1038/ncb1665. [DOI] [PubMed] [Google Scholar]
- 53.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5(5):e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Nitta R, Kikkawa M, Okada Y, Hirokawa N. KIF1A alternately uses two loops to bind microtubules. Science. 2004;305(5684):678–683. doi: 10.1126/science.1096621. [DOI] [PubMed] [Google Scholar]
- 56.Larson AG, Naber N, Cooke R, Pate E, Rice SE. The conserved L5 loop establishes the pre-powerstroke conformation of the Kinesin-5 motor, eg5. Biophys J. 2010;98(11):2619–2627. doi: 10.1016/j.bpj.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hizlan D, Mishima M, Tittmann P, Gross H, Glotzer M, Hoenger A. Structural analysis of the ZEN-4/CeMKLP1 motor domain and its interaction with microtubules. J Struct Biol. 2006;153(1):73–84. doi: 10.1016/j.jsb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Maddika S, Sy SM, Chen J. Functional interaction between Chfr and Kif22 controls genomic stability. J Biol Chem. 2009;284(19):12998–13003. doi: 10.1074/jbc.M900333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. All kinesins contain significant, discrete regions of intrinsic disorder. The disorder predictions for all 43 human kinesin molecules are shown. The x-axis corresponds to the residue number, and the y-axis corresponds to the number of intrinsic disorder prediction algorithms (0-7) indicating that a region is likely to be disordered. The colored bars below the graphs represent the boundaries of the predicted motor domains, coiled-coils, and known globular domains.