Abstract
Mutations in FLNC cause two distinct types of myopathy. Disease associated with mutations in filamin C rod domain leading to expression of a toxic protein presents with progressive proximal muscle weakness and shows focal destructive lesions of polymorphous aggregates containing desmin, myotilin and other proteins in the affected myofibres; these features correspond to the profile of myofibrillar myopathy. The second variant associated with mutations in the actin-binding domain of filamin C is characterized by weakness of distal muscles and morphologically by non-specific myopathic features. A frameshift mutation in the filamin C rod domain causing haploinsufficiency was also found responsible for distal myopathy with some myofibrillar changes but no protein aggregation typical of myofibrillar myopathies. Controversial data accumulating in the literature require re-evaluation and comparative analysis of phenotypes associated with the position of the FLNC mutation and investigation of the underlying disease mechanisms. This is relevant and necessary for the refinement of diagnostic criteria and developing therapeutic approaches. We identified a p.W2710X mutation in families originating from ethnically diverse populations and re-evaluated a family with a p.V930_T933del mutation. Analysis of the expanded database allows us to refine clinical and myopathological characteristics of myofibrillar myopathy caused by mutations in the rod domain of filamin C. Biophysical and biochemical studies indicate that certain pathogenic mutations in FLNC cause protein misfolding, which triggers aggregation of the mutant filamin C protein and subsequently involves several other proteins. Immunofluorescence analyses using markers for the ubiquitin–proteasome system and autophagy reveal that the affected muscle fibres react to protein aggregate formation with a highly increased expression of chaperones and proteins involved in proteasomal protein degradation and autophagy. However, there is a noticeably diminished efficiency of both the ubiquitin–proteasome system and autophagy that impairs the muscle capacity to prevent the formation or mediate the degradation of aggregates. Transfection studies of cultured muscle cells imitate events observed in the patient’s affected muscle and therefore provide a helpful model for testing future therapeutic strategies.
Keywords: myofibrillar myopathy, filaminopathy, filamin C mutation, immunoglobulin-like domain, limb-girdle myopathy
Introduction
Filamin C-related myopathies specify diseases caused by mutations in the FLNC gene located within the chromosomal band 7q32–q35 and expressed predominantly in skeletal and cardiac muscles. The encoded filamin C protein (FLNC) contains an N-terminal actin-binding domain followed by a semiflexible rod comprising 24 highly homologous immunoglobulin-like domains (Xie et al., 1998; van der Flier and Sonnenberg, 2001). The carboxy-terminal immunoglobulin-like domain is required and sufficient for dimerization (Himmel et al., 2003; Pudas et al., 2005; Sjekloća et al., 2007), which is the molecular basis for filamin’s actin cross-linking activity. In the sarcomere, FLNC cross-links actin in the Z-disc region and additionally binds myotilin, FATZ/calsarcin/myozenin, myopodin and Xin (Faulkner et al., 2000; van der Ven et al., 2000; Gontier et al., 2005; van der Ven et al., 2006; Linnemann et al., 2010). At the sarcolemma, FLNC interacts with sarcoglycan γ and -δ, two components of the dystrophin–dystroglycan complex (Thompson et al., 2000). Deficiency of FLNC in mice results in severe muscle defects and perinatal lethality, indicating a critical role for FLNC in muscle development and maintenance of muscle structural integrity (Dalkilic et al., 2006).
The first filamin C-related myopathy was described in 2005 when a nonsense mutation (c.G8130A, p.W2710X) in the FLNC dimerization domain was shown to cause a disease in a large German family characterized by muscle weakness of predominantly limb-girdle distribution and typical myofibrillar myopathy features on muscle biopsy (Vorgerd et al., 2005). Myofibrillar myopathy is a clinically and genetically diverse group of progressive devastating hereditary skeletal and cardiac myopathies. Thus far myofibrillar myopathy has been associated with mutations in seven genes (DES, MYOT, LDB3, CRYAB, BAG3, FLNC and FHL1), but a significant number of patients with myofibrillar myopathy failed to show disease-causing mutations in these genes (Selcen, 2008; Olivé et al., 2011; Selcen et al., 2011a, b). The most important common feature of these diseases is the disintegration of myofibrils and formation of desmin-positive protein aggregates within muscle fibres (Nakano et al., 1996; Schröder and Schoser, 2009). The p.W2710X mutation in FLNC impedes its ability to dimerize (Vorgerd et al., 2005; Löwe et al., 2007). Instead, the mutant protein acquires a strong tendency for uncontrolled aggregation, resulting in the deposition of massive protein aggregates that, in addition to FLNC, attract multiple other proteins including desmin and other Z-disc-associated proteins. These events ultimately lead to disintegration of myofibrils (Vorgerd et al., 2005; Löwe et al., 2007). A haplotype-sharing set of further German families also carrying the p.W2710X FLNC mutation were described soon after the first report (Kley et al., 2007), and an identical mutation was found in three kinships of the Mayo myofibrillar myopathy cohort that were not described in detail (Selcen, 2011a).
More recently, two families with filamin C-related myopathy harbouring mutations in FLNC rod domains have been reported: an internal 12-nucleotide deletion (c.2997_3008del, p.V930_T933del) (Shatunov et al., 2009) and an 18-nucleotide deletion/6 nucleotide insertion (p.K899_V904del/V899_C900ins) (Luan et al., 2010), both in immunoglobulin-like repeat 7. Clinical and pathological characteristics of the disease associated with these mutations were similar to the p.W2710X German phenotype, including characteristic features of myofibrillar myopathy, but no exhaustive comparison was provided and the disease pathomechanisms were not investigated.
Additionally, three distantly related families were described, in which a deletion (c.5160delC, p.F1720LfsX63) in exon 30 encoding FLNC immunoglobulin-like domain 15 triggers a frameshift, nonsense-mediated decay and haploinsufficiency, affecting primarily distal muscles of the upper and lower limbs. Although histological evaluation indicated disease-associated myofibrillar abnormalities, desmin-positive protein aggregates required for diagnosis of myofibrillar myopathy were not detected, probably because no truncated mutant protein is expressed. Muscle imaging revealed a pattern of muscle involvement different from that associated with p.W2710X FLNC mutation (Guergueltcheva et al., 2011). Finally, mutations located in the N-terminal actin-binding domain of FLNC (p.A193T and p.M251T) have also been associated with a distal myopathy characterized by a disease onset in the third decade of life, distal muscle weakness (primarily intrinsic hand muscles), and non-specific myopathic abnormalities with no features of myofibrillar myopathy (Duff et al., 2011). Although aggregation of mutant FLNC with F-actin was observed in transfection experiments in cultured cells, no aggregates were found in muscle biopsies of patients carrying these mutations.
This clearly differentiates the two latter syndromes from myofibrillar myopathy-associated pathologies caused by mutations occurring in FLNC immunoglobulin-like domains 7 and 24 and indicates that, as has previously been shown for FLNA and FLNB, different mutations in FLNC can lead to distinct disease phenotypes.
This requires a critical re-evaluation of the class of diseases caused by mutations in immunoglobulin-like domains of FLNC. We present here a comparative analysis of filamin C myopathy phenotypes associated with myofibrillar myopathy pathology and caused by mutations in FLNC rod domains including two novel p.W2710X families of distinct ethnic backgrounds and the re-evaluated p.V930_T933del family (Shatunov et al., 2009). We also assessed the patho-mechanisms of these phenotypes by analysing protein degradation machineries and investigating pathogenic effects of transient expression of FLNC mutants in murine myoblast cell lines.
Materials and methods
Newly identified and re-evaluated filaminopathy kindreds
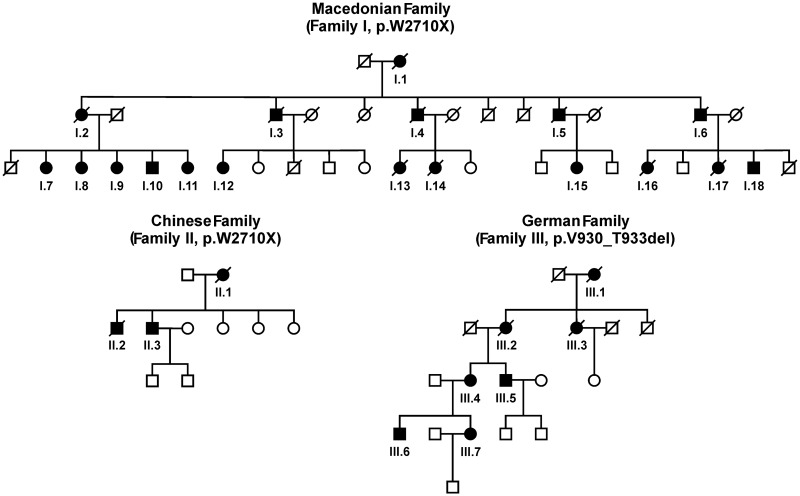
We identified a large East European kindred originating from a rural region of Macedonia in which 18 members were known to be affected with a skeletal and cardiac myopathy. This kindred is referred to as Family I (Fig. 1). The second family with three affected individuals is of Chinese origin and referred to as Family II (Fig. 1). Using complementary DNA sequencing, a mutation in exon 48 of FLNC identical to the c.G8130A, p.W2710X previously detected in four German families (Vorgerd et al., 2005; Kley et al., 2007) was uncovered in Families I and II. In addition, three patients from a German family carrying the p.V930_T933del mutation in FLNC first described by Shatunov et al. (2009) were re-examined for this report (Family III in Fig. 1). Basic information on clinical examination, neurophysiological studies and muscle imaging is provided in the online Supplementary material. Genetic studies were approved by the Institutional Review Board of the National Institute of Neurological Disorders and Stroke in Bethesda, Maryland. Diagnostic criteria and investigation procedures were agreed upon by the participating neuromuscular centres under the leadership of the European Neuromuscular Centre (Goebel and Fardeau, 2004; Goebel et al., 2008).
Figure 1.
Pedigrees of three filaminopathy families. Affected members of the families from Macedonia (Family I) and China (Family II) harboured the p.W2710X mutation in FLNC. The German family with p.V930_T933del mutation in FLNC (Family III) was first described by Shatunov et al. (2009) (pedigree extended). Individuals with proven mutation and deceased family members who had suffered from muscle weakness are represented by filled symbols.
Myopathological and double immunofluorescence analyses
Skeletal muscle biopsies were performed in two patients from Family I, one patient from Family II, three patients from Family III and 10 existing muscle samples from the previously reported German p.W2710X families (Kley et al., 2007) were re-examined and re-evaluated for comparative analysis. Previously established procedures for light and electron microscopy analyses (Olivé et al., 2005) were used. Double immunofluorescence staining was performed on 4 µm frozen serial sections of muscle biopsies from two patients from Family III with p.V930_T933del mutation and two patients with p.W2710X mutation. Primary antibodies used in this study are listed in Table 1. A newly developed FLNC-specific rabbit polyclonal antibody and a mouse monoclonal myotilin antibody were used to localize areas of protein aggregation in double staining experiments with mouse antibodies and rabbit or guinea pig antibodies, respectively. Isotype-specific secondary antibodies conjugated with fluorescein isothiocyanate (Dako), DyLight® 488, Cy3 or Texas Red® (Dianova) were applied according to the recommendations of the manufacturer. Adjacent serial sections were stained with Gomori trichrome and Haematoxylin and Eosin.
Table 1.
Primary antibodies used in double immunofluorescence studies
| Antigen | Clone/code | Source | Company | Dilution |
|---|---|---|---|---|
| 19S regulator | AH1.1 | Mouse (mAb) | Abcam | 1/100 |
| 20S subunits | PA1-979 | Rabbit (pAb) | Thermo Scientific | 1/20 |
| Atrogin-1 (Fbx32) | ab74023 | Rabbit (pAb) | Abcam | 1/100 |
| BAG3 | ab47124 | Rabbit (pAb) | Abcam | 1/2000 |
| CHIP | PC711 | Rabbit (pAb) | Calbiochem | 1/1000 |
| Filamin C | FLNC d16-20 | Rabbit (pAb) | Custom-made (BioGenes) | 1/1000 |
| HDAC6 | BV-48 | Rabbit (pAb) | BioVision | 1/50 |
| Hsp20 | HSP20-11 | Mouse (mAb) | BioTrend | 1/100 |
| Hsp22 | 520817 | Mouse (mAb) | R+D Systems | 1/50 |
| Hsp27 | 2B4 | Mouse (mAb) | Novocastra | 1/500 |
| Hsp40 | C64B4 | Rabbit (pAb) | Cell Signalling | 1/50 |
| Hsp60 | D307 | Rabbit (pAb) | Cell Signalling | 1/50 |
| Hsp70 | 4872 | Rabbit (pAb) | Cell Signalling | 1/50 |
| Hsp90 | E289 | Rabbit (pAb) | Cell Signalling | 1/25 |
| LAMP2 | H4B4 | Mouse (mAb) | DSHB | 1/10 |
| LC3 | 2775 | Rabbit (pAb) | Cell Signalling | 1/200 |
| Myotilin | RS034 | Mouse (mAb) | Novocastra | 1/20 |
| p53 | PAb 240 | Mouse (mAb) | Abcam | 1/250 |
| p62 | GP62-N | Guinea pig (pAb) | Progen | 1/100 |
| Ubiquitin | ab7780 | Rabbit (pAb) | Abcam | 1/50 |
| Ubiquitin +1 | 40B3 | Mouse (mAb) | Abcam | 1/25 |
| VCP | 5 | Mouse (mAb) | ABR | 1/500 |
mAb = monoclonal antibodies; pAb = polyclonal antibodies.
Cloning of full length and truncated filamin C constructs
Full-length FLNC complementary DNA clones in pEGFP-N3 or pEGFP-C2 (Clontech) were obtained as described (Duff et al., 2011). The c.2997_3008del (p.V930_T933del, d7ΔVKYT) mutation was introduced in the complementary DNA encoding full-length FLNC in pEGFP-N3 using the QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene) and the following oligonucleotides: ACAACCATGACTACTCCTACACTGCTGTCCAGCAG and CTGCTGGACAGCAGTGTAGGAGTAGTCATGGTTGT. The c.8130G>A (p.W2710X) mutation was introduced into full-length FLNC clones by exchanging the complementary DNA encoding wild-type immunoglobulin-like domains 23–24 with the truncated variant (Löwe et al., 2007) using a unique BspEI restriction site within the complementary DNA sequence encoding immunoglobulin-like domain 23.
For biochemical analyses complementary DNA fragments encoding wild-type and mutant c.2997_3008del, d7ΔVKYT immunoglobulin-like domains 7–8 or 5–9 were amplified from the patient’s complementary DNA using primers F(GTGCGGGACTTTGAGATCAT) and R(ATTCACCACAAAGGGGCTCT) or F(CACATCCTGCCCGCCCCACCT) and R(AGGCCGAATGGTGGCTTTGAA), respectively. Fragments were cloned into prokaryotic expression vectors pET23-EEF or pET23-T7 enabling expression of fusion proteins carrying a C-terminal His-tag and a C-terminal EEF- or N-terminal T7-immunotag, and into pGEX-6P3 enabling expression of glutathione S-transferase-fusion proteins (Amersham). Integrity of all constructs was verified by sequencing (LCG Genomics).
Expression and purification of recombinant proteins
Protein expression in Escherichia coli BL21(DE3) CodonPlus cells (Stratagene) and purification of His-tagged proteins was performed essentially as described previously (Linnemann et al., 2010). Glutathione S-transferase-tagged proteins were purified using Protino® Glutathione Agarose 4B according to the manufacturer’s instructions (Macherey-Nagel). After purification, the glutathione S-transferase tag was removed using PreScission Protease (Amersham Biosciences) according to the recommendation of the manufacturer. Subsequently, the glutathione S-transferase tag was removed from the protein solution together with the glutathione S-transferase-tagged PreScission Protease, by affinity chromatography using Protino® Glutathione Agarose 4B.
Biophysical characterization of recombinant proteins
Circular dichroism spectroscopy was performed subsequent to dialysis of purified proteins (wild-type d7–8 and mutant d7–8ΔVKYT) against 0.1 M potassium chloride and 10 mM potassium phosphate, pH 7.3. The samples at a concentration of 0.5 mg/ml were added to 1 mm quartz cuvettes and circular dichroism spectra recorded in the spectral range from 195 to 260 nm. For differential scanning fluorimetry (thermofluor) stability assays, SYPRO® Orange dye (40×, Invitrogen) was added to 5 µg protein sample in a total volume of 25 µl. Assays were carried out in an iQTM5 real-time PCR detection system (BIO-RAD). Temperatures varied from 20 to 95°C with a fluorescence excitation at 470 nm and detection at 555 nm.
Structural analysis of recombinant proteins
To understand the effect of mutations at the molecular level, we searched for closest structural homologues in the protein structure database (PDB) employing PSI-Search, and the PHYRE server (Kelley and Sternberg, 2009). Three structural homologues were identified: immunoglobulin-like domain 13 of human FLNC (PDB code: 2DJ4; sequence ID: 38%), immunoglobulin-like domain 14 of human FLNC (PDB code: 2D7M; sequence ID: 38%) and immunoglobulin-like domains 5–6 of Dictyostelium discoideum FLNC (PDB code: 1QFH; sequence ID: 23%). The structure of 2D7M was used for subsequent structural analyses of the deletion mutant FLNC d7ΔVKYT.
Limited proteolysis
Proteolytic susceptibility was investigated using the endopeptidase thermolysin (Sigma). Recombinant proteins were diluted to 10 µM in 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0, 10 µg/ml thermolysin was added and the mixture was incubated at 37°C. At each incubation interval, the reaction was stopped by adding 0.2 vol. 5× SDS sample buffer. The samples were analysed by SDS–PAGE on 10% polyacrylamide gels.
Cell culture and transfection studies
C2 and C2C12 cells were cultured to 60–80% confluence in six-well plates (TPP) in Dulbecco’s modified Eagle medium supplemented with 15% foetal calf serum, 4 mM l-glutamine, 1% non-essential amino acids and 2 mM sodium pyruvate (Invitrogen). Subsequently, the cells were transfected with wild-type or mutant FLNC constructs using Lipofectamine® LTX and Plus Reagent according to the manufacturer’s instructions (Invitrogen). Aggregate formation was evaluated 12 and 24 h after transfection by live cell imaging using an IX51 microscope (Olympus) or an LSM710 confocal microscope (Zeiss). Unpaired t-tests were used for statistical analysis.
Results
Myofibrillar myopathy patients with filamin C mutations exhibit a uniform phenotype
A comparative analysis of clinical data of the 25 newly identified patients (18 from Family I, three from Family II and four from Family III) is presented and compared with data on 41 previously reported patients (Table 2). The average age of disease onset was 39–56 years and did not vary significantly between families. The almost uniform initial symptom was bilateral weakness in the proximal lower limb muscles manifesting as difficulty with rising from a chair and climbing stairs. Other muscle groups became involved upon disease progression, primarily proximal muscles of the upper limbs and later distal muscles, neck flexors, abdominal/paraspinal and rarely, facial muscles. Muscle weakness slowly progressed to inability to walk. Respiratory weakness developed with disease progression and was a frequent cause of death. A number of patients in each studied family had cardiac abnormalities. Sudden cardiac arrest was presumed in at least three patients from the Chinese family (Luan et al., 2010) and in two from the Macedonian family. Three patients from the same Chinese family suffered from unexplained chronic gastroenteritis before the development of neurological symptoms (Luan et al., 2010). A similar phenomenon was noted in one of the newly identified Macedonian patients. Creatine kinase levels were elevated in the majority of cases.
Table 2.
Comparative clinical analysis of 66 filaminopathy patients with myofibrillar myopathy phenotype
| Reference | Family I | Family II | Family III | Kley et al., 2007 | Luan et al., 2010 |
|---|---|---|---|---|---|
| FLNC mutation (domain) | p.W2710X (24th Ig-like) | p.W2710X (24th Ig-like) | p.V930_T933del (7th Ig-like) | p.W2710X (24th Ig-like) | p.K899_V904del/V899_ C900ins (7th Ig-like) |
| Inheritance pattern | AD | AD | AD | AD | AD |
| Country of origin | Macedonia | China | Germany | Germany | China |
| Studied patients | 18 | 3 | 4 | 31 | 10 |
| Onset age, mean (range), years | 41 ± 4 (34–52) | 56 ± 1 (55–57) | 48 ± 12 (34–60) | 44 ± 6 (24–57) | 39 ± 3 (35–40) |
| Gender (female/male) | 12/6 | 1/2 | 3/1 | 22/9 | 3/7 |
| Initial symptoms | |||||
| Back pain | 0/9 | 0/1 | 0/4 | 11/26 | 5/10 |
| Proximal LL weakness | 8/8 | 3/3 | 4/4 | 23/26 | 10/10 |
| Advanced illness | |||||
| Skeletal myopathy | |||||
| Limb girdle weakness | |||||
| LL alone | 0/8 | 3/3 | 0/4 | 4/28 | 0/10 |
| P > D, LL > UL | 3/8 | 0/3 | 4/4 | 20/28 | 10/10 |
| P > D, LL = UL | 0/8 | 0/3 | 0/4 | 2/28 | 0/10 |
| P > D, LL < UL | 3/8 | 0/3 | 0/4 | 1/28 | 0/10 |
| D > P, LL | 2/8 | 0/3 | 0/4 | 1/28 | 0/10 |
| Trunk/abdominal muscle weakness | 6/8 | 0/3 | 2/3 | 10/28 | 2/10 |
| Winged scapula | 4/9 | 0/1 | 3/3 | 6/28 | n.r. |
| Facial weakness | 1/8 | 0/3 | 0/3 | 5/28 | 0/10 |
| Muscle contracture | 2/8 | 0/3 | 1/3 | 0/28 | n.r. |
| Respiratory weakness | 2/8 | 0/3 | 2/3 | 14/30 | n.e. |
| Cardiac findings | |||||
| Conduction blocks | 0/8 | 1/3 | 0/3 | 3/9 | 1/6 |
| LV hypertrophy | 0/8 | 0/3 | 1/3 | 3/9 | n.r. |
| Diastolic dysfunction | 0/8 | 0/3 | 1/3 | 2/9 | n.r. |
| CM (not specified) | 0/8 | 0/3 | 0/3 | 2/9 | 3/6 |
| Sudden cardiac arrest | 2/8 | n.k. | n.k. | n.k. | 3/6* |
| Other** | 0/8 | 0/3 | 1/3 | 4/9 | 2/6 |
| Normal | 6/8 | 2/3 | 2/3 | 16/25 | 1/6 |
| Laboratory studies | |||||
| Elevated CK | 2/8 | 1/1 | 3/3 | 22/26 | 2/2 |
| EMG: myopathic | 4/4 | 0/1 | 2/2 | 16/16 | 2/2 |
| NCS abnormality | n.d. | 0/1 | 0/2 | 0/14 | 0/2 |
| Outcome | |||||
| Wheelchair dependency | 2/8 | 0/3 | 2/4 | 4/31 | n.r. |
| Respirator dependency | 1/8 | 0/3 | 0/4 | 3/31 | n.r. |
| Death before age 65 (range, years) | 8/9 (47–61) | 1/2 (62) | n.k. | 4/9 (54–64) | 6/6 (59–60) |
AD = autosomal dominant; CK = creatine kinase; CM = cardiomyopathy; D = distal; Ig = immunoglobulin; LL = lower limbs; LV = left ventricular; NCS = nerve conduction study; P = proximal; nd = not done; n.r. = not reported; n.e. = not extractable; n.k. = not known.
*presumed; **includes atrial flutter, decreased ejection fraction, mitral/aortic valve regurgitation, palpitations, ST segment depression; number of studied patients is indicated as denominator.
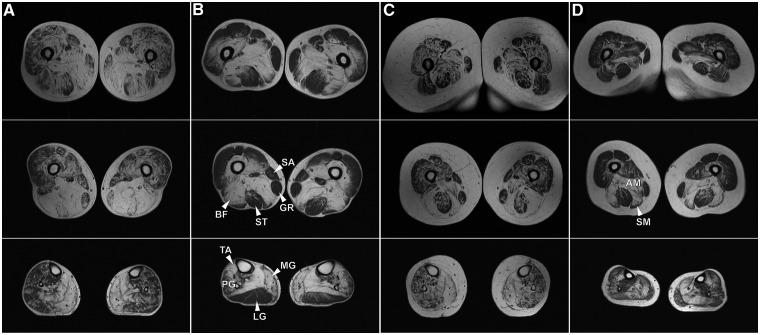
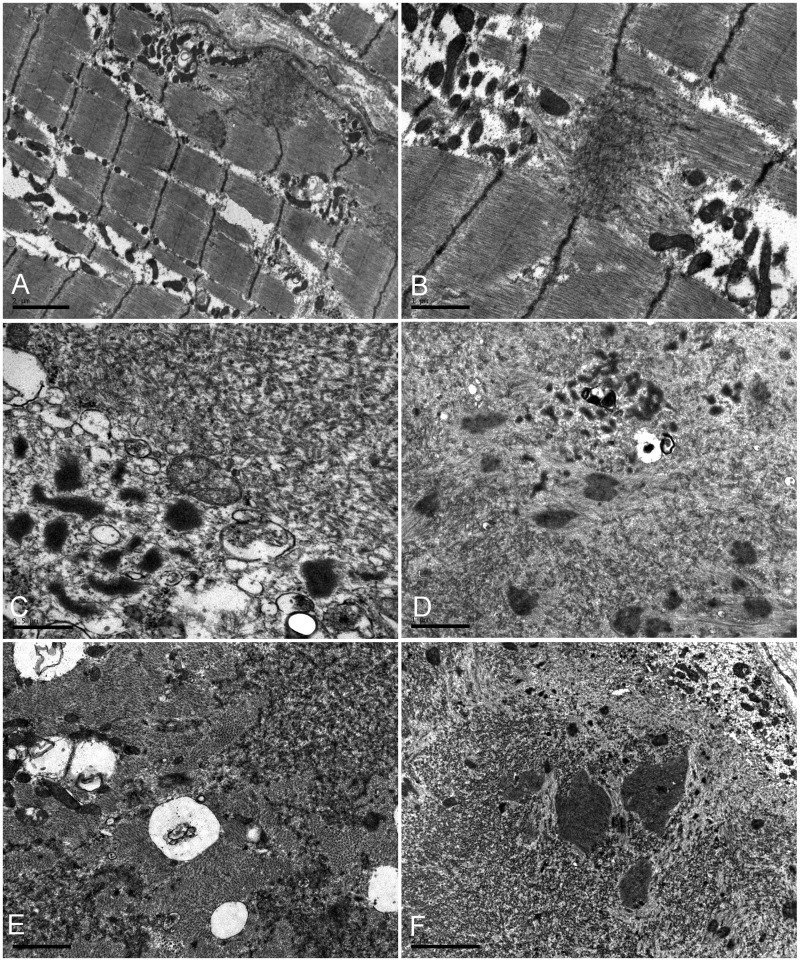
Muscle imaging pattern of lipomatous alterations in lower limb muscles in three patients from Family III carrying the p.V930_T933del mutation (Fig. 2) was similar to that in 10 previously reported p.W2710X patients (Fischer et al., 2008). On the thigh level, adductor longus and adductor magnus, semimembranosus and biceps femoris were most severely affected in the p.W2710X patients, although the involvement of rectus femoris was more pronounced in the carriers of the p.V930_T933del mutation. In the lower legs, the soleus showed distinct alterations in all patients irrespective of the mutation and the medial head of the gastrocnemius was always more affected than the lateral head. Table 3 provides a comparison of myopathological findings in patients with p.W2710X, V930_T933del and p.K899_V904del/V899_C900ins mutations in FLNC. The histological features are similar and seem to be independent of the individual mutation. Some phenomena such as rimmed vacuoles may not be detected in early stages of the disease. The most important finding is the presence of myofibrillar polymorphous aggregates (Supplementary Fig. 1) appearing as single or multiple plaque-like formations within the cytoplasm, as convoluted serpentine inclusions of varying thickness or spheroid bodies. Details of histopathological findings are provided in the online Supplementary material. Ultrastructural examination reveals major myofibrillar abnormalities, including accumulation of fine thin filaments emanating from the Z-disc that coalesce into electron dense inclusions underneath the sarcolemma or between the myofibrils often surrounded by groups of mitochondria (Fig. 3). Many fibres contain large areas occupied by granulofilamentous material interspersed with remnants of filaments and small nemaline rods. Additional findings include tubulofilaments measuring 18 nm and large autophagic vacuoles containing myelin-like figures and cellular debris. In general, the range of myopathological changes in these filaminopathy patients overlap significantly with phenomena described in other subtypes of myofibrillar myopathy.
Figure 2.
MRI (T1-weighted) of lower extremities in filaminopathy patients (first row: proximal upper legs, second row: distal upper legs, third row: lower legs). (A and C) Patients with p.V930_T933del mutation in FLNC (A: a 48-year-old male, disease duration 10 years; C: a 45-year-old female, disease duration 10 years). (B and D) Patients harbouring the p.W2710X mutation in FLNC (B: a 58-year-old male, disease duration 13 years; D: a 54-year-old female, disease duration 5 years). Hyperintensities in muscles denote lipomatous alterations. The relative involvement of marked muscles is important for separation of filaminopathy from other myofibrillar myopathy subtypes. AM = adductor magnus; BF = biceps femoris; GR = gracilis; LG = lateral gastrocnemius; MG = medial gastrocnemius; PG = peroneal group muscles; SA = sartorius; SM = semimembranosus; ST = semitendinosus; TA = tibialis anterior.
Table 3.
Myopathological findings in 17 filaminopathy patients with myofibrillar myopathy phenotype
| Reference | Family I | Family II | Family III | Kley et al., 2007 | Luan et al., 2010 |
|---|---|---|---|---|---|
| FLNC mutation | p.W2710X | p.W2710X | p.V930_T933del | p.W2710X | p.K899_V904del/V899_C900ins |
| Country of origin | Macedonia | China | Germany | Germany | China |
| Number of studied patients | 2 | 1 | 3 | 10 | 1 |
| Light microscopy | |||||
| Variation of fibre size | 2/2 | 1/1 | 3/3 | 10/10 | 1/1 |
| Necrosis/phagocytosis | 0/2 | 0/1 | 2/3 | 8/10 | 1/1 |
| Rimmed vacuoles | 2/2 | 1/1 | 3/3 | 4/10 | 1/1 |
| Polymorphous cytoplasmic inclusions | 2/2 | 1/1 | 2/2 | 9/10 | 1/1 |
| Core-like lesions | 2/2 | 1/1 | 2/2 | 9/10 | 1/1 |
| Type I fibre predominance | 2/2 | 1/1 | 2/2 | 7/10 | n.r. |
| Electron microscopy | |||||
| Granulofilamentous material | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 |
| Z-line streaming | 2/2 | 1/1 | n.r. | 1/1 | 1/1 |
| Tubulofilamentous inclusions | 2/2 | 1/1 | 1/1 | 1/1 | n.r. |
| Nemaline rods | 2/2 | 1/1 | 1/1 | 1/1 | 1/1 |
| Autophagic vacuoles | 2/2 | 1/1 | 1/1 | n.r. | n.r. |
n.r. = not reported.
Figure 3.
Ultrastructural analysis of skeletal muscle from the proband of the Macedonian family. (A and B) Fine filaments emanating at the Z-line level that coalesced into electron-dense inclusions under the sarcolemma or between the myofibrils are surrounded by groups of mitochondria (B). (C–E) Collections of tubulofilaments (right upper corner in C), granulofilamentous material and small vacuoles (left lower corner in C); small rod bodies at the periphery of a fibre region harbouring granulofilamentous material (D), fine electron-dense granulofilamentous material, vacuoles and sparse mitochondria between normal myofibrils (E). (F) Abnormal fibre region containing remnants of filaments, small rod bodies, and prominent electrondense inclusions. Scale bars: C = 0.5 µm; B, D and E = 1 µm; A, F = 2 µm.
Double immunofluorescence studies indicate an impairment of protein degradation in abnormal fibres
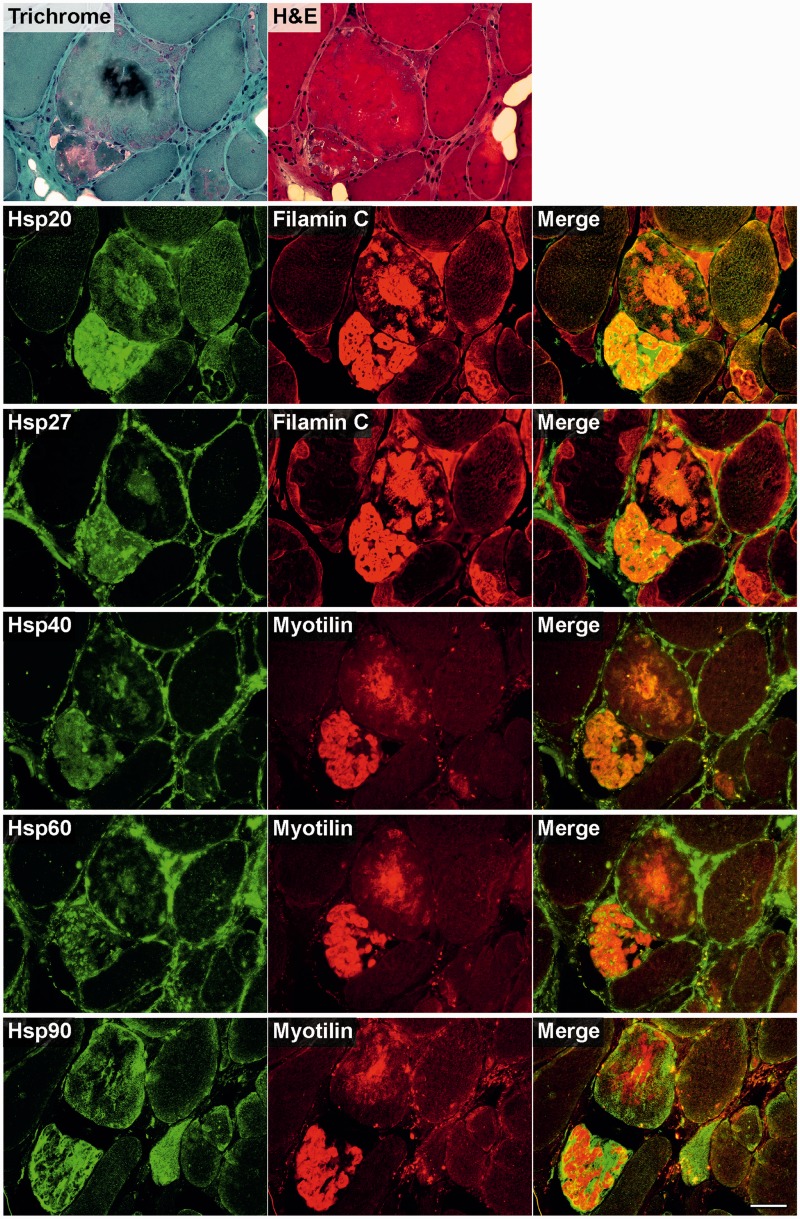
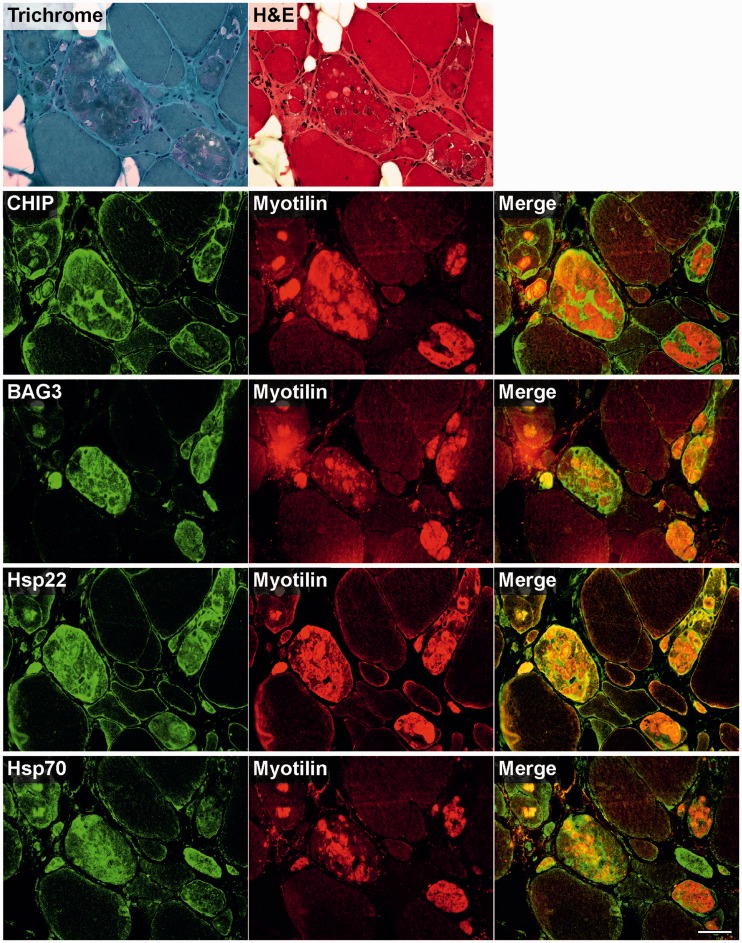
To analyse skeletal muscle samples from patients with myofibrillar myopathy with FLNC mutations for disturbances of protein quality control mechanisms, we studied the expression and distribution patterns of heat shock proteins (Fig. 4), markers for a BAG3-mediated degradation pathway called chaperone-assisted selective autophagy (Fig. 5), the autophagic-lysosomal pathway (Fig. 6), and the ubiquitin–proteasome system (Fig. 7). Findings of our immunolocalization studies were similar in all patients independent of the FLNC mutation (p.V930_T933del or p.W2710X). Figures 4–7 show the results in a patient with p.V930_T933del mutation. It is noteworthy that increased immunoreactivity of all proteins listed above was only detected in abnormal fibres containing protein aggregates (identified by modified Gomori Trichrome staining and immunostaining for FLNC or myotilin) in at least a subset of serial sections, but not in normal looking myofibres.
Figure 4.
Immunolocalization of heat shock proteins in skeletal muscle cryosections from a filaminopathy patient from Family III (p.V930_T933del mutation). Serial cryosections were double-stained with antibodies recognizing the indicated heat shock proteins (Hsp) and either FLNC (Filamin C) or myotilin to localize protein aggregates. For comparison, trichrome and haematoxylin and eosin (H&E) stained sections are shown at the top. Scale bar = 50 µm.
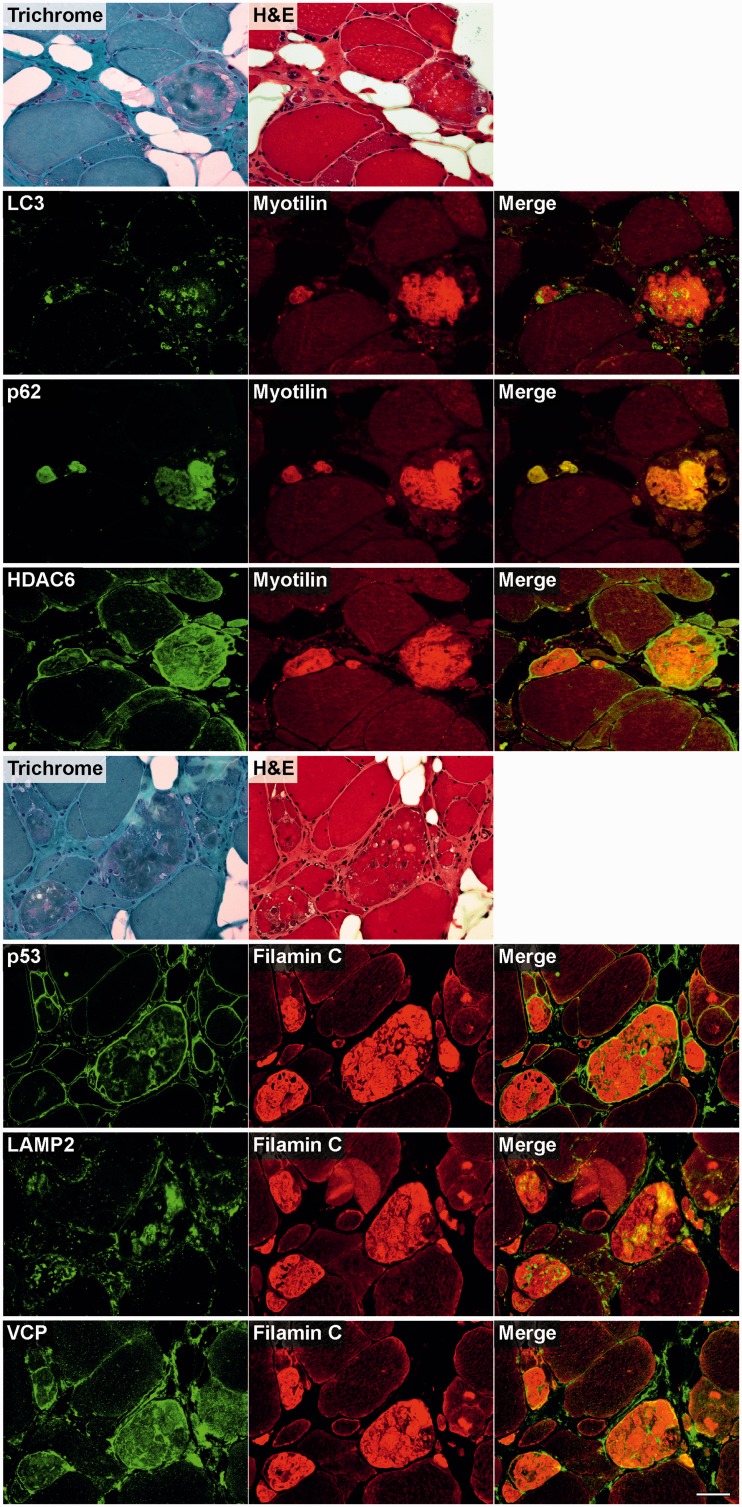
Figure 5.
Immunolocalization of the components of chaperone complexes involved in chaperone-assisted selective autophagy in skeletal muscle cryosections from a filaminopathy patient (p.V930_T933del mutation). Serial cryosections were double-stained with antibodies recognizing the indicated chaperone-assisted selective autophagy pathway markers and either FLNC or myotilin to localize protein aggregates. For comparison, trichrome and haematoxylin and eosin (H&E) stained sections are also shown. Scale bar = 50 µm.
Figure 6.
Immunolocalization of markers of autophagy and associated proteins in skeletal muscle cryosections from a filaminopathy patient with p.V930_T933del mutation. Serial sections were double-stained with antibodies recognizing the indicated heat shock proteins and either FLNC or myotilin to localize protein aggregates. For comparison, trichrome and haematoxylin and eosin (H&E) stained sections are also shown. Scale bar = 50 µm.
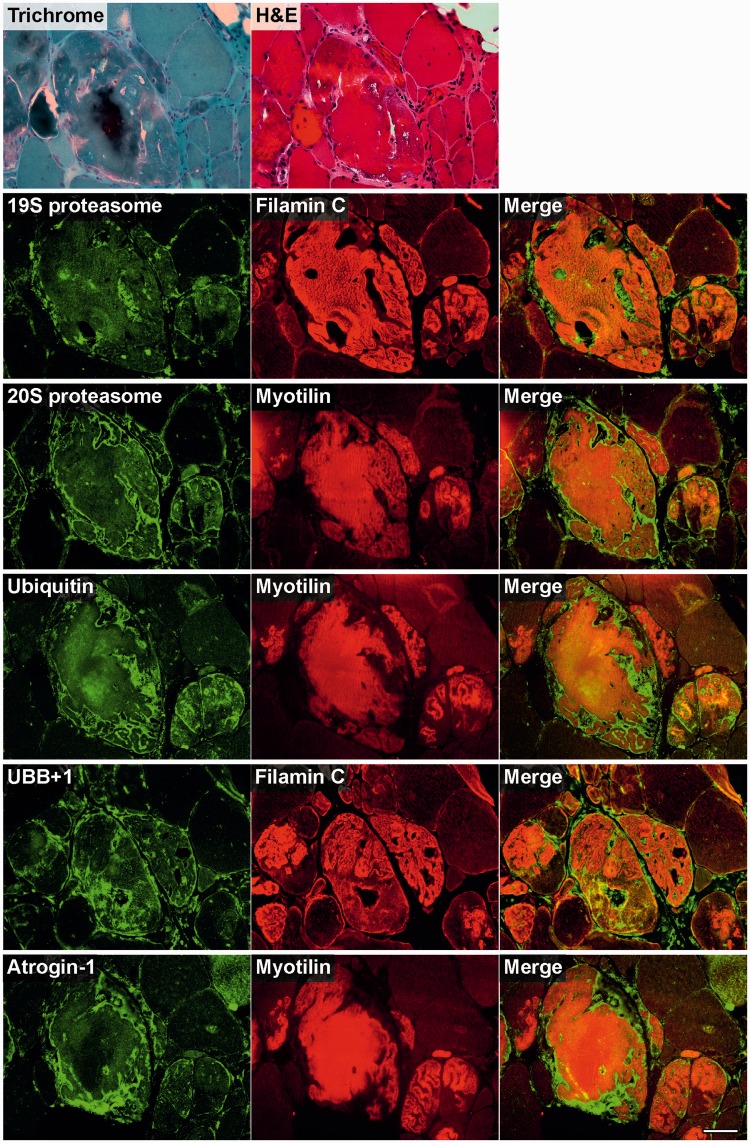
Figure 7.
Immunolocalization of markers of the ubiquitin proteasome system and associated proteins in skeletal muscle cryosections from a filaminopathy patient (p.V930_T933del mutation). Serial sections were double-stained with antibodies recognizing the indicated ubiquitin–proteasome system components and either FLNC or myotilin to localize protein aggregates. For comparison, trichrome and haematoxylin and eosin (H&E) stained sections are also shown. Scale bar = 50 µm.
Immunofluorescence studies revealed increased immunoreactivity for heat shock proteins Hsp20, Hsp27, Hsp40 and Hsp60 in abnormal fibres, predominantly located within protein aggregates. In contrast, immunoreactivity for Hsp90 was especially enhanced in the periphery of aggregates (Fig. 4). Chaperone-assisted selective autophagy components (Hsp22, Hsp70 and p62) showed strong immunoreactivity either within or outside of the protein aggregates (CHIP), while BAG3 was found at both locations (Figs 5 and 6).
Strong immunoreactivity for the autophagy markers LC3 and LAMP2 was more frequently observed between rather than within the aggregates. HDAC6 and VCP also displayed increased immunoreactivity in abnormal fibres, particularly around and in the space between protein deposits. The most distinctly enhanced p53 immunoreactivity was detected between aggregates and in subsarcolemmal regions. Immunoreactivity for p62 was markedly increased within protein aggregates (Fig. 6). Abundant reactivity for 19S proteasome, 20S proteasome, ubiquitin, mutant ubiquitin (UBB+1) and atrogin 1, all markers of ubiquitin–proteasome system, was observed around and between aggregates, but also, especially ubiquitin, within protein deposits (Fig. 7).
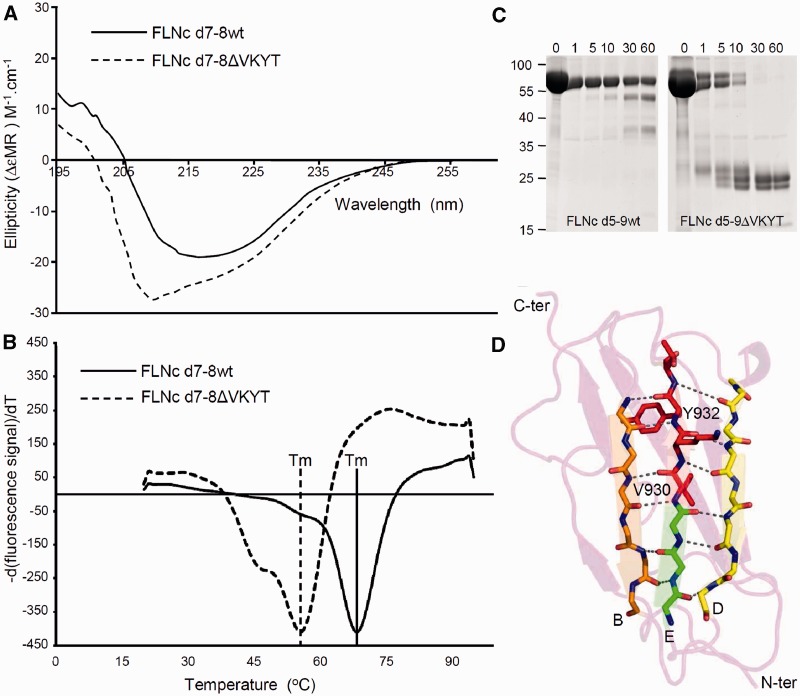
Biophysical characteristics
In order to investigate potential alterations of biophysical and biochemical properties caused by the p.V930_T933del mutation, we first analysed the secondary structure of the mutant d7-8ΔVKYT by circular dichroism spectroscopy of the bacterially expressed recombinant protein. In accordance with the established crystal structures of immunoglobulin-like domains of filamins (Djinović-Carugo and Carugo, 2010; Nakamura et al., 2011), the wild-type proteins exhibited a high proportion of β-strands, as evident by a minimum in the circular dichroism spectra at 218 nm and a positive ellipticity at ∼205 nm. The spectrum of d7-8ΔVKYT was significantly different from that of the wild-type protein, in that the mutant showed a blue shift, displaying a minimum in circular dichroism spectra at ∼208 nm and a positive ellipticity at 200 nm (Fig. 8A). Apart from that, the maximum amplitude of the mutant protein was highly increased. These changes in circular dichroism spectra are a characteristic sign of a higher proportion of unfolded or disordered structures, indicating significant changes in the 3D structure of the mutant domain. Apart from that, the spectrum displays an increase of magnitude in the region typical for α-helices, with the exception of the high-energy spectral range, suggesting formation of a short α-helical region.
Figure 8.
Biophysical characterization and limited proteolysis of the FLNC p.V930_T933del mutation. (A) Circular dichroism spectroscopy performed with purified, bacterially expressed wild-type and deletion mutant FLNC d7-8ΔVKYT constructs. The mean residue ellipticity (ΔεMR) of the wild-type shows a minimum in the spectrum at 218 nm and a positive value at 205 nm, while the deletion mutant displays a blue shift with minimum at 208 nm and positive ellipticity at 200 nm, a characteristic sign of the presence of unstructured regions. Solid line = wild-type; dashed line = FLNC d7-8ΔVKYT construct. (B) Differential scanning fluorimetry. Temperature denaturation experiments for FLNC d7–8 and deletion mutant FLNC d7–8ΔVKYT. The melting temperature (Tm) is determined at the inflection point of the fluorescence signal, the first derivative of which is reported here. Solid line = wild-type; dashed line = deletion mutant, scaled to the wild-type. (C) Protein stability analysed by thermolysin digestion. Wild-type and mutant FLNC d5-9ΔVKYT were treated with the protease thermolysin for 1 to 60 min, as indicated above each lane. Samples were analysed by PAGE. Note that the mutant protein was completely digested after 30 min, while a significant portion of the wild-type variant was still intact after 60 min of incubation, indicating less stable folding of the mutant protein. (D) Ribbon diagram of homology model of immunoglobulin-like domain 7 of human FLNC. β-strands B, E and D forming a β-sheet are coloured orange, green and yellow, respectively. Main chain atoms of residues involved hydrogen bonds indicated by dashed lines, stabilizing the β-sheet are depicted in stick model, with N and O coloured in blue and red. Residues 930VKYT933 that are missing in the deletion mutant are shown as full-atom stick model and coloured in red.
To test whether this alteration undermines protein stability, we compared thermal stability (thermofluor) of a recombinant bacterially expressed fragment comprising immunoglobulin-like domains 7–8 to the corresponding fragment harbouring the deletion mutant (d7–8ΔVKYT) using fluorescence-based thermal shift assays. This revealed a melting temperature of 69°C for the wild-type fragment, whereas the mutant variant showed a considerably decreased average melting temperature of 56°C (Fig. 8B).
The second test for protein stability, limited thermolysin protease digestion, revealed a significantly increased susceptibility of mutant recombinant FLNC fragments d7–8ΔVKYT (not shown) and d5–9ΔVKYT (Fig. 8C). Whereas a major fraction of the wild-type fragments was still intact after 60 min of incubation with the enzyme, the mutant proteins were already almost completely digested after 5–10 min, similarly to our previously described experiments with the p.W2710X mutant FLNC fragments (Löwe et al., 2007).
Actin-binding capacity
Since FLNA was recently shown to bind to F-actin not only through its actin-binding domain, but also through its first rod domain (immunoglobulin-like domains 1–15) (Nakamura et al., 2007), we hypothesized that the mutation in immunoglobulin-like domain 7 might alter actin-binding capacities of FLNC. In our actin co-sedimentation assays using FLNC immunoglobulin-like domains 5–9 we could not, however, detect any binding of either the wild-type or the mutant variant to F-actin (results not shown). This renders it highly improbable that an alteration of the binding capacity of the p.V930_T933del mutant to F-actin plays a significant role in the pathomechanism of filaminopathy caused by this deletion mutation or any other mutation in this part of FLNC.
Transfection studies
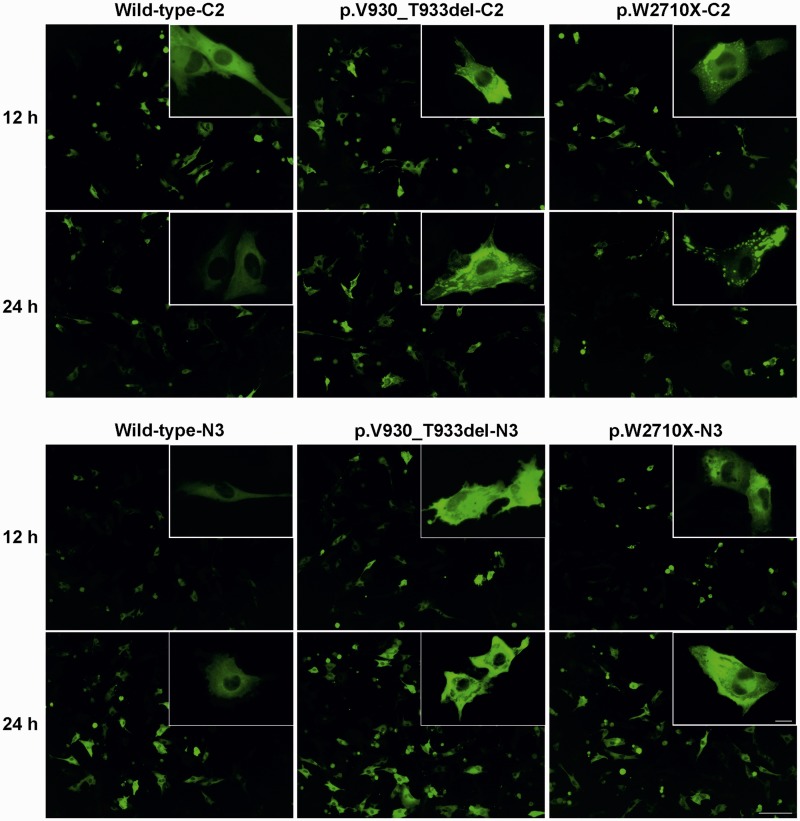
We have previously shown that expression of mutant p.W2710X variants of mini-FLNC (actin-binding domain + immunoglobulin-like domains 15–24) result in spontaneous aggregation of the mutant but not the wild-type protein (Löwe et al., 2007). These experiments were performed in non-muscle cells. To investigate the behaviour of mutant FLNC proteins in muscle cells, we transiently transfected C2 and C2C12 cell lines with constructs encoding N- or C-terminal EGFP-fusion proteins of full-length wild-type, p.W2710X or p.V930_T933del FLNC variants and analysed aggregation at different time points after transfection by using live cell microscopy. In both cell lines the expression of mutant FLNC induced the development of cytoplasmic aggregates (Fig. 9). Analysis of the number of transfected cells containing filamin aggregates showed that in comparison with cells transfected with wild-type FLNC, transfection with p.V930_T933del FLNC yielded 1.6–3.7 times more cells with aggregates, while transfection with p.W2710X FLNC resulted in an 8 to ∼30 times increased number of cells containing aggregates (Table 4). All differences were statistically significant, indicating that expression of mutant FLNC is sufficient for the increased formation of protein aggregates in muscle cells. Noteworthy, transfection with the p.W2710X mutant resulted in up to 11.4 times more cells with aggregates than the p.V930_T933del mutant.
Figure 9.
Transient expression of full-length wild-type, p.W2710X or p.V930_T933del FLNC variants in C2C12 cells. C2C12 cells were transiently transfected with FLNC variants cloned in the pEGFPC2 (−C2) or pEGFPN3 (−N3) vectors and cells were photographed 12 h and 24 h after transfection. Scale bar = 100 µm; insert = 12.5 µm.
Table 4.
Statistical analyses of data from transfection studies
| Cell line | Vector | FLNC insert | 12 h after transfection |
24 h after transfection |
||||
|---|---|---|---|---|---|---|---|---|
| GFP expressing cells (%) | → | Cells harbouring aggregates (%) | GFP expressing cells (%) | → | Cells harbouring aggregates (%) | |||
| C2 | pEGFPC2 | Wild-type | 12.3 | 4.9 | 25.4 | 3.4 | ||
| C2 | pEGFPC2 | p.V930_T933del | 12.6 | 10.5** | 26.4 | 9.3** | ||
| C2 | pEGFPC2 | p.W2710X | 11.4 | 45.4** | 19.7 | 50.3** | ||
| C2 | pEGFPN3 | Wild-type | 25.5 | 0.9 | 36.3 | 2.7 | ||
| C2 | pEGFPN3 | p.V930_T933del | 27.1 | 2.3** | 39.3 | 4.3* | ||
| C2 | pEGFPN3 | p.W2710X | 18.5 | 26.3** | 29.3 | 41.3** | ||
| C2C12 | pEGFPC2 | Wild-type | 29.8 | 4.1 | 32.1 | 13.1 | ||
| C2C12 | pEGFPC2 | p.V930_T933del | 27.3 | 14.8** | 30.5 | 30.7** | ||
| C2C12 | pEGFPC2 | p.W2710X | 20.8 | 47.7** | 21.6 | 60.0** | ||
| C2C12 | pEGFPN3 | Wild-type | 33.9 | 0.8 | 16.9 | 0.9 | ||
| C2C12 | pEGFPN3 | p.V930_T933del | 34.0 | 1.4* | 18.4 | 1.6* | ||
| C2C12 | pEGFPN3 | p.W2710X | 23.7 | 6.4** | 13.6 | 15.4** | ||
*P < 0.05; **P < 0.001 (comparison p.V930_T933del/wild-type and p.W2710X/wild-type). More than 50 000 green fluorescent protein (GFP) expressing cells have been evaluated for statistical analyses.
Discussion
We provide here analysis of phenotypic characteristics and the pathomechanisms of myofibrillar myopathy caused by mutations in immunoglobulin-like domains of FLNC in new and newly re-evaluated families and compare our results to recently reported data. This analysis is needed for the progress of future studies in the field of filaminopathies, including prospects in differential diagnostics and treatment strategies.
Comparative clinical and myopathological characteristics of myofibrillar myopathy associated with filamin C rod mutations
The initial identification of a mutation in FLNC causing myofibrillar myopathy (Vorgerd et al., 2005) induced worldwide genetic testing for FLNC mutations in patients with various types of muscle disease. Subsequently, families with filamin C myopathy were identified in Europe, Asia, USA and Australia (Kley et al., 2007; Shatunov et al., 2009; Luan et al., 2010; Duff et al., 2011; Guergueltcheva et al., 2011; Selcen, 2011a). We include in this report new families from Macedonia and China harbouring the p.W2710X mutation in the dimerization domain of FLNC that is identical to the mutation previously described in German families (Kley et al., 2007). These observations have now established that the p.W2710X mutation is the cause of filaminopathy in genetically unrelated families originating from different ethnic groups, which implies that FLNC codon 2710 is a mutational hotspot. We also report the results of re-evaluation of a German family with an FLNC p.V930_T933del mutation. The phenotypic features of myofibrillar myopathy caused by FLNC rod mutations, either p.W2710X (occurring in immunoglobulin-like domain 24) or p.V930_T933del (occurring in immunoglobulin-like domain 7), are markedly homogeneous, presenting with limb-girdle muscular dystrophy distribution of muscle weakness, involving distal muscles in the course of illness, and showing pathological phenomena similar to the previously described in other myofibrillar myopathy subtypes (Vorgerd et al., 2005). Less than 5% of patients show distal lower limb weakness as the first clinical sign. This phenotype differs, however, from those caused by c.577G>A (p.A193T) and c.752T>C (p.M251T) mutations occurring in the actin-binding domain, or the c.5160delC (p.F1720LfsX63) mutation in the rod domain of FLNC leading to haploinsufficiency; both account for distal myopathy regularly starting in the upper limbs without obvious histological changes typical of myofibrillar myopathy (Duff et al., 2011; Guergueltcheva et al., 2011).
New data on p.W2710X and p.V930_T933del mutations strongly confirm cardiac and respiratory muscle involvement that shortens life expectancy to a significant degree. Diagnostic criteria of this type of disease, in comparison with data available for FLNC-associated distal myopathy, are summarized in Table 5. If these criteria are applicable to a patient under evaluation, it is advisable to perform sequence analysis of FLNC. Since exon 48 is a hot spot for mutations, we recommend sequencing this exon first, followed by a full FLNC sequencing in case of a negative result. It is very important to use appropriate primers to avoid the interference with a pseudogene (pseFLNC) that may lead to misinterpretation of sequencing data (Odgerel et al., 2010; van der Ven et al., 2010). A mutation in the actin-binding domain of FLNC should be considered in patients with distal myopathy, especially if thenar atrophy is the first clinical symptom, and the family history is consistent with an autosomal dominant pattern of inheritance.
Table 5.
Main characteristics of filaminopathies
| FLNC rod domain mutations associated with MFM phenotype | Distal myopathy caused by FLNC actin-binding domain mutations | |
|---|---|---|
| Inheritance pattern | Autosomal dominant | Autosomal dominant |
| Age at onset | Fourth to sixth decade of life | Third to fourth decade of life |
| Initial symptoms | Proximal lower limb weakness | Thenar muscle weakness |
| Advanced illness | Involvement (weakness) of proximal upper limbs, distal limbs and trunk muscles | Calf muscle weakness, proximal muscle weakness |
| Cardiac involvement | Frequent | Insufficient data (reported in two out of 13 patients) |
| Respiratory weakness | Regular in advanced illness | No |
| Muscle imaging of lower limbs (most affected muscles) | Thigh: semimembranosus, adductor magnus and longus, biceps femoris, vastus intermedius and vastus medialis | Thigh: semimembranosus, semitendinosus, biceps femoris, adductor magnus |
| Lower leg: soleus, gastrocnemius medialis, tibialis anterior | Lower leg: soleus, gastrocnemius lateralis and medialis, peroneal muscles | |
| Creatine kinase level | Normal up to 10× increased | Up to 2.5× increased |
| Muscle biopsy | Non-specific changes, dystrophic pattern in advanced disease | Non-specific changes, dystrophic pattern in advanced disease |
| Polymorphous cytoplasmic protein aggregates (plaque-like formations, convoluted serpentine inclusions, spheroid bodies) | No myofibrillar myopathy-typical protein aggregation | |
| Rimmed vacuoles | No rimmed vacuoles | |
| Core-like lesions, Type I fibre predominance | Areas lacking oxidative enzyme activity (moth-eaten appearance) | |
| EM: myofibrillar disintegration, deposits of granulofilamentous material, tubulofilamentous inclusions, nemaline rods, autophagic vacuoles | EM: no myofibrillar pathology |
EM = electron microscopy.
A further helpful contribution to differential diagnosis in this group of disorders is the characterization of muscle imaging. Lipomatous muscle alterations in the lower extremities observed in patients with p.W2710X and p.V930_T933del mutations closely resemble the patterns previously described in patients with myofibrillar myopathy and allow the use of an algorithm we recently developed for the differentiation of myofibrillar myopathy subtypes (Kley et al., 2007; Fischer et al., 2008; Wattjes et al., 2010). The muscle imaging pattern in patients with p.W2710X and p.V930_T933del mutations differs from that reported in patients with the distal myopathy phenotype caused by p.F1720LfsX63 or actin-binding domain FLNC mutations (Duff et al., 2011; Guergueltcheva et al., 2011). Muscle imaging is also useful for selecting an affected muscle for a diagnostic biopsy.
The most characteristic myopathological finding in myofibrillar myopathy-associated filaminopathy is the presence of polymorphous aggregates appearing at light microscopy as single or multiple plaque-like formations, convoluted serpentine inclusions or spheroid bodies within the cytoplasm of affected myofibres and, at the ultrastructural level, as electron-dense inclusions or larger areas of granulofilamentous material interspersed with remnants of filaments and small nemaline rods. Accumulating aggregates eventually leading to myofibrillar destruction is a major feature of myofibrillar myopathy (Nakano et al., 1996; Schröder and Schoser, 2009; Selcen, 2011a).
Insights into pathomechanisms of myofibrillar myopathy associated with filamin C mutations
Mutant FLNC interferes with protein homeostasis thus launching a process that results in the formation of massive protein aggregates. It has been shown that the cell reacts to this process in multiple ways. Molecular chaperones bind non-native proteins and facilitate folding of newly translated or damaged proteins into protein complexes (Bukau and Horwich, 1998; Frydman, 2001; Young et al., 2004; Hartl and Hayer-Hartl, 2009). Other chaperones determine whether misfolded or aggregated proteins are subsequently degraded by the ubiquitin–proteasome machinery or through the autophagy pathway (Massey et al., 2006; Arndt et al., 2007, 2010; Carra et al., 2008; Gamerdinger et al., 2009). Impairment of protein degradation plays a pivotal pathogenic role in protein aggregate diseases including many neurodegenerative diseases that are characterized by intracellular accumulation of altered or misfolded proteins (de Pril et al., 2006; Rubinsztein, 2006; Lehman, 2009; Xilouri and Stefanis, 2011). Abnormal expression of proteins involved in protein degradation was observed in some subtypes of myofibrillar myopathy (Olivé et al., 2008), but specific and detailed knowledge of the mechanisms of disease in FLNC-associated myofibrillar myopathy is still lacking. We performed extensive immunofluorescence studies of several protein degradation components in muscle tissue sections of patients with p.W2710X and p.V930_T933del FLNC mutations.
Our results indicate that muscle fibres react to aggregate formation with a highly increased expression and accumulation of chaperones and proteins involved in proteasomal protein degradation and autophagy including chaperone-assisted selective autophagy. However, these compensatory responses do not warrant efficient protein degradation. The detection of abundant mutant ubiquitin UBB+1 in abnormal fibres suggests ubiquitin–proteasome system dysfunction, as noted in other subtypes of myofibrillar myopathy (Fischer et al., 2003; Ferrer et al., 2004; Janué et al., 2007; Olivé et al., 2008; van Tijn et al., 2010).
Another compensatory response to proteasome impairment caused by misfolded or aggregated proteins is upregulation of autophagy mediated by HDAC6, as has been shown in neurodegenerative diseases (Iwata et al., 2005; Pandey et al., 2007). The highly increased immunoreactivity for HDAC6 in abnormal muscle fibres of our patients therefore can be interpreted as an attempt to increase autophagy. Ubiquitin–proteasome system inhibition by itself is capable of inducing autophagy through p53-mediated mechanisms (Du et al., 2009; Vousden and Ryan, 2009); however, the role of p53 critically depends on its subcellular localization. Nuclear p53 potently induces autophagy through transcriptional activation of DRAM (Crighton et al., 2006) and inactivation of the mTOR (mammalian target of rapamycin) pathway (D’Amelio and Cecconi, 2009; Maiuri et al., 2009). In contrast, cytoplasm p53 suppresses autophagy (Tasdemir et al., 2008). Our detection of p53 in the sarcoplasm of abnormal muscle fibres therefore suggests inhibition of autophagy. This assumption is supported by the markedly increased immunoreactivity for p62, since inhibition of autophagy leads to accumulation of p62 (Korolchuk et al., 2009). In agreement with this finding, the observed accumulation of LC3 in abnormal fibres also indicates that autophagy is inhibited (Masiero et al., 2009).
Our data suggest therefore that the diminished functionality of both ubiquitin–proteasome system and autophagy impairs the capacity of molecular chaperones, whose expression levels are increased in abnormal fibres, to prevent the formation or mediate degradation of aggregates. This also seems to be true of chaperone-assisted selective autophagy, an autophagy pathway dedicated to the degradation of Z-disk proteins like FLNC. This chaperone machinery consists of BAG3, Hsp22, Hsp70 and CHIP and is essential for Z-disk maintenance (Arndt et al., 2010).
In addition to the p.W2710X mutation, we present here for the first time data providing insights into the pathomechanisms of the p.V930_T933del mutation located in immunoglobulin-like domain 7 of FLNC. Structural analysis based on the similarly structured immunoglobulin-like domain 14 of human FLNC (PDB code: 2D7M) identified the location and function of the deleted residues (930VKYT933) within immunoglobulin-like domain 7. These residues reside in β-strand E, which is sandwiched by β-strands B and D, and are involved in a network of interactions stabilizing the immunoglobulin-like fold. The hydrophobic residues V (930) and Y (932) normally stabilize the core of the domain through hydrophobic interactions with neighbouring buried residues. The main chain atoms of residues VKY are also involved in a hydrogen bonding network that stabilizes the β-sheet formed by β-strands B, E and D. Deletion of these four residues therefore has a dual destabilizing effect: first, it disrupts the hydrophobic core of the immunoglobulin-like fold and concomitantly loosens the β-sheet that is critical for immunoglobulin-like domain integrity; this is clearly reflected in the circular dichroism spectra and thermal stability assays. Second, the deletion results in exposure of the hydrophobic core of the domain and leads to non-specific protein–protein interactions resulting in protein aggregation.
These results, together with the previously obtained biochemical and biophysical data on the effects of the p.W2710X mutation (Vorgerd et al., 2005; Löwe et al., 2007), uncover the pathway by which both the p.W2710X and the p.V930_T933del mutations cause a moderate to pronounced spontaneous aggregation of mutant FLNC upon transient expression in murine myoblasts. The efficiency of the transfection of C2 and C2C12 cells and the high proportion of transfected cells that show aggregates make them an excellent model for studies of the effects of pharmacological substances on protein aggregation in muscle cells. The establishment of stably transfected cells might further improve the applicability of this cell culture model for these purposes.
To summarize, our results of functional and immunolocalization studies suggest that pathogenic myofibrillar myopathy-causing mutations in the rod of FLNC promote misfolding of affected immunoglobulin-like domains, thereby inducing aggregation of the mutant protein. Our clinical and histological data, especially the late disease onset and the finding that patients in early disease stages show only mild protein aggregation, indicate that muscle fibres can compensate for this effect for long periods of time by mechanisms including the aforementioned protein degradation strategies. Proteasomal and autophagic degradation pathways were shown to decline in relation to oxidative stress and mitochondrial alterations accruing with ageing (Combaret et al., 2009; Wohlgemuth et al., 2010). Both will attenuate protective mechanisms, resulting in manifest aggregation of mutated FLNC and triggering the accumulation of other proteins. As a consequence, Z-disk proteins entrapped in aggregates will disturb general Z-disk protein homeostasis and lead to focal myofibril destruction. These assumptions are consistent with findings in other protein aggregation disorders (Grune et al., 2004; Lehman, 2009; Riederer et al., 2011).
Implications for future therapeutic strategies
At present, therapy of filaminopathy is limited to symptomatic treatment. Data presented here underline the importance of regular cardiac and pulmonary evaluation in order to prevent complications and premature death by timely pacemaker implantation or respiratory support, as indicated. We also advise our patients to perform regular aerobic exercise training and to try creatine supplementation since this has proven to be safe and beneficial in various muscle diseases (Olsen et al., 2005; Ørngreen et al., 2005; Dawes et al., 2006; Jeppesen et al., 2006; Sveen et al., 2007, 2008; Kley et al., 2011).
The finding that increased immunoreactivity of various components of the protein degradation machinery occurs only in muscle fibres harbouring protein aggregates raises one important possibility: early induction of e.g. chaperones or specific E3 ligases may be able to prevent protein aggregation. If confirmed, this may offer new therapeutic approaches. Results of studies in other myofibrillar myopathy subtypes support this hypothesis: over-expression of αβ-crystallin significantly reduced aberrant protein aggregation in cultured HEK293 cells expressing mutant desmin (Wang et al., 2003). Hsp70 and αβ-crystallin also significantly reduced desmin aggregation and attenuated the induction of ubiquitin–proteasome system malfunction in cardiomyocytes expressing mutant desmin (Liu et al., 2006). In cell models of αβ-crystallinopathy, over-expression of Hsp22, Hsp25, Hsp27, Hsp70 and wild-type αβ-crystallin efficiently prevented aggregate formation (Chávez Zobel et al., 2003; Sanbe et al., 2007). Finally, in a mouse model of αβ-crystallinopathy (HSPB5 R120G transgenic mice), treatment with geranylgeranylacetone, a potent heat shock protein inducer, and over-expression of Hsp22 reduced aggregate formation and improved cardiac function and survival (Sanbe et al., 2009).
A simple over-expression of proteins involved in protein degradation may, however, not be sufficient to eliminate formed aggregates, as implied by the massive expression of such proteins in abnormal fibres (Figs 4–7). The cell culture models presented here currently seem to be most appropriate to test such strategies before testing them in animal models or patients.
Funding
The German Research foundation [KL 2487/1-1 to R.A.K., FOR1228 to Y.H., M.V., D.O.F., FOR1352 to J.H., K.Dj.-C., D.O.F.]; the German Ministry of Education and Research [01GM0887 to R.A.K., P.F.M.v.d.V., M.V., D.O.F.]; the Ruhr-University Bochum [FoRUM K042-09 to R.A.K.]; and the Spanish Instituto de Salud Carlos III [PI08-574 to M.O.]. This research was supported in part by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH.
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
We thank the patients for participation in this study. We also thank Mrs A. Schreiner and Mrs J. Mertens-Rill for technical assistance.
Glossary
Abbreviation
- PDB
protein structure database
References
- Arndt V, Rogon C, Höhfeld J. To be, or not to be–molecular chaperones in protein degradation. Cell Mol Life Sci. 2007;64:2525–41. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–8. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–66. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–44. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- Chávez Zobel AT, Loranger A, Marceau N, Thériault JR, Lambert H, Landry J. Distinct chaperone mechanisms can delay the formation of aggresomes by the myopathy-causing R120G alphaB-crystallin mutant. Hum Mol Genet. 2003;12:1609–20. doi: 10.1093/hmg/ddg173. [DOI] [PubMed] [Google Scholar]
- Combaret L, Dardevet D, Béchet D, Taillandier D, Mosoni L, Attaix D. Skeletal muscle proteolysis in aging. Curr Opin Clin Nutr Metab Care. 2009;12:37–41. doi: 10.1097/MCO.0b013e32831b9c31. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–34. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amelio M, Cecconi F. A novel player in the p53-mediated autophagy: Sestrin2. Cell Cycle. 2009;8:1467. [PubMed] [Google Scholar]
- Dawes H, Korpershoek N, Freebody J, Elsworth C, van Tintelen N, Wade DT, et al. A pilot randomised controlled trial of a home-based exercise programme aimed at improving endurance and function in adults with neuromuscular disorders. J Neurol Neurosurg Psychiatr. 2006;77:959–62. doi: 10.1136/jnnp.2006.087726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pril R, Fischer DF, van Leeuwen FW. Conformational diseases: an umbrella for various neurological disorders with an impaired ubiquitin-proteasome system. Neurobiol Aging. 2006;27:515–23. doi: 10.1016/j.neurobiolaging.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Djinović -Carugo K, Carugo O. Structural portrait of filamin interaction mechanisms. Curr Protein Pept Sci. 2010;11:639–50. doi: 10.2174/138920310794109111. [DOI] [PubMed] [Google Scholar]
- Du Y, Yang D, Li L, Luo G, Li T, Fan X, et al. An insight into the mechanistic role of p53-mediated autophagy induction in response to proteasomal inhibition-induced neurotoxicity. Autophagy. 2009;5:663–75. doi: 10.4161/auto.5.5.8377. [DOI] [PubMed] [Google Scholar]
- Duff RM, Tay V, Hackman P, Ravenscroft G, McLean C, Kennedy P, et al. Mutations in the N-terminal actin-binding domain of Filamin C cause a distal myopathy. Am J Hum Genet. 2011;88:729–40. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G, Pallavicini A, Comelli A, Salamon M, Bortoletto G, Ievolella C, et al. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J Biol Chem. 2000;275:41234–42. doi: 10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Martín B, Castaño JG, Lucas JJ, Moreno D, Olivé M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J Neuropathol Exp Neurol. 2004;63:484–98. doi: 10.1093/jnen/63.5.484. [DOI] [PubMed] [Google Scholar]
- Fischer D, Kley RA, Strach K, Meyer C, Sommer T, Eger K, et al. Distinct muscle imaging patterns in myofibrillar myopathies. Neurology. 2008;71:758–65. doi: 10.1212/01.wnl.0000324927.28817.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer DF, de Vos RAI, van Dijk R, de Vrij FMS, Proper EA, Sonnemans MAF, et al. Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. FASEB J. 2003;17:2014–24. doi: 10.1096/fj.03-0205com. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–47. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel H, Fardeau M. 121st ENMC international workshop on desmin and protein aggregate myopathies. 7-9 November 2003, Naarden, The Netherlands. Neuromuscul Disord 2004; 14: 767–73. [DOI] [PubMed]
- Goebel HH, Fardeau M, Olivé M, Schröder R. 156th ENMC international Workshop: desmin and protein aggregate myopathies, 9-11 November 2007, Naarden, The Netherlands. Neuromuscul Disord. 2008;18:583–92. doi: 10.1016/j.nmd.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Gontier Y, Taivainen A, Fontao L, Sonnenberg A, van der Flier A, Carpén O, et al. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J Cell Sci. 2005;118:3739–49. doi: 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJA. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and 'aggresomes' during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–30. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Guergueltcheva V, Peeters K, Baets J, Ceuterick-de Groote C, Martin JJ, Suls A, et al. Distal myopathy with upper limb predominance caused by filamin C haploinsufficiency. Neurology. 2011;77:2105–14. doi: 10.1212/WNL.0b013e31823dc51e. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct Mol Biol. 2009;16:574–81. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Himmel M, van der Ven PFM, Stöcklein W, Fürst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry. 2003;42:430–9. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–92. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Janué A, Olivé M, Ferrer I. Oxidative stress in desminopathies and myotilinopathies: a link between oxidative damage and abnormal protein aggregation. Brain Pathol. 2007;17:377–88. doi: 10.1111/j.1750-3639.2007.00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen TD, Schwartz M, Olsen DB, Wibrand F, Krag T, Dunø M, et al. Aerobic training is safe and improves exercise capacity in patients with mitochondrial myopathy. Brain. 2006;129:3402–12. doi: 10.1093/brain/awl149. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kley RA, Hellenbroich Y, van der Ven PFM, Fürst DO, Huebner A, Bruchertseifer V, et al. Clinical and morphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130:3250–64. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- Kley RA, Tarnopolsky MA, Vorgerd M. Creatine for treating muscle disorders. Cochrane Database Syst Rev. 2011 doi: 10.1002/14651858.CD004760.pub3. Issue 2. Art. No.: CD004760. DOI: 10.1002/14651858.CD004760.pub3. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–47. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann A, van der Ven PFM, Vakeel P, Albinus B, Simonis D, Bendas G, et al. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and alpha-actinin. Eur J Cell Biol. 2010;89:681–92. doi: 10.1016/j.ejcb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–4. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Löwe T, Kley RA, van der Ven PFM, Himmel M, Huebner A, Vorgerd M, et al. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet. 2007;16:1351–8. doi: 10.1093/hmg/ddm085. [DOI] [PubMed] [Google Scholar]
- Luan X, Hong D, Zhang W, Wang Z, Yuan Y. A novel heterozygous deletion-insertion mutation (2695-2712 del/GTTTGT ins) in exon 18 of the filamin C gene causes filaminopathy in a large Chinese family. Neuromuscul Disord. 2010;20:390–6. doi: 10.1016/j.nmd.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel P, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–6. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–35. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–25. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell Adh Migr. 2011;5:160–9. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Engel AG, Waclawik AJ, Emslie-Smith AM, Busis NA. Myofibrillar myopathy with abnormal foci of desmin positivity. I. Light and electron microscopy analysis of 10 cases. J Neuropathol Exp Neurol. 1996;55:549–62. doi: 10.1097/00005072-199605000-00008. [DOI] [PubMed] [Google Scholar]
- Odgerel Z, van der Ven PFM, Fürst DO, Goldfarb LG. DNA sequencing errors in molecular diagnostics of filamin myopathy. Clin Chem Lab Med. 2010;48:1409–14. doi: 10.1515/CCLM.2010.272. [DOI] [PubMed] [Google Scholar]
- Olivé M, Goldfarb LG, Shatunov A, Fischer D, Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain. 2005;128:2315–26. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- Olivé M, van Leeuwen FW, Janué A, Moreno D, Torrejón-Escribano B, Ferrer I. Expression of mutant ubiquitin (UBB+1) and p62 in myotilinopathies and desminopathies. Neuropathol Appl Neurobiol. 2008;34:76–87. doi: 10.1111/j.1365-2990.2007.00864.x. [DOI] [PubMed] [Google Scholar]
- Olivé M, Odgerel Z, Martínez A, Poza JJ, Bragado FG, Zabalza RJ, et al. Clinical and myopathological evaluation of early- and late-onset subtypes of myofibrillar myopathy. Neuromuscul Disord. 2011;21:533–42. doi: 10.1016/j.nmd.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DB, Ørngreen MC, Vissing J. Aerobic training improves exercise performance in facioscapulohumeral muscular dystrophy. Neurology. 2005;64:1064–6. doi: 10.1212/01.WNL.0000150584.45055.27. [DOI] [PubMed] [Google Scholar]
- Ørngreen MC, Olsen DB, Vissing J. Aerobic training in patients with myotonic dystrophy type 1. Ann Neurol. 2005;57:754–7. doi: 10.1002/ana.20460. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pudas R, Kiema T, Butler PJG, Stewart M, Ylänne J. Structural basis for vertebrate filamin dimerization. Structure. 2005;13:111–9. doi: 10.1016/j.str.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Riederer BM, Leuba G, Vernay A, Riederer IM. The role of the ubiquitin proteasome system in Alzheimer's disease. Exp Biol Med. 2011;236:268–76. doi: 10.1258/ebm.2010.010327. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Yamauchi J, Miyamoto Y, Fujiwara Y, Murabe M, Tanoue A. Interruption of CryAB-amyloid oligomer formation by HSP22. J Biol Chem. 2007;282:555–63. doi: 10.1074/jbc.M605481200. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Daicho T, Mizutani R, Endo T, Miyauchi N, Yamauchi J, et al. Protective effect of geranylgeranylacetone via enhancement of HSPB8 induction in desmin-related cardiomyopathy. PLoS One. 2009;4:e5351. doi: 10.1371/journal.pone.0005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder R, Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol. 2009;19:483–92. doi: 10.1111/j.1750-3639.2009.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D. Myofibrillar myopathies. Curr Opin Neurol. 2008;21:585–9. doi: 10.1097/WCO.0b013e32830a752b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D. Myofibrillar myopathies. Neuromuscul Disord. 2011a;21:161–71. doi: 10.1016/j.nmd.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D, Bromberg MB, Chin SS, Engel AG. Reducing bodies and myofibrillar myopathy features in FHL1 muscular dystrophy. Neurology. 2011b;77:1951–9. doi: 10.1212/WNL.0b013e31823a0ebe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatunov A, Olivé M, Odgerel Z, Stadelmann-Nessler C, Irlbacher K, van Landeghem F, et al. In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet. 2009;17:656–63. doi: 10.1038/ejhg.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjekloća L, Pudas R, Sjöblom B, Konarev P, Carugo O, Rybin V, et al. Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer. J Mol Biol. 2007;368:1011–23. doi: 10.1016/j.jmb.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Sveen M, Jeppesen TD, Hauerslev S, Krag TO, Vissing J. Endurance training: an effective and safe treatment for patients with LGMD2I. Neurology. 2007;68:59–61. doi: 10.1212/01.wnl.0000250358.32199.24. [DOI] [PubMed] [Google Scholar]
- Sveen ML, Jeppesen TD, Hauerslev S, Køber L, Krag TO, Vissing J. Endurance training improves fitness and strength in patients with Becker muscular dystrophy. Brain. 2008;131:2824–31. doi: 10.1093/brain/awn189. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson TG, Chan YM, Hack AA, Brosius M, Rajala M, Lidov HG, et al. Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J Cell Biol. 2000;148:115–26. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- van der Ven PFM, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, et al. Indications for a novel muscular dystrophy pathway. gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J Cell Biol. 2000;151:235–48. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven PFM, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, et al. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp Cell Res. 2006;312:2154–67. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- van der Ven PFM, Odgerel Z, Fürst DO, Goldfarb LG, Kono S, Miyajima H. Dominant-negative effects of a novel mutation in the filamin myopathy. Neurology. 2010;75:2137–38. doi: 10.1212/WNL.0b013e3182031bb3. [DOI] [PubMed] [Google Scholar]
- van Tijn P, Verhage MC, Hobo B, van Leeuwen FW, Fischer DF. Low levels of mutant ubiquitin are degraded by the proteasome in vivo. J Neurosci Res. 2010;88:2325–37. doi: 10.1002/jnr.22396. [DOI] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PF, Bruchertseifer V, Löwe T, Kley RA, Schröder R, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Wang X, Klevitsky R, Huang W, Glasford J, Li F, Robbins J. AlphaB-crystallin modulates protein aggregation of abnormal desmin. Circ Res. 2003;93:998–1005. doi: 10.1161/01.RES.0000102401.77712.ED. [DOI] [PubMed] [Google Scholar]
- Wattjes MP, Kley RA, Fischer D. Neuromuscular imaging in inherited muscle diseases. Eur Radiol. 2010;20:2447–60. doi: 10.1007/s00330-010-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–48. doi: 10.1016/j.exger.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Xu W, Davie EW, Chung DW. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun. 1998;251:914–9. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- Xilouri M, Stefanis L. Autophagic pathways in Parkinson disease and related disorders. Expert Rev Mol Med. 2011;13:e8. doi: 10.1017/S1462399411001803. [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–91. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.