Abstract
The most common progressive myoclonus epilepsies are the late infantile and late infantile-variant neuronal ceroid lipofuscinoses (onset before the age of 6 years), Unverricht–Lundborg disease (onset after the age of 6 years) and Lafora disease. Lafora disease is a distinct disorder with uniform course: onset in teenage years, followed by progressively worsening myoclonus, seizures, visual hallucinations and cognitive decline, leading to a vegetative state in status myoclonicus and death within 10 years. Biopsy reveals Lafora bodies, which are pathognomonic and not seen with any other progressive myoclonus epilepsies. Lafora bodies are aggregates of polyglucosans, poorly constructed glycogen molecules with inordinately long strands that render them insoluble. Lafora disease is caused by mutations in the EPM2A or EPM2B genes, encoding the laforin phosphatase and the malin ubiquitin ligase, respectively, two cytoplasmically active enzymes that regulate glycogen construction, ensuring symmetric expansion into a spherical shape, essential to its solubility. In this work, we report a new progressive myoclonus epilepsy associated with Lafora bodies, early-onset Lafora body disease, map its locus to chromosome 4q21.21, identify its gene and mutation and characterize the relationship of its gene product with laforin and malin. Early-onset Lafora body disease presents early, at 5 years, with dysarthria, myoclonus and ataxia. The combination of early-onset and early dysarthria strongly suggests late infantile-variant neuronal ceroid lipofuscinosis, not Lafora disease. Pathology reveals no ceroid lipofuscinosis, but Lafora bodies. The subsequent course is a typical progressive myoclonus epilepsy, though much more protracted than any infantile neuronal ceroid lipofuscinosis, or Lafora disease, patients living into the fourth decade. The mutation, c.781T>C (Phe261Leu), is in a gene of unknown function, PRDM8. We show that the PRDM8 protein interacts with laforin and malin and causes translocation of the two proteins to the nucleus. We find that Phe261Leu-PRDM8 results in excessive sequestration of laforin and malin in the nucleus and that it therefore likely represents a gain-of-function mutation that leads to an effective deficiency of cytoplasmic laforin and malin. We have identified a new progressive myoclonus epilepsy with Lafora bodies, early-onset Lafora body disease, 101 years after Lafora disease was first described. The results to date suggest that PRDM8, the early-onset Lafora body disease protein, regulates the cytoplasmic quantities of the Lafora disease enzymes.
Keywords: Lafora disease, progressive myoclonus epilepsy, genetic study, PRDM8, Lafora bodies
Introduction
Lafora disease is the principal form of adolescence-onset progressive myoclonus epilepsy. Most patients are completely normal in childhood, with the exception of early learning difficulties in some (Ganesh et al., 2002a). Earliest symptoms are headaches, decline in school performance, spontaneous and induced myoclonus, and convulsive seizures, with EEG showing background slowing and occipital-based irregular spike-wave discharges (Lafora and Glueck, 1911; Minassian, 2001). Subsequent symptoms include dysarthria and ataxia, both strongly accentuated by the myclonus, and visual hallucinations, of both epileptic and psychotic origin (Tinuper et al., 1983; Andrade et al., 2005). All these symptoms worsen over time and within 5 years become intractable. The myoclonus becomes extremely frequent and is associated with alteration of consciousness interfering with every thought, resulting in frustration, itself worsening the myoclonus. The mental confusion is exacerbated by a progressive disinhibited dementia. Convulsive seizures need not be frequent but are never fully controlled. Patients maintain awareness of self and family until very late in the disease and then enter an extended period of vegetative state in status myoclonicus. Death occurs through convulsive status epilepticus and aspiration pneumonia, usually by 10 years from onset. Pathologically, Lafora disease is characterized by increased glycogen content in tissues and presence of malformed glycogen molecules called polyglucosans. The latter accumulate over time into masses called Lafora bodies. Although present in multiple tissues, including skeletal muscle, heart, liver, skin and brain, Lafora bodies are not pathogenic outside the brain, at least not prior to the patient succumbing to the neurological disease (Lafora and Glueck, 1911; Minassian, 2001; Tagliabracci et al., 2007).
Glycogen synthesis begins with the protein glycogenin glycosylating itself to generate a short glucosaccharide, which is then extended by glycogen synthase until a 13-mer glycogenin-bound glucose polymer is produced. Glycogen branching enzyme then detaches the last six glucoses as a hexamer, which it reattaches to the side of the remaining heptamer, thus converting the original linear polymer to a fork with two ends. Glycogen synthase now extends each end, and branching enzyme removes and branches each new hexamer as above, the process continuing until the tightly packed glycogen sphere, containing up to 55 000 glucose units, is formed. Spherical structure provides glycogen solubility, preventing it from precipitating (Roach, 2002). Polyglucosans have excessively long strands and inadequate branching, and lack spherical form. They are more similar to plant starch than to glycogen, and like starch they precipitate, aggregate and accumulate. In the brain, for reasons unknown, this takes place mainly in dendrites, initiating severe progressive myoclonus epilepsy after a threshold accumulation is reached (Sakai et al., 1970; Van Heycop Ten Ham, 1975; Cavanagh, 1999; Minassian, 2001; Turnbull et al., 2011). One other neurological disease, adult polyglucosan body disease, is also characterized by polyglucosan formation (Robitaille et al., 1980). In adult polyglucosan body disease, polyglucosans accumulate exclusively in axons, again for reasons unclear, and the disease is an axonopathy with upper and lower motor neuron signs, sensory neuropathy and bladder control deficits; there is no epilepsy. Adult polyglucosan body disease is caused by mutations in the branching enzyme gene resulting in decreased branching enzyme activity and leading to polyglucosan formation due to inability of branching enzyme to maintain pace with glycogen synthase (Lossos et al., 1998).
Lafora disease is inherited in an autosomal recessive fashion. The disease is caused by mutations in either the EPM2A gene encoding the laforin phosphatase or the EPM2B gene encoding the malin ubiquitin E3 ligase (Minassian et al., 1998; Chan et al., 2003b). Functional studies to date have generated two hypotheses of pathogenesis. Laforin and malin were shown to form a complex that interacts with protein targeting to glycogen (PTG, now known as PPP1R3C), a protein that targets the pleiotropic phosphatase protein phosphatase 1 (PP1) to dephosphorylate and activate glycogen synthase (Fernandez-Sanchez et al., 2003; Solaz-Fuster et al., 2008; Worby et al., 2008). In cell culture experiments, the absence of laforin or malin is associated with increased PTG, most likely through loss of malin-mediated ubiquitination and degradation of glycogen synthase and PTG, and resultant increased glycogen synthase activity (Vilchez et al., 2007). In this hypothesis, increased glycogen synthase activity without increased branching enzyme activity imbalances glycogen elongation and branching, leading to formation of polyglucosans. The second hypothesis stems from the observation that the glycogen synthase reaction is subject to enzymatic error. Episodically, glycogen synthase adds phosphoglucose to glycogen instead of glucose, and laforin corrects this by removing the phosphates (Tagliabracci et al., 2011). When laforin is absent, phosphates accumulate on glycogen and through their high charge disturb glycogen’s spherical structure, rendering it insoluble and leading it to precipitate (Tagliabracci et al., 2008). While the two hypotheses are disparate, they are not mutually exclusive and may be reconciled with future work. Lafora bodies, in any case, have a major pathogenic role in Lafora disease as evidenced by the demonstration that preventing Lafora bodies formation, by reducing glycogen synthesis in laforin-deficient mice, leads to disappearance of myoclonus and neurodegeneration, curing these mice from Lafora disease (Turnbull et al., 2011).
In 2004, we reported preliminary description of a consanguineous family with a progressive myoclonus epilepsy resembling Lafora disease. The proband had Lafora bodies in skeletal muscle but neither she nor her affected siblings had mutations in EPM2A or EPM2B, and the disease did not segregate with EPM2A or EPM2B markers (Chan et al., 2004b). In this work, we detail the clinical and pathological features of this progressive myoclonus epilepsy, identify the underlying disease gene (PRDM8) and describe functional characteristics of the gene’s protein product and its interactions with and effects on laforin and malin.
Materials and methods
Histology and electron microscopy
This study was approved by the Research Ethics Board of the Hospital for Sick Children, and informed consent was obtained from all subjects or their guardians.
Tissues collected at biopsy were fixed in either 10% neutral buffered formalin or minced into pieces ∼1 mm3 in 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. Formalin-fixed materials were embedded in paraffin, sectioned and mounted on glass slides. They were then stained with periodic acid-Schiff reagent following diastase treatment, coverslipped and examined under the light microscope. Material collected in glutaraldehyde was rinsed thoroughly in phosphate buffer, post-fixed in OsO4 and dehydrated and infiltrated in Embed 812. The tissues were then embedded and polymerized at 60°C overnight in flat embedding moulds to insure proper orientation. Areas of interest were then chosen using 0.5 µm toluidine blue-stained sections. Ultrathin sections were then cut, mounted on grids and stained with uranyl acetate and lead citrate prior to examination in a transmission electron microscope (JEOL 1011).
Genome analysis, mutation screening and expression study
Microsatellite linkage excluding the EPM2A and EPM2B loci and mutation screening of both genes was previously described (Chan et al., 2004b). For linkage analysis, autosomal recessive inheritance was specified, with a disease allele frequency of 0.001. Pairwise linkage and simulated maximum logarithm of odds (MAX LOD) scores were calculated using MLINK (Lathrop and Lalouel, 1984; Lathrop et al., 1984). Parametric multipoint linkage analysis was performed using GeneHunter (Kruglyak et al., 1996) with the MAX LOD score calculated using Allegro (Gudbjartsson et al., 2000). Both were performed as previously described (Chan et al., 2003a). Primers for custom microsatellite markers were designed using the Primer3 software (Rozen and Skaletsky, 2000) and flanking dinucleotide repeat sequences with a >20 repeat length.
Single nucleotide polymorphisms were studied using the 10 K Affymetrix GeneChip® SNP array to check for smaller areas of interest that may have been missed from the microsatellite screen. Analysis of copy number variations was performed using the Illumina Human 660 W-Quad beadchip. Pairwise and multipoint parametric linkage analysis was carried out using MERLIN (Abecasis et al., 2002). Regions of interest were determined from MERLIN using a cut-off LOD score of 0.25 and a P-value of ≤0.15. For each region of interest, both microsatellite and single nucleotide polymorphism data were analysed. Haplotypes were constructed and checked for homozygosity and/or shared alleles among the affected members of the family that were not shared with the unaffected siblings.
Genes for mutation screening were prioritized based on function as well as tissue expression. Primers for mutation screening were designed using Primer3 software (Rozen and Skaletsky, 2000). PCR was performed to amplify exons and separated on 1–1.5% agarose gels. PCR products were either purified directly using microCLEAN (Microzone) or gel-extracted and purified using QIAquick® gel extraction kit (QIAGEN). All were directly sequenced for mutations in exons and splice junctions. At least 50 nucleotides on each side of an intron/exon junction were analysed for each exon. Primer sequences are available upon request. Controls were screened for the Phe261Leu-PRDM8 variant using a TaqMan® single nucleotide polymorphism genotyping assay for allelic discrimination (Applied Biosystems).
For semi-quantitative reverse transcriptase-PCR, total RNA (Clontech) from indicated tissues was transcribed to complementary DNA using SuperScript® II (Invitrogen) and random hexamers for amplification.
Histone methyltransferase assays
We performed enzymatic assays of PRDM8 in vitro and in vivo as previously described (Eom et al., 2009) with minor modifications. In vitro, we incubated wild-type or mutant PRDM8 (mammalian-expressed and immunoprecipitated or bacterially expressed and purified) at 30°C overnight in 35 μl reactions including 5× histone methyltransferase assay buffer (50 mM Tris, pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 1.25 M sucrose), 1 μg/μl core histones (Roche) and 200 nCi of S-[methyl-3H]-adenosyl-l-methionine (GE Healthcare Sciences). SET7 histone methyltransferase immunoprecipitated from transfected COS7 cells was used as positive control. Reactions were analysed by autoradiography or scintillation counting. For the latter, reactions were filtered on P81 filter papers (Millipore Inc.) and retained radiolabelled histones washed three times with cold 10% teichloroacetic acid and 70% ethanol for 5 min at room temperature and air-dried. S-[methyl-3H]-adenosyl-l-methionine was quantified by scintillation counting and values normalized to glutathione S-transferase alone.
In the in vivo experiment, we transfected COS7 cells with wild-type or mutant HA-tagged PRDM8, following which we assessed the amounts of endogenous dimethylated histone H3’s using western blotting with specific antibodies.
Immunoprecipitation, immunofluorescence and western blotting
COS7 cells and/or HeLa cells were transfected using Lipofectamine 2000™ (Invitrogen) with 1 µg total DNA for both immunoprecipitation and immunofluorescence experiments and used 48 h after transfection. For immunoprecipitation experiments, cells were lysed using RIPA buffer (Sigma). Lysates were incubated with primary antibody overnight at 4°C, then added to pre-washed protein G-Sepharose beads (Sigma) and incubated for at least 2 h. Beads were washed a minimum of three times with RIPA buffer before eluting by boiling in 10× loading dye.
For immunofluorescence experiments, cells were grown on glass cover slips or chamber slides and fixed in 4% paraformaldehyde. Fixed cells were permeabilized using 0.5% Triton™ X-100 and washed. Cells were blocked in 5% bovine serum albumin/PBS. Primary and secondary antibodies were diluted in 5% bovine serum albumin/PBS. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma), cells were mounted using N-propyl gallate and imaged using a confocal microscope (Zeiss). Images were collected sequentially and then merged. Nuclear co-localization foci sizes were calculated using Volocity 6.0.1 software (PerkinElmer).
For western blotting, immunoprecipitates and/or lysates were run on 12% SDS-PAGE gels before transfer and detection. Blots were probed with antibodies specific for the tagged proteins followed by incubation with either anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology).
Rabbit anti-FLAG, mouse anti-MYC, rabbit anti-haemagglutinin, mouse anti-haemagglutinin, anti-glyceraldehyde 3-phosphate dehydrogenase, and all secondary horseradish peroxidase-conjugated antibodies were from Santa Cruz. Mouse anti-FLAG and rabbit anti-MYC antibodies were from Sigma. Anti-PRDM8 antibody was a kind gift from Dr C. Jung. Anti-laforin antibody was from Abnova. Anti-H3 histone primary antibodies were from Active Motif. Anti-mouse and anti-rabbit fluorescent secondary antibodies were from Jackson Labs. Anti-glycogen synthase and phospho-glycogen synthase antibodies were from Cell Signalling.
Results
The clinical syndrome
The family is of Pakistani origin living in England. Parents are first-degree cousins. Three children are affected, and two, now in their late twenties, are completely normal (Fig. 1A). Table 1 summarizes the clinical features.
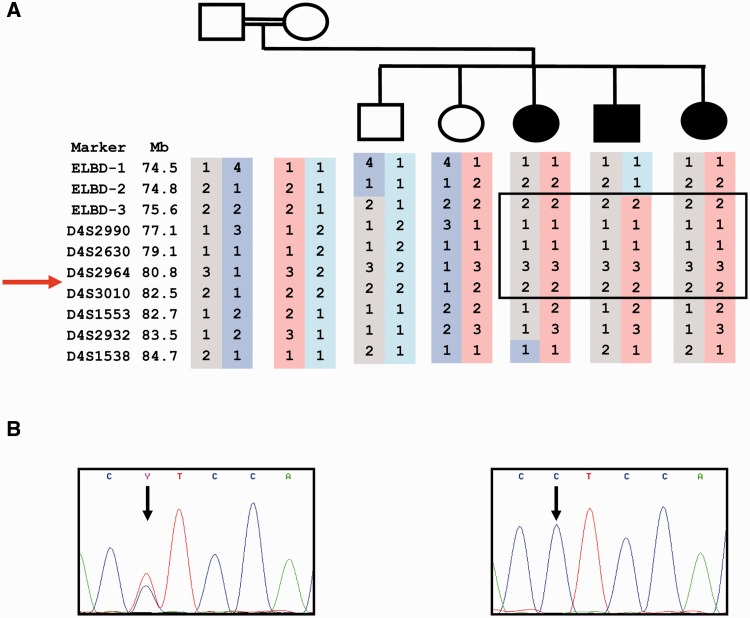
Figure 1.
Gene locus and mutation. (A) Pedigree and locus in chromosome 4q21.21; microsatellite haplotype shown; ELBD-1, 2, 3, un-named microsatellites identified in-house; arrow indicates position of the PRDM8 gene. (B) Non-synonymous single nucleotide replacement is present in homozygous state in all three affected (proband’s electropherogram shown, right panel) and in none of the unaffected individuals (heterozygous change in the father shown, left panel). ELBD = early-onset Lafora body disease.
Table 1.
Clinical features of the three affected patients
| Patient | Onset age (years) | Onset symptoms | Symptoms in late teenage years | Present state |
|---|---|---|---|---|
| 1 | 5 | Cognitive decline, myoclonus | Severely ataxic, wheelchair-dependent, incontinent, myoclonus, seizures, mute | Bedridden, unresponsive, frequent myoclonus, rare seizures, spastic-quadriparetic (age 34) |
| 2 | 5 | Dysarthria, myoclonus, ataxia | Cognitive decline, ataxia, seizures, myoclonus | Severely ataxic, confused, paranoid, screaming outbursts, myoclonus, spastic-hyperreflexic, rare seizures (age 30) |
| 3 | 5 | Dysarthria | Dysarthria | Severely ataxic, confused, paranoid, incontinent, screaming outbursts, myoclonus, spastic-hyperreflexic (age 29) |
The proband’s early development was unremarkable. She began exhibiting school difficulties, dysarthria and myoclonus at age 5 and the next year had her first generalized convulsion. Over the following years, she became increasingly ataxic, lost bladder control, became mute by age 12 and wheelchair-bound by age 14. At present, she is 34-years-old, bedridden, has frequent myoclonus and is unresponsive except for smiling when the family interacts with her. On a combination of levetiracetam, valproic acid and clonazepam, she has generalized seizures only several times a year usually with infections or menses. Examination shows her to be spastic-quadriparetic with bilaterally extensor plantar responses.
The second patient has a milder presentation. He developed dysarthria, myoclonus and gait ataxia since primary school, and a generalized seizure at age 13. He reached secondary school but failed his GCSE (General Certificate of Secondary Education). Presently 30 years old, he is mobile, though severely ataxic and frequently falling. He ventures outdoors and gets lost, and is paranoid with frequent screaming outbursts. He remains continent. He seizes with variable frequency on valproic acid. He married at age 18 (now divorced) and has a daughter. On examination, he is markedly ataxic, spastic and hyperreflexic.
The third affected member, 29 years old, also attained secondary school, failed the GCSE, married and had two children. She had been dysarthric since childhood but apparently was otherwise healthy until age 21 when she developed progressive myoclonus and ataxia and worsening of her dysarthria. She started experiencing hallucinations and paranoia, lost sphincter control and independent ambulation, became mute and at present commonly spends the entire night screaming loudly. Like her affected siblings, she exhibits spasticity, brisk reflexes, severe ataxia and has myoclonus, which is not as frequent as in the other two. She has not had generalized seizures to date.
Neuroimaging and EEG studies were available only from the proband. CT scan at age 19 showed non-specific generalized atrophy. EEG at age 14 showed diffuse slowing with no epileptiform discharges and at age 24 greater slowing with generalized interictal and myoclonus-associated spike-wave discharges.
Pathological findings
Skin biopsy in Lafora disease is commonly performed in the axilla because skin Lafora bodies are present only in cell types related to sweat glands, specifically duct cells of eccrine glands and myoepithelial cells surrounding apocrine glands (Carpenter and Karpati, 1981; Busard et al., 1987; Andrade et al., 2003). Glycogen, in normal or polyglucosan form, stains with periodic acid-Schiff. Normal glycogen is readily digested by diastase (amylase), whereas polyglucosans, because they are insoluble and concretized, are not. Figure 2A shows diastase-resistant periodic acid-Schiff-positive inclusions, Lafora bodies, in the myoepithelia of apocrine glands from a patient with Lafora disease (EPM2B mutation). Analysis of sections through the depth of the dermis of axillary skin biopsies of the proband from the present family showed no Lafora bodies (Fig. 2B). Similar extensive search for skin Lafora bodies in her sister was likewise negative.
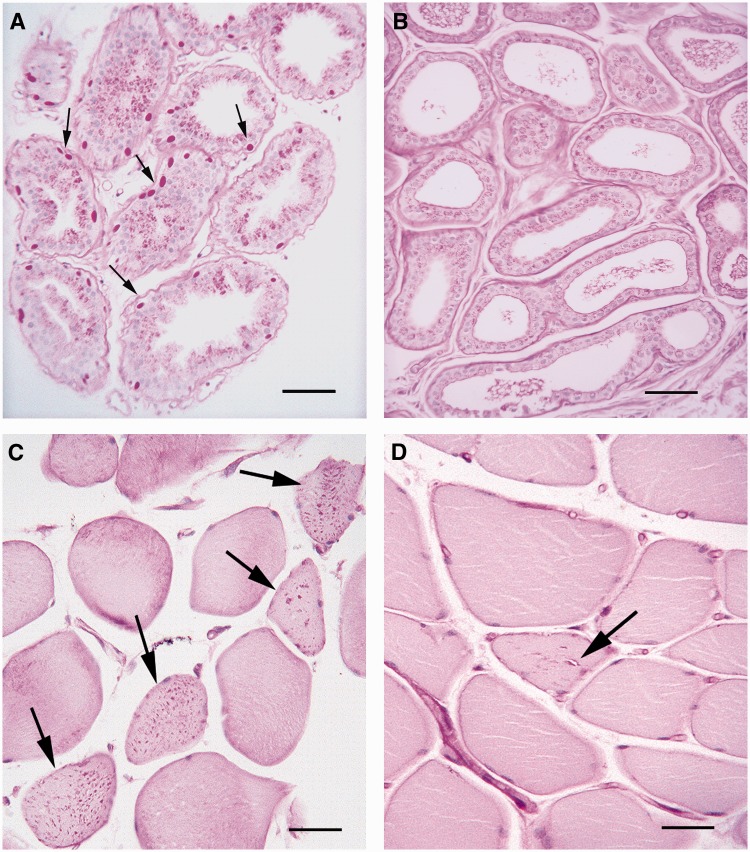
Figure 2.
Absence of Lafora bodies in skin and the presence in skeletal muscle. (A) Apocrine glands in skin from a patient with typical Lafora disease; note numerous Lafora bodies (arrows) in the myoepithelia surrounding the glands. Scale bar = 50 µm. (B) Comparable apocrine glands from the proband of the present family, no Lafora bodies are seen. Scale bar = 50 µm. (C) Skeletal muscle from the proband; numerous Lafora bodies are present throughout the sarcoplasm of several muscle fibres (arrows). Scale bar = 100 µm. (D) Vacuolation in skeletal muscle from the proband. Arrow indicates a sarcoplasmic vacuole surrounded by Lafora bodies. All slides stained with periodic acid-Schiff following diastase treatment. Scale bar = 100 µm.
Skeletal muscle biopsy in both sisters showed numerous Lafora bodies (Fig. 2C). In many cases, these were associated with vacuolar change of the myofibre cytoplasm in proximity to the Lafora bodies, a feature not reported in Lafora disease. These vacuoles were sometimes visible through the light microscope (Fig. 2D) but were better appreciated under electron microscopy, which showed them to consist of whorled membranes. The Lafora bodies themselves, under electron microscopy, were densely packed masses of fibrils (Fig. 3A–C) indistinguishable from Lafora bodies in Lafora disease (e.g. Fig. 3D from a case with Lafora disease and EPM2A mutations). Finally, the Lafora bodies in the present cases differed from those in Lafora disease in that they were never membrane-bound, whereas in Lafora disease, skeletal muscle Lafora bodies are frequently enclosed within a membrane (Carpenter et al., 1974) as in the example in Fig. 3D.
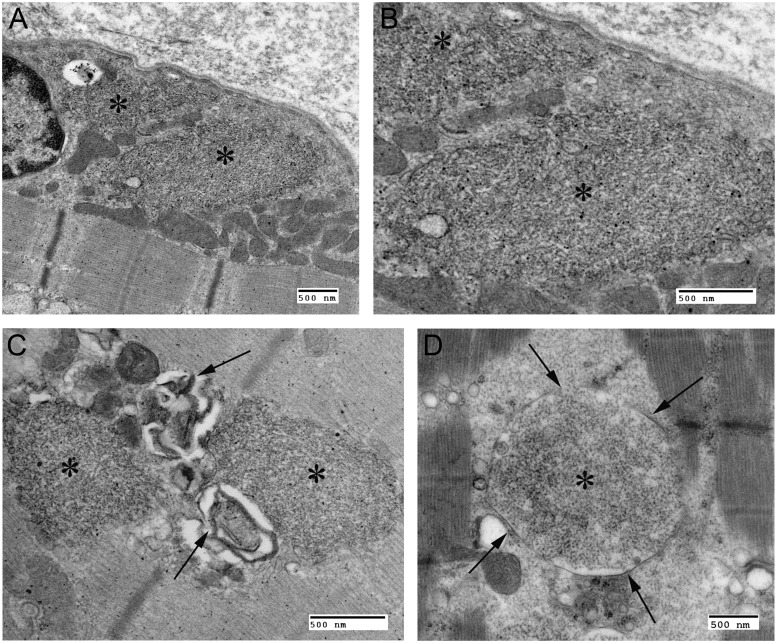
Figure 3.
Electron microscopy of skeletal muscle Lafora bodies. (A) Transmission electron micrograph from the proband: asterisks indicate Lafora bodies in perinuclear sarcoplasm. (B) Higher magnification of A: note filamentous nature of Lafora bodies and absence of any surrounding membrane. (C) Vacuolation (arrows) in association with Lafora bodies in the proband. (D) Transmission electron microscope image of skeletal muscle from a patient with Lafora disease; note membrane-bound Lafora bodies (arrows and asterisk). All scale bars = 500 nm.
The combination of progressive myoclonus epilepsy with Lafora bodies has to date been pathognomonic of Lafora disease. The present family has this combination, but both the progressive myoclonus epilepsy and the Lafora bodies have clear differences from Lafora disease. We propose calling this new disease early-onset Lafora body disease.
Genetic mapping
Two separate genome-wide scans, one using microsatellites and the other single nucleotide polymorphisms, identified a 6.9 Mb homozygous region of interest on chromosome 4 (75.8–82.7 Mb) (Fig. 1A), encompassing 40 genes. In the microsatellite scan, maximum pairwise LOD score in this region was 1.96 (the simulated maximum LOD in this family was 1.45), and multipoint linkage analysis in the region yielded a LOD score of 2.56. No other genomic region had a multipoint LOD score higher than 1.1. In the single nucleotide polymorphism scan, maximum LOD in the region was 0.93 (simulated maximum LOD = 0.94). No other region had a LOD score >0.69. No previously undocumented copy number variations were found in this region or in the whole genome. We analysed for homozygosity in every genomic region with LOD ≥ 0.25 and found that none other than the chromosome 4 region had homozygosity in affected and lack of homozygosity in unaffected siblings. These results indicate that the 6.9 Mb chromosome 4 region is the early-onset Lafora body disease locus.
Exclusion of functional and positional candidate genes
The 6.9 Mb chromosome 4 region contains 40 genes, two of which, STBD1 and SCARB2, are strong functional candidates. The function of STBD1 is not known, but it contains a particular type of carbohydrate binding domain, CBM20, found in laforin. CBM20 is utilized by hundreds of plant and prokaryote starch binding proteins but is extremely rare in mammalian proteomes, having been found in only three proteins (Christiansen et al., 2009). The second functional candidate gene, SCARB2, encodes the membrane-integral protein that transports the glucocerebrosidase enzyme to lysosomes (Reczek et al., 2007). Mutations of this gene cause a progressive myoclonus epilepsy called action myoclonus–renal failure characterized by adult and rarely late-teenage onset progressive tremor, myoclonus (severe), ataxia, dysarthria and infrequent seizures. Most but not all patients have renal failure. All are reported normal in childhood. With the exception of two with mild cognitive changes, the great majority have no cognitive decline or psychosis. Pathology reveals membrane-bound granules containing autofluorescent lipopigments and stacked lamellae, and no polyglucosans or Lafora bodies (Badhwar et al., 2004; Berkovic et al., 2008; Dibbens et al., 2009).
We sequenced all exons of STBD1 and SCARB2 and their flanking intronic and 5′- and 3′-untranslated regions in our patients and found no mutations. We next measured the amounts of STBD1 and SCARB2 messenger RNA using quantitative reverse transcriptase-PCR in fibroblasts and frozen muscle samples from the patients and using western blots quantified STBD1 and SCARB2 proteins in skeletal muscle and found no differences from controls (data not shown). Absence of coding sequence mutations and normal protein quantities in an affected tissue (muscle) do not support a role for STBD1 or SCARB2 in early-onset Lafora body disease.
Of the 38 remaining genes in the region, we excluded 12 based on causative association with other completely distinct diseases in humans or animal models or based on clear lack of expression in brain and/or skeletal muscle. Finally, we sequenced all exons and exon-flanking regions of the remaining 26 genes (Supplementary Table 1). No frameshift, deletion, nonsense or non-synonymous sequence change not present in the single nucleotide polymorphism database (dbSNP) was present except in one gene, PRDM8.
Sequence variant of a highly conserved nucleotide in the PRDM8 gene in patients with early-onset Lafora body disease
We identified a non-synonymous coding sequence change in the PRDM8 gene, c.781T>C (Phe261Leu), present in homozygous state in the three affected patients and in neither unaffected sibling. The parents were heterozygous for the change (Fig. 1B). The sequence alteration is not present in dbSNP, nor did we find it in 100 CEPH controls. We proceeded to screen 500 ethnically and geographically matched individuals (Pashtun from the Attock district, Punjab, northwestern Pakistan). None had the variant in either heterozygous or homozygous state.
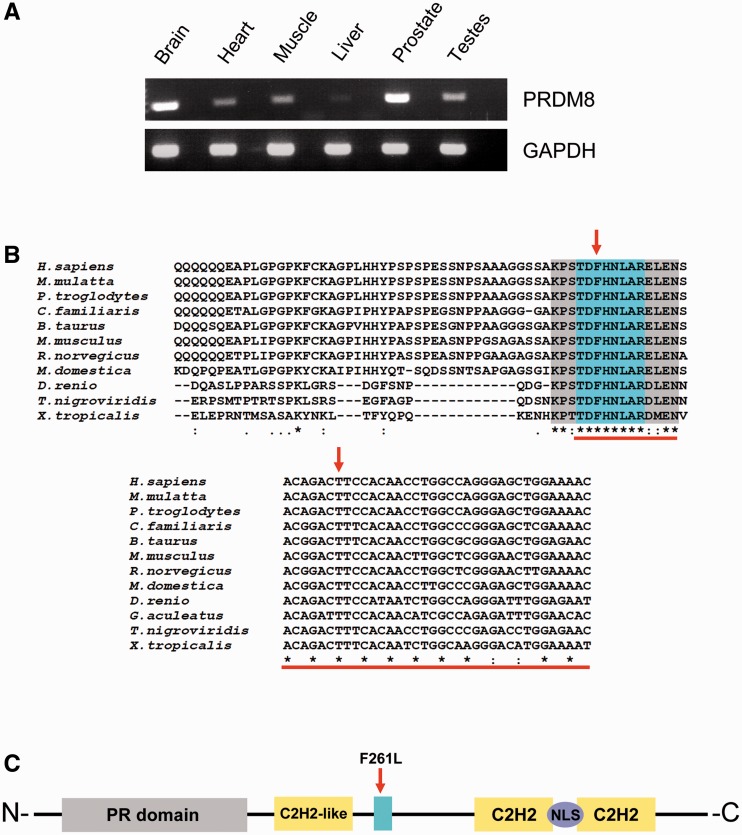
PRDM8 is expressed in brain, skeletal muscle and other tissues (Fig. 4A). Evolutionarily, it is only found in vertebrates (like EPM2A and EPM2B). Comparing the human PRDM8 primary structure to all sequenced orthologues shows that the Phe261 amino acid is part of a block of eight amino acids that is completely conserved across vertebrates. The exact c.781T nucleotide is itself conserved throughout vertebrate evolution (Fig. 4B).
Figure 4.
PRDM8 expression, domain conservation and structure. (A) Semiquantitative reverse transcriptase-PCR using human tissue complementary DNAs and primers amplifying across the first PRDM8 exon–exon boundary: the gene is expressed in brain and skeletal muscle, among other human tissues; lanes between bands are reverse transcriptase-PCRs of preceding tissues with reverse transcriptase omitted. (B) ClustalW alignments of PRDM8 protein and gene sequences from different vertebrate species show strict evolutionary conservation of the substituted amino acid, the protein domain containing this amino acid and the mutated nucleotide. (C) Five conserved domains with predicted functions are present in PRDM8, the PR domain, three zinc finger domains (C2H2 and C2H2-like) and the nuclear localization signal (NLS); blue highlight indicates position of the conserved domain containing the replaced amino acid.
PRDM8 does not have methyltransferase activity
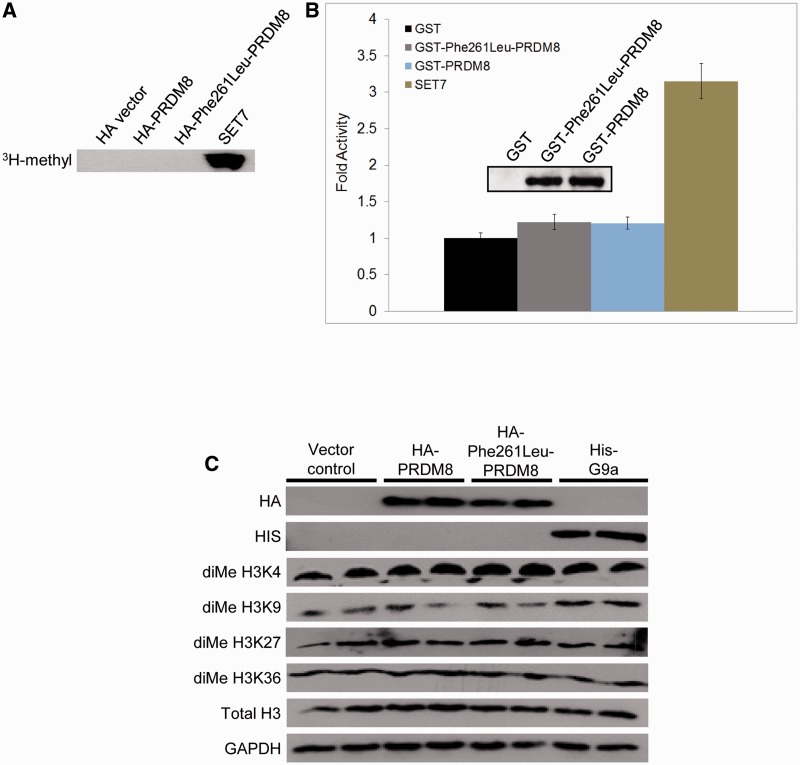
Bioinformatic analysis could not assign a predicted function to the conserved eight-amino acid block containing the sequence change. The protein contains other conserved domains, five with predicted functions: three zinc finger domains, commonly utilized in protein–DNA or protein–protein interaction (Brayer and Segal, 2008), a nuclear localization signal and a PR domain (Fig. 4C). The PR domain has 20–30% similarity to the SET histone methyltransferase domain. Although this similarity does not include a key histidine required by SET for methyltransferase activity (Rea et al., 2000; Davis et al., 2006), to date 3 of 17 PR containing proteins have been shown to possess histone methyltransferase activity (Kim et al., 2003; Hayashi et al., 2005; Eom et al., 2009), including PRDM8, which was reported to increase levels of histone H3 dimethylated at lysine 9 (diMe H3K9) (Eom et al., 2009). To determine whether the Phe261Leu substitution affects this activity, we first tested the methyltransferase activity reported in wild-type PRDM8. We were unable to confirm this activity, using the same methods as in the earlier report (Eom et al., 2009). We find no methyltransferase activity of wild-type PRDM8 on core histones generally (Fig. 5A and B), nor increased dimethylation of lysines 4, 9, 27 or 36 of histone H3 specifically (Fig. 5C). Expectedly, Phe261Leu-mutated PRDM8 also has no histone methyltransferase activity (Fig. 5).
Figure 5.
PRDM8 histone methyltransferase assays. (A and B) Wild-type and mutant PRDM8 have no above-baseline methyltransferase activities against mixed core histones. Haemagglutinin (HA)-tagged proteins were expressed in mammalian cells and activities detected using autoradiography; glutathione S-transferase (GST)-tagged proteins were bacterially expressed/purified and activities measured by scintillation counting, expressed as fold increase normalized to glutathione S-transferase alone. SET7 = positive control. Inset in (B) shows western blot of purified PRDM8 proteins used in the assay. (C) Levels of endogenous histone H3 dimethylated at lysines 4, 9, 27 and 36 are not increased following transfection with wild-type (or mutant) haemagglutinin-tagged PRDM8 (each lane represents a separate transfection); first two rows, levels of transfected proteins; ‘Total H3’ row, levels of total histone H3; bottom row, GAPDH loading control; pcDNA-His-G9a is a specific methyltransferase positive control for diMe H3K9.
PRDM8 relocates cytoplasmic laforin and malin to distinct foci in the nucleus
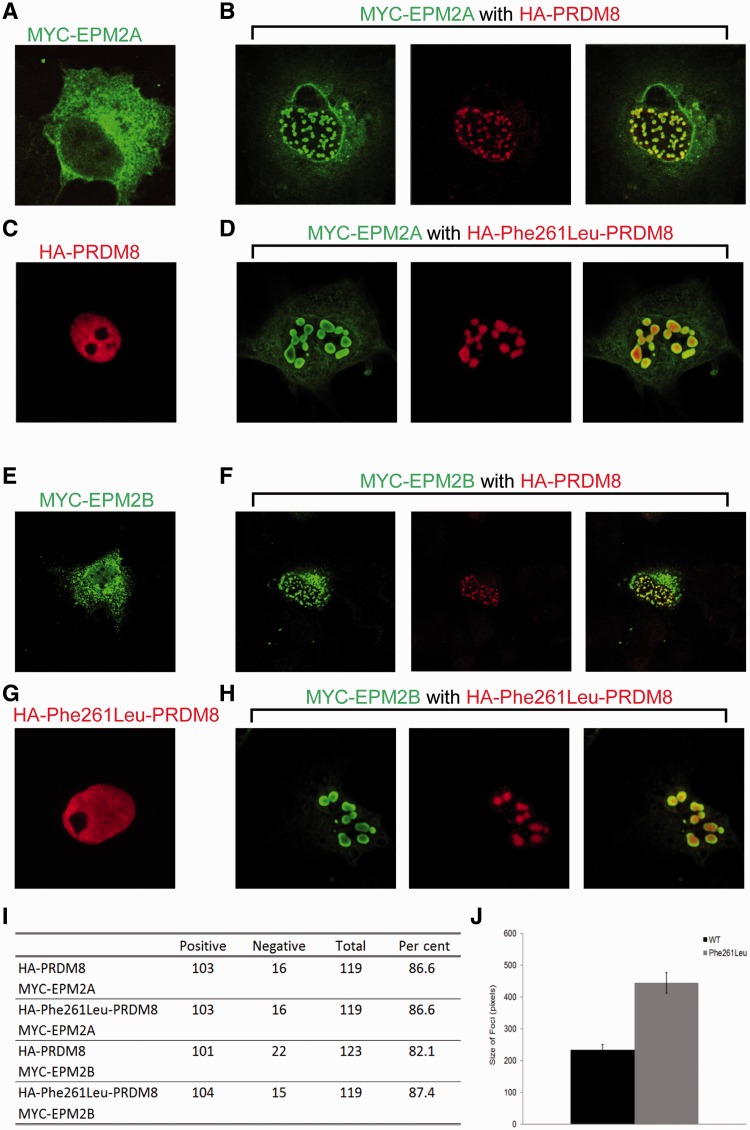
To date, due to lack of suitable antibodies, the subcellular locations of the EPM2A and EPM2B gene products (laforin and malin) have been studied through epitope-tagging complementary DNAs of each gene, transfection and overexpression in cells, and staining with epitope-specific antibodies. Both proteins under these conditions are predominantly cytoplasmic, with a minor portion of laforin, and a somewhat larger portion of malin localizing in the nucleus (Fig. 6A and E) (Chan et al., 2003b; Cheng et al., 2007; Singh et al., 2008). The nuclear localization signal of PRDM8 predicts that PRDM8 is nuclear, and indeed epitope-tagged PRDM8 localized wholly in the nucleus (Fig. 6C). To assess whether the small nuclear fractions of laforin and malin co-localize with PRDM8, we expressed each with PRDM8. We observed that not only did nuclear laforin co-localize with PRDM8, but that in the presence of PRDM8 a large portion of cytoplasmic laforin relocated to the nucleus. In the nucleus, PRDM8, which when alone is diffusely distributed (Fig. 6C), in the presence of laforin was no longer so and instead localized intensely with laforin in distinctive punctate foci (Fig. 6B). These foci are not associated with DNA (Supplementary Video 1). The same results were obtained with malin (Fig. 6E and F), and also when both laforin and malin were expressed with PRDM8 (result not shown). The relocation of laforin or malin from cytoplasm to the nucleus by PRDM8 was not unspecific, as PRDM8 did not alter the location of two other cytoplasmic proteins (glycogen synthase and PTG); it was not cell-specific, as we observed it in two cell types used (COS7 and HeLa), and expression of a separate nuclear protein, NKX6.1, did not cause any nuclear relocation of laforin and malin (not shown).
Figure 6.
PRDM8 modifies the subcellular distributions of laforin (EPM2A) and malin (EPM2B). (A) Representative image of subcellular distribution of overexpressed laforin; note predominant cytoplasmic expression in reticular pattern and minor nuclear presence. (B) PRDM8 and laforin co-expression: green signal, laforin; red, PRDM8. (C) Nuclear distribution of overexpressed PRDM8. (D) Mutant Phe261Leu-PRDM8 and laforin co-expression: green, laforin; red, PRDM8. (E) Representative image of subcellular distribution of overexpressed malin: cytoplasmic expression in reticular pattern and diffuse nuclear expression. (F) PRDM8 and malin co-expression: green, malin; red, PRDM8. (G) Nuclear distribution of overexpressed Phe261Leu-PRDM8. (H) Mutant Phe261Leu-PRDM8 and malin co-expression: green, malin; red, PRDM8. (I) Percentage of transfected cells with nuclear sequestration foci of laforin and malin by wild-type versus Phe261Leu-PRDM8. (J) Size of nuclear foci with laforin, malin and either wild-type PRDM8 or Phe261Leu-PRDM8. Size is shown as means ± SEM and significance calculated using an unpaired student’s t-test (P < 2.54 × 10−9).
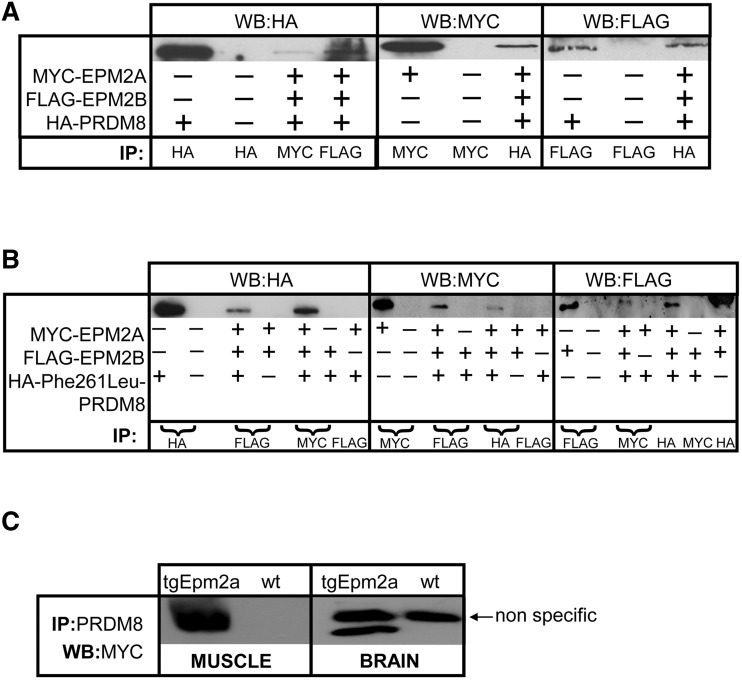
When laforin, malin and PRDM8 were expressed together, immunoprecipitating one co-precipitated the other two (Fig. 7A), indicating that the three proteins form a complex. The same results were obtained with PRDM8 with laforin alone or PRDM8 with malin alone (not shown). These, we note, though reproduced many times, were more difficult to obtain than the triple expression experiments, which readily replicate, suggesting greater stability of the complexes when all three proteins are present together.
Figure 7.
Laforin and malin co-precipitate with both PRDM8 and mutant Phe261Leu-PRDM8. (A and B) The three proteins co-expressed, immunoprecipitating one co-precipitates the other two. MYC-EPM2A = myc-tagged laforin; FLAG-EPM2B = FLAG-tagged malin; HA-PRDM8 = haemagglutinin-tagged PRDM8; HA-Phe261Leu-PRDM8 = haemagglutinin-tagged mutant Phe261Leu PRDM8; IP = antibody used for imunoprecipitation; WB = antibody used for western blot; (+) indicates that the particular plasmid is transfected and (–) that it is not. (C) PRDM8 immunoprecipitates myc-tagged laforin from skeletal muscle and brain of a transgenic mouse (tgEpm2a) overexpressing myc laforin; wt = wild-type mouse tissues.
Although we still do not have antibodies that immunoprecipitate endogenous laforin or malin from animal tissues, we were able to obtain a polyclonal antibody that precipitates endogenous PRDM8. We had previously generated a transgenic mouse line that overexpresses a myc-tagged form of laforin (Chan et al., 2004a). Immunoprecipitating PRDM8 from skeletal muscle and brain from these mice co-precipitated myc-laforin (Fig. 7C).
Repeating the co-localization and co-immunoprecipitation experiments with Phe261Leu-mutated PRDM8, we obtained the same results as with the wild-type protein, i.e. Phe261Leu-PRDM8 relocates laforin and malin to the nucleus in distinctive large punctuate foci and co-immunoprecipitates with these two proteins (Figs 6D, G and H and 7B). To determine whether there is any difference in the nuclear sequestration of laforin and malin by mutant PRDM8 versus wild-type, we first measured the percentage of co-transfected cells exhibiting the intense nuclear co-localization puncta and found no differences (Fig. 6I). We next measured sizes of the co-localization foci and found they are highly significantly larger with mutant than with wild-type (Fig. 6J), suggesting a greater nuclear relocation or confinement of laforin and malin by mutant than by wild-type PRDM8.
Polyglucosan formation in early-onset Lafora body disease appears not to be associated with increased glycogen synthase
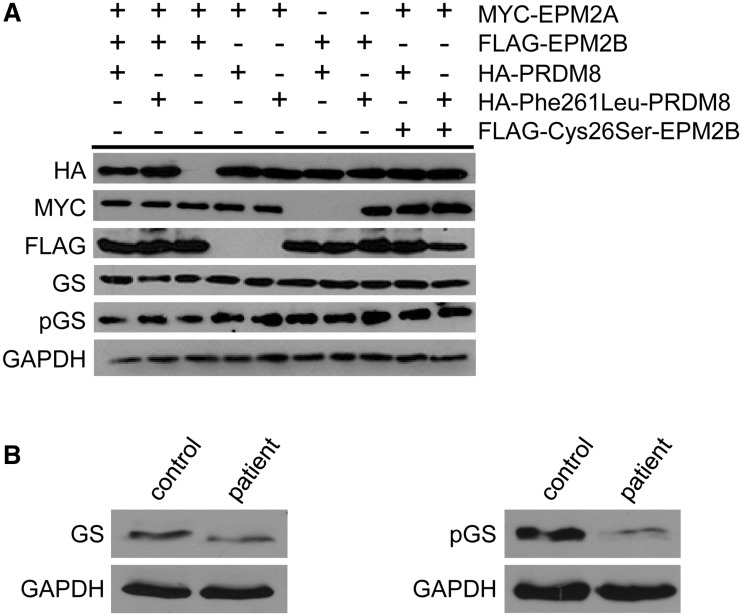
As mentioned, the two leading hypotheses of polyglucosan formation in Lafora disease, i.e. in the absence of cytoplasmic laforin or malin, are as follows: (i) increased glycogen synthase activity (due to increased glycogen synthase protein and decrease of its phosphorylation); and (ii) increased glycogen phosphorylation. Testing hypothesis (ii) is not possible in cell culture (glycogen quantities are inadequate for phosphate content measurement) and requires whole tissues. We tested hypothesis (i) by western blotting for total glycogen synthase and glycogen synthase phosphorylated at Ser640 (which reflects the overall glycogen synthase phosphorylation state; Skurat and Roach, 2004) in PRDM8- and Phe261Leu-PRDM8-transfected cells. Total glycogen synthase was not increased nor was phosphorylated glycogen synthase decreased with mutant PRDM8, indicating no effect of the mutation on glycogen synthase activity (Fig. 8A). Performing these western blots in a small amount of muscle biopsy tissue still available from one of the patients with early-onset Lafora body disease, we found that the quantity of phosphorylated glycogen synthase was decreased. However, the total quantity of glycogen synthase was also decreased, suggesting no likely net change in total glycogen synthase activity (Fig. 8B). Combining the cell culture experiments with this single patient sample assay, our preliminary exploration of the mechanisms of Lafora body formation in early-onset Lafora body disease does not support hypothesis (i), i.e. increased glycogen synthase activity, in the generation of polyglucosans in this disease.
Figure 8.
Total and phosphorylated glycogen synthase levels with Phe261Leu-PRDM8 overexpression in early-onset Lafora body disease. (A) Western blots of extracts of cell cultures overexpressing indicated (+) proteins. GS = glycogen synthase; pGS = phosphorylated glycogen synthase; FLAG-Cys26Ser-EPM2B = a ubiquitin ligase-inactive malin mutant, used as a control; GAPDH = loading control; note that neither glycogen synthase nor phosphorylated glycogen synthase are altered with Phe261Leu-PRDM8 compared with wild-type PRDM8. (B) Western blots of extracts from skeletal muscle from a patient with early-onset Lafora body disease and a normal control with antibodies against total (left) and phosphorylated (right) glycogen synthase.
Discussion
We have identified a new progressive myoclonus epilepsy, the core features of which are childhood onset progressive dysarthria, myoclonus, ataxia, cognitive decline, psychosis, dementia, spasticity and a protracted course so far into the fourth decade of life. There is variability between patients. While all have early speech difficulties, in one, the rest of the symptoms were delayed until adulthood, after which they followed a course similar to her siblings.
Early-onset Lafora body disease differs from the progressive myoclonus epilepsies of Unverricht–Lundborg disease, Lafora disease and sialidosis. Its onset is earlier, one of its presenting symptoms, dysarthria, is not part of early phases of these diseases, and its myoclonus is somewhat less severe. Its cognitive decline is much more severe than in Unverricht–Lundborg disease and sialidosis, where cognitive loss is minimal (Lehesjoki and Koskiniemi, 1999; Canafoglia et al., 2011). Its psychosis differs from that of Lafora disease, where patients have visual hallucinations (Andrade et al., 2005) but do not exhibit outbursts of prolonged agitation and screaming. It does not stabilize in adulthood, as occurs with Unverricht–Lundborg disease, but does not progress quite as rapidly as Lafora disease where most patients are deceased by age 30.
Clinically, early-onset Lafora body disease would be closest to certain cases of neuronal ceroid lipofuscinosis. CLN1 (now known as PPT1; infantile ceroid lipofuscinosis), CLN2 (now known as TPP1; late-infantile ceroid lipofuscinosis), and CLN5, 6, 7 (now known as MFSD8) and 8 (late-infantile variant ceroid lipofuscinosis) all include cases where the progressive myoclonus epilepsy presents later than late infancy, after age 5, with dysarthria, myoclonus, ataxia and seizures (Bessa et al., 2006, 2008; Cannelli et al., 2009; Kousi et al., 2009; Reinhardt et al., 2010; Perez-Poyato et al., 2011). Early-onset Lafora body disease and these cases diverge in the subsequent course of the progressive myoclonus epilepsy, which is more protracted in early-onset Lafora body disease than most, though not all, of these cases. They also diverge in visual loss, which is mainstay in these neuronal ceroid lipofuscinosis and not present in early-onset Lafora body disease.
Early-onset Lafora body disease differs from the remaining known progressive myoclonus epilepsies. The progressive myoclonus epilepsy due to SCARB2 mutations has adult onset and little to no cognitive decline (Berkovic et al., 2008). The disease due to PRICKLE1 mutation does start in childhood but has a particularly prolonged period of ataxia preceding other progressive myoclonus epilepsy symptoms and manifests little or no cognitive loss (Bassuk et al., 2008). Finally, the progressive myoclonus epilepsy caused by GOSR2 mutation also begins in early childhood but is distinguished by the presence of drop attacks, the absence of cognitive problems till age 25 and scoliosis (Corbett et al., 2011).
Pathologically, early-onset Lafora body disease is now the only known progressive myoclonus epilepsy, other than Lafora disease, with Lafora bodies, at least in skeletal muscle. So far, we cannot find Lafora bodies in skin. Whether there are Lafora bodies in brain, and to what extent they contribute to the neurodegeneration and clinical symptoms in this disease remain to be addressed in future studies. In the clinic, patients with a disease resembling childhood-onset comparatively mild neuronal ceroid lipofuscinosis without visual disturbance and without CLN mutations should be tested for PRDM8 mutations or by muscle biopsy for Lafora bodies. It is in these patients that we expect additional cases of early-onset Lafora body disease will be found. PRDM8 is not likely a gene for typical Lafora disease. At present, in our collection of >120 patients with the classical Lafora disease phenotype, including onset after age 10 and Lafora bodies in skin, every patient has mutations in either EPM2A or EPM2B.
We identified the causative defect of early-onset Lafora body disease in the PRDM8 gene. We showed that the normal protein product removes laforin and malin from the cytoplasm, their normal locale of activity. We therefore hypothesize that the PRDM8 missense mutation in the early-onset Lafora body disease family is a gain-of-function alteration that over-sequesters laforin and malin in the nucleus or prevents their timely or sufficient release to the cytoplasm for adequate activity. Consistent with this hypothesis, we find that Phe261Leu-PRDM8 confines larger amounts of laforin and malin nucleoplasmically than does the wild-type protein. Precisely how this results in generation of polyglucosans in the cytoplasm remains unknown but does not appear to involve glycogen synthase overactivity. The gain-of-function hypothesis predicts that the absence of PRDM8 should not result in Lafora bodies. While this article was in review, a preliminary description of PRDM8-deficient mice was published, and no Lafora bodies were mentioned (Ross et al., 2012). It must be noted, however, that these mice were not studied after 2 months of age when Lafora bodies in Lafora disease mouse models become evident (Ganesh et al., 2002a; Turnbull et al., 2010), nor were their skeletal muscles analysed. The mice did have neurological symptoms, including abnormal excessive scratching behaviour of neurological origin and in some an abnormal gait (Ross et al., 2012). Future experiments replacing Phe261Leu-mutated PRDM8 in this strain of mice should serve as a powerful animal model of early-onset Lafora body disease.
Is the nucleus only a sequestration and storage zone for laforin and malin, or do these proteins have nuclear functions? Recent work has shown that under heat shock laforin and malin translocate to the nucleus and participate in the cytoprotective heat shock response (Sengupta et al., 2011). It is possible that the intricate transcriptional regulation of heat shock proteins, in which laforin and malin appear to participate, involves PRDM8, their new nuclear partner identified in the present study.
One hundred and one years ago, Lafora described the inclusions (Lafora bodies) that distinguished the second and more severe progressive myoclonus epilepsy, Lafora disease, from the first, Unverricht–Lundborg disease, described by Unverricht 20 years prior (Unverricht, 1891; Lafora and Glueck, 1911). We now describe a second progressive myoclonus epilepsy with Lafora bodies, early-onset Lafora body disease, intermediate in severity between Unverricht–Lundborg disease and Lafora disease. Recent studies have shown that Lafora bodies are causative of Lafora disease. Preventing their formation in Lafora disease mice completely prevents the disease, including its neurodegeneration and progressive myoclonus epilepsy (Turnbull et al., 2011). PRDM8 is now a third starting point after laforin and malin towards uncovering the biochemical basis of Lafora bodies generation in severe and fatal childhood-onset epilepsy with neurodegeneration.
Funding
This work was supported by the Canadian Institutes for Health Research (B.A.M. - MOP14667) and The National Institutes of Health (Y.L. - 1R21NS062391). J.T. was supported by a National Sciences and Engineering Research Council of Canada, Canada Graduate Scholarship.
Supplementary material
Supplementary material is available at Brain online.
Glossary
Abbreviation
- LOD
logarithm of odds
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Andrade DM, Ackerley CA, Minett TS, Teive HA, Bohlega S, Scherer SW, et al. Skin biopsy in Lafora disease: genotype-phenotype correlations and diagnostic pitfalls. Neurology. 2003;61:1611–4. doi: 10.1212/01.wnl.0000096017.19978.cb. [DOI] [PubMed] [Google Scholar]
- Andrade DM, del Campo JM, Moro E, Minassian BA, Wennberg RA. Nonepileptic visual hallucinations in Lafora disease. Neurology. 2005;64:1311–2. doi: 10.1212/01.WNL.0000156907.49247.05. [DOI] [PubMed] [Google Scholar]
- Badhwar A, Berkovic SF, Dowling JP, Gonzales M, Narayanan S, Brodtmann A, et al. Action myoclonus-renal failure syndrome: characterization of a unique cerebro-renal disorder. Brain. 2004;127:2173–82. doi: 10.1093/brain/awh263. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008;83:572–81. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovic SF, Dibbens LM, Oshlack A, Silver JD, Katerelos M, Vears DF, et al. Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet. 2008;82:673–84. doi: 10.1016/j.ajhg.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa C, Teixeira CA, Dias A, Alves M, Rocha S, Lacerda L, et al. CLN2/TPP1 deficiency: the novel mutation IVS7-10A>G causes intron retention and is associated with a mild disease phenotype. Mol Genet Metab. 2008;93:66–73. doi: 10.1016/j.ymgme.2007.08.124. [DOI] [PubMed] [Google Scholar]
- Bessa C, Teixeira CA, Mangas M, Dias A, Sa Miranda MC, Guimaraes A, et al. Two novel CLN5 mutations in a Portuguese patient with vLINCL: insights into molecular mechanisms of CLN5 deficiency. Mol Genet Metab. 2006;89:245–53. doi: 10.1016/j.ymgme.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein–protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50:111–31. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- Busard HL, Gabreels-Festen AA, Renier WO, Gabreels FJ, Stadhouders AM. Axilla skin biopsy: a reliable test for the diagnosis of Lafora’s disease. Ann Neurol. 1987;21:599–601. doi: 10.1002/ana.410210613. [DOI] [PubMed] [Google Scholar]
- Canafoglia L, Franceschetti S, Uziel G, Ciano C, Scaioli V, Guerrini R, et al. Characterization of severe action myoclonus in sialidoses. Epilepsy Res. 2011;94:86–93. doi: 10.1016/j.eplepsyres.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Cannelli N, Garavaglia B, Simonati A, Aiello C, Barzaghi C, Pezzini F, et al. Variant late infantile ceroid lipofuscinoses associated with novel mutations in CLN6. Biochem Biophys Res Commun. 2009;379:892–7. doi: 10.1016/j.bbrc.2008.12.159. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Karpati G. Sweat gland duct cells in Lafora disease: diagnosis by skin biopsy. Neurology. 1981;31:1564–8. doi: 10.1212/wnl.31.12.1564. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Karpati G, Andermann F, Jacob JC, Andermann E. Lafora’s disease: peroxisomal storage in skeletal muscle. Neurology. 1974;24:531–8. doi: 10.1212/wnl.24.6.531. [DOI] [PubMed] [Google Scholar]
- Cavanagh JB. Corpora-amylacea and the family of polyglucosan diseases. Brain Res Brain Res Rev. 1999;29:265–95. doi: 10.1016/s0165-0173(99)00003-x. [DOI] [PubMed] [Google Scholar]
- Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, et al. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum Mol Genet. 2004a;13:1117–29. doi: 10.1093/hmg/ddh130. [DOI] [PubMed] [Google Scholar]
- Chan EM, Bulman DE, Paterson AD, Turnbull J, Andermann E, Andermann F, et al. Genetic mapping of a new Lafora progressive myoclonus epilepsy locus (EPM2B) on 6p22. J Med Genet. 2003a;40:671–5. doi: 10.1136/jmg.40.9.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, et al. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology. 2004b;63:565–7. doi: 10.1212/01.wnl.0000133215.65836.03. [DOI] [PubMed] [Google Scholar]
- Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003b;35:125–7. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease. Genes Dev. 2007;21:2399–409. doi: 10.1101/gad.1553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen C, Abou Hachem M, Janecek S, Vikso-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20—diversity, structure, and function. FEBS J. 2009 doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- Corbett MA, Schwake M, Bahlo M, Dibbens LM, Lin M, Gandolfo LC, et al. A mutation in the Golgi Qb-SNARE Gene GOSR2 causes progressive myoclonus epilepsy with early ataxia. Am J Hum Genet. 2011;88:657–63. doi: 10.1016/j.ajhg.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, et al. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–36. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Michelucci R, Gambardella A, Andermann F, Rubboli G, Bayly MA, et al. SCARB2 mutations in progressive myoclonus epilepsy (PME) without renal failure. Ann Neurol. 2009;66:532–6. doi: 10.1002/ana.21765. [DOI] [PubMed] [Google Scholar]
- Eom GH, Kim K, Kim SM, Kee HJ, Kim JY, Jin HM, et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem Biophys Res Commun. 2009;388:131–6. doi: 10.1016/j.bbrc.2009.07.134. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, Garcia-Fojeda B, Medrano-Fernandez I, Gomez-Garre P, et al. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum Mol Genet. 2003;12:3161–71. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, et al. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet. 2002a;11:1251–62. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- Ganesh S, Delgado-Escueta AV, Suzuki T, Francheschetti S, Riggio C, Avanzini G, et al. Genotype-phenotype correlations for EPM2A mutations in Lafora's progressive myoclonus epilepsy: exon 1 mutations associate with an early-onset cognitive deficit subphenotype. Hum Mol Genet. 2002b;11:1263–71. doi: 10.1093/hmg/11.11.1263. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–3. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–8. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Kim KC, Geng L, Huang S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003;63:7619–23. [PubMed] [Google Scholar]
- Kousi M, Siintola E, Dvorakova L, Vlaskova H, Turnbull J, Topcu M, et al. Mutations in CLN7/MFSD8 are a common cause of variant late-infantile neuronal ceroid lipofuscinosis. Brain. 2009;132:810–9. doi: 10.1093/brain/awn366. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- Lafora GR, Glueck B. Beitrag zur Histopathologie der myoklonischen Epilepsie. Zeitschrift Gesamte Neurologische Psychiatrie. 1911;6:1–14. [Google Scholar]
- Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–5. [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA. 1984;81:3443–6. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehesjoki AE, Koskiniemi M. Progressive myoclonus epilepsy of Unverricht–Lundborg type. Epilepsia. 1999;40(Suppl 3):23–8. doi: 10.1111/j.1528-1157.1999.tb00895.x. [DOI] [PubMed] [Google Scholar]
- Lossos A, Meiner Z, Barash V, Soffer D, Schlesinger I, Abramsky O, et al. Adult polyglucosan body disease in Ashkenazi Jewish patients carrying the Tyr329Ser mutation in the glycogen-branching enzyme gene. Ann Neurol. 1998;44:867–72. doi: 10.1002/ana.410440604. [DOI] [PubMed] [Google Scholar]
- Minassian BA. Lafora’s disease: towards a clinical, pathologic, and molecular synthesis. Pediatr Neurol. 2001;25:21–9. doi: 10.1016/s0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–4. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- Perez-Poyato MS, Mila Recansens M, Ferrer Abizanda I, Montero Sanchez R, Rodriguez-Revenga L, Cusi Sanchez V, et al. Juvenile neuronal ceroid lipofuscinosis: clinical course and genetic studies in Spanish patients. J Inherit Metab Dis. 2011;34:1083–93. doi: 10.1007/s10545-011-9323-7. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reczek D, Schwake M, Schroder J, Hughes H, Blanz J, Jin X, et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–83. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Reinhardt K, Grapp M, Schlachter K, Bruck W, Gartner J, Steinfeld R. Novel CLN8 mutations confirm the clinical and ethnic diversity of late infantile neuronal ceroid lipofuscinosis. Clin Genet. 2010;77:79–85. doi: 10.1111/j.1399-0004.2009.01285.x. [DOI] [PubMed] [Google Scholar]
- Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002;2:101–20. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- Robitaille Y, Carpenter S, Karpati G, DiMauro SD. A distinct form of adult polyglucosan body disease with massive involvement of central and peripheral neuronal processes and astrocytes: a report of four cases and a review of the occurrence of polyglucosan bodies in other conditions such as Lafora's disease and normal ageing. Brain. 1980;103:315–36. doi: 10.1093/brain/103.2.315. [DOI] [PubMed] [Google Scholar]
- Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000a;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form): II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20:160–76. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Badhwar I, Upadhyay M, Singh S, Ganesh S. Malin and laforin are essential components of a protein complex that protects cells from thermal stress. J Cell Sci. 2011;124:2277–86. doi: 10.1242/jcs.082800. [DOI] [PubMed] [Google Scholar]
- Singh S, Satishchandra P, Shankar SK, Ganesh S. Lafora disease in the Indian population: EPM2A and NHLRC1 gene mutations and their impact on subcellular localization of laforin and malin. Hum Mutat. 2008;29:E1–12. doi: 10.1002/humu.20737. [DOI] [PubMed] [Google Scholar]
- Skurat AV, Roach PJ. Regulation of glycogen synthesis. In: Leroith D, Taylor SI, editors. Diabetes mellitus: a fundamental and clinical text. 3 edn. Philadelphia: Lippincott Williams & Wilkins; 2004. [Google Scholar]
- Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, et al. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17:667–78. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, et al. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283:33816–25. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, et al. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–82. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, et al. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104:19262–6. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinuper P, Aguglia U, Pellissier JF, Gastaut H. Visual ictal phenomena in a case of Lafora disease proven by skin biopsy. Epilepsia. 1983;24:214–8. doi: 10.1111/j.1528-1157.1983.tb04881.x. [DOI] [PubMed] [Google Scholar]
- Turnbull J, Depaoli-Roach AA, Zhao X, Cortez MA, Pencea N, Tiberia E, et al. PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS Genet. 2011;7:e1002037. doi: 10.1371/journal.pgen.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull J, Wang P, Girard JM, Ruggieri A, Wang TJ, Draginov AG, et al. Glycogen hyperphosphorylation underlies lafora body formation. Ann Neurol. 2010;68:925–33. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- Unverricht H. Franz Deuticke: 1891. Die myoklonie. Leipzig and Vienna. [Google Scholar]
- Van Heycop Ten Ham M. Amsterdam: Elsevier; 1975. Lafora disease, a form of progressive myoclonus epilepsy. [Google Scholar]
- Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10:1407–13. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of PTG. J Biol Chem. 2008;283:4069–76. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.