Abstract
Numerous software packages exist to provide support for quantifying peptides and proteins from mass spectrometry (MS) data. However, many support only a subset of experimental methods or instrument types, meaning that laboratories often have to use multiple software packages. The Progenesis LC-MS software package from Nonlinear Dynamics is a software solution for label-free quantitation. However, many laboratories using Progenesis also wish to employ stable isotope-based methods that are not natively supported in Progenesis. We have developed a Java programming interface that can use the output files produced by Progenesis, allowing the basic MS features quantified across replicates to be used in a range of different experimental methods. We have developed post-processing software (the Progenesis Post-Processor) to embed Progenesis in the analysis of stable isotope labeling data and top3 pseudo-absolute quantitation. We have also created export ability to the new data standard, mzQuantML, produced by the Proteomics Standards Initiative to facilitate the development and standardization process. The software is provided to users with a simple graphical user interface for accessing the different features. The underlying programming interface may also be used by Java developers to develop other routines for analyzing data produced by Progenesis.
Introduction
A variety of experimental methods have been described for quantitative MS-based proteomics research, in which mass spectrometry (MS) is used to quantify peptides and proteins. These can be categorized into two general principles: label-based methods and label-free methods. Label-based methods commonly involve either differential stable isotope labeling through metabolic labeling in vivo, or by chemical modification in vitro. In the former approach, intact proteins are labeled in vivo through the incorporation of stable isotopes into the media on which cells are growing, for example incorporation of 13C into specific amino acids, such as stable isotope labeling of amino acids in cell culture (SILAC; Ong et al., 2002). Chemical labeling techniques include protein-level tagging such as with isotope-coded affinity tags (ICAT) applied to cysteine residues (Gygi et al., 1999), and isobaric tag for relative and absolute quantitation (iTRAQ), which uses a multiplexed set of isobaric reagents that yield amine-derivatized peptides for quantitation (Ross et al., 2004). Label-free quantitation methods that directly use raw spectral data from parallel MS runs to determine relative protein abundance are increasing in popularity. Spectral counting and intensity-based methods are among the most commonly used approaches. Spectral counting relates protein abundance to the number of peptide-spectrum matches (PSMs) in a given run for each protein. Intensity-based methods align precursor ion spectra of the same peptide from parallel runs according to their retention times (RT); protein quantitation is obtained by summation of ion intensities that have been matched to peptides for a given protein.
The Progenesis LC-MS software from Nonlinear Dynamics (Newcastle upon Tyne, U.K.) has been designed to perform label-free quantitation. A variety of other software packages exist, as described in another article in this issue (Gonzalez-Galarza et al., 2012). However, Progenesis is capable of processing a large number of replicates, and has an accessible graphical user interface allowing users to view their MS data in two- or three-dimensional (2D or 3D) maps to verify if features have been quantified accurately; a “feature” is defined as a peptide signal in 2D (retention time versus mass/charge space). The basic steps the software performs are as follows.
Map alignment to warp the 2D maps of different replicate runs to remove effects derived from peptides eluting at different times from the liquid chromatography (LC) stage in different runs, ensuring that like-for-like peptide signals are compared;
Feature detection, comprising the identification of the isotope pattern of the peptide, including the separation of overlapping peptides, and quantitation using an area-under-the-curve method;
Peptide identification using an external search engine; and
Peptide and protein quantitation using the abundance signals derived from the features to which identifications have been matched.
Data are output for the user as tables of values for raw features, peptides, and proteins. As such, since quantitation is performed by simple analysis of the signal present in a given feature, the basic feature-level quantitation performed by the software could be used for label-based or other types of experimental workflows in which quantitation is performed at the MS1 level.
At present, it is not possible to use Progenesis LC-MS for techniques other than label-free. We have developed a Java programming interface to enable the feature-level quantitation performed by Progenesis to be used for some other techniques, such as SILAC. The software searches the table of features to find pairs of features nearby in the RT dimension of the 2D map (since heavy:light peptide pairs tend to co-elute), separated by the mass shift in the m/z space due to the heavy label being incorporated into lysine or arginine. For example, typical labels incorporate 13C and/or 15N into lysine and arginine to produce known mass shifts of +6 Da, +8 Da, +10 Da, and so on. There are no built-in limits on the mass shift that is permissible, for example overlapping features (say +2 Da) are in theory identifiable, so long as Progenesis LC-MS has correctly separated features in the 2D map, although this feature has not yet been comprehensively tested. We have also produced a simple graphical interface for capturing the parameters required for such an analysis (the mass shift on a particular residue), such as the number of missed cleavages, mass/charge tolerance for the peptide pair match, and RT tolerance for the pair matching. Minor updates will be made in future releases to capture triple SILAC and labeling on any amino acid.
A similar method of label-free analysis performed by Progenesis is the top3 protocol, developed by Waters for the SYNAPT range of instruments (Silva et al., 2006). The basic premise of this protocol is to find and sum the signal of the three highest-intensity peptides matched to each protein in a given run. A correlation has been observed between the sum of the three most intense peptide ions and the absolute abundance of a given protein. The summed (or average) intensity over the top3 can be converted into a value of absolute protein abundance by spiking in one or more proteins of known abundance and scaling protein abundance values accordingly. Top3 quantitation is not natively supported by Progenesis, and as such the Progenesis Post-Processor incorporates a module for top3 quantitation (in fact topX, where X is any integer).
Finally, we are contributing to the development of a new data standard from the Proteomics Standards Initiative (PSI), called mzQuantML, which captures the output of quantitation software used in proteomics. It captures values about MS features, peptides, and proteins or groups of proteins, when there is ambiguity in peptide to protein inference (http://code.google.com/p/mzquantml/). This standard is complementary to the recently reported PSI standards for raw or processed MS data, mzML (Martens et al., 2011), and for peptide and protein identification data, mzIdentML (Jones et al., 2012). During the development of a data standard, it is important to develop software for creating genuine example files from software routinely used in laboratories. This toolkit was initially conceived to enable results from Progenesis to be converted into mzQuantML for label-free methodologies, and subsequently extended to cover label-based and top3 quantitation.
We demonstrate that the software is effective for SILAC and top3 label-free quantitation of a dataset created in-house (Supplementary Tables S2 and S3; see online supplementary material at http://www.liebertonline.com). In the first experiment, a SILAC test dataset, created by mixing digests of unlabeled and labeled yeast lysates in known ratios, was prepared and analyzed by ion mobility-coupled data-independent LCMS. The processed data was input into the Progenesis Post-Processor and the reported ratios compared to the actual ratios. We have also used a standard data set created by CPTAC (Paulovich et al., 2009), in which 48 human proteins (SIGMA UPS1 standard) were spiked into an equimolar yeast background in five different concentrations, which can be used for label-free and top3 benchmarking. We have performed various comparisons and benchmarks with these data sets to demonstrate the effectiveness of the software. The software toolkit and data sets are available in the public domain under a permissive license at http://code.google.com/p/progenesis-post-processor.
Materials and Methods
CPTAC study 6
Five sample conditions were created by splitting a single yeast culture and spiking it with different levels of a standard protein mix: Sigma48 UPS proteins at 0.25, 0.74, 2.2, 6.7, and 20 fmol/μL (Paulovich et al., 2009). These samples were designated A–E and run in triplicate on various MS platforms. It was reported in the original study that the UPS proteins could not be detected in the A sample (hence only samples B–E are used in our analysis). For this study, the dataset resulting from analysis using a Thermo LTQ Orbitrap mass spectrometer (LTQ-OrbitrapO@65 dataset) was used, as this was seen to have the highest number of peptide-spectrum matches for yeast proteins, and as this is a popular instrument platform within the field. The Thermo raw files were loaded into the Progenesis LC-MS software (version 1.0.0.1), aligned using automatic alignment, and the resulting aggregate spectrum filtered to include +1, +2, and +3 charge states only. The samples were grouped according to experimental conditions (B–E). Features without MS2 spectra were deleted from the analysis, as this information is necessary for peptide/protein identification. An .mgf file representing the aggregate spectrum was exported and searched using Mascot (1 missed cleavage, fixed modification: carbamidomethyl [C]; variable modifications: oxidation [M]; peptide tolerance:±10 ppm; MS/MS tolerance:±0.6 Da), and the resulting .xml file was re-imported to assign peptides to features, using the following thresholds: Mascot score>40, hits>2, to ensure that only high-quality identifications were included. The spectra were searched against the reference strain yeast database, downloaded from the Saccharomyces Genome Database (orf_trans.fasta, version 03-Feb-2011), concatenated with the Sigma UPS 48 proteins sequences, and a full reverse database (Yeast and UPS sequences reversed) for decoy searching.
SILAC dataset
To create a SILAC test sample, Saccharomyces cerevisiae (EUROSCARF accession number Y11335 BY4742; MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YJL088w::KanMX4) was grown in minimal media (Supplementary Table S1; see online supplementary material at http://www.liebertonline.com) using 10 g/L of glucose as the sole carbon source. The minimal media was supplemented with 0.5 mM arginine (unlabeled or 13[C]6 arginine), and 1 mM lysine (unlabeled or 13[C]6 lysine), to meet the auxotrophic requirements of the strain. Cultures of 200 mL were grown in flasks, and aliquots (15 mL) of the culture were centrifuged (4000 rpm at 4°C for 10 min). The supernatant was discarded and the pellet was immediately frozen at −80°C and stored at that temperature for subsequent protein extraction. Proteins were extracted according to a previously described method (Carroll et al., 2011). An amount of unlabeled or labeled lysate representing 100 μg of protein was dispensed into low-protein-binding microcentrifuge tubes and digested according to the previously described method. A 1:1 mixture of unlabeled and labeled digest was analyzed by LC-MS using a nanoACQUITY UPLC™ system (Waters MS Technologies, Manchester, U.K.), coupled to a Synapt G2 mass spectrometer (Waters MS Technologies). The sample (2 μL corresponding to approximately 1 μg of protein) was loaded onto the trapping column (Waters MS Technologies C18; 180 μm×20 mm) using partial loop injection for 3 min at a flow rate of 5 μL/min with 0.1% (v/v) TFA. The sample was resolved on an analytical column (nanoACQUITY UPLC HSS T3 C18; 75 μm×150 mm×1.7 μm column), using a gradient of 97% A (0.1% formic acid), 3% B (99.9% ACN and 0.1% formic acid), to 60% A 40% B over 120 min at a flow rate of 300 nL/min. The mass spectrometer acquired data using an ion-mobility-coupled data-independent program with 1 sec scan times, and a collision energy ramp of 15–40 eV for elevated energy scans. The mass spectrometer was calibrated before use with the fragment ions of glufibrinopeptide, and throughout the analytical run at 1-min intervals using the NanoLockSpray source with glufibrinopeptide. The data were processed and the database was searched using ProteinLynx Global Server v2.5 (hereafter referred to as PLGS; Waters MS Technologies). The data were processed using a low-energy threshold of 100 (counts), and an elevated energy threshold of 20, and the processed spectra were searched against the complete proteome set of S. cerevisiae from Uniprot (reference strain, downloaded June 2010, 6560 proteins). Two searches were applied. The first search was targeted to identify unlabeled peptides using the following parameters: fixed carbamidomethyl modification for cysteine and variable oxidation modification for methionine were specified; one trypsin miscleavage was allowed; and the automatic settings in PLGS for the precursor ion and fragment ion mass tolerance were used. The search thresholds used were: minimum fragment ion matches per peptide 3; minimum fragment ion matches per protein 7; minimum peptides per protein 1; and a false-positive value of 4. The threshold score/expectation value for accepting individual spectra was the default value in the program, such that the false-positive value was 4. The second search targeted labeled peptides using identical search parameters, but with additional fixed modifications of 13C6 lysine and 13C6 arginine. The average light:heavy ratio was determined from this single analytical run, and then further ratios of 100:1, 50:1, 25:1, 10:1, 5:1, 3:1, 2:1, 1:1, 1:2, 1:3, 1:5, 1:10, 1:25, 1:50, and 1:100 were prepared. These samples were analyzed by the same workflow, and technical variability was assessed by three replicate injections. Quantification was performed by Progenesis (version 4, Nonlinear Dynamics, Newcastle upon Tyne, U.K.). Samples were aligned according to RT using a combination of manual and automatic alignment. Maximum peak picking parameters were applied with identifications imported from PLGS, and features with charges from 1+ to 4+ featuring three or more isotope peaks were retained. Identifications were imported from PLGS, and peptides were filtered on a PLGS score of 7. The resulting feature set was exported to the Progenesis Post-Processor, which was used with the following settings: a mass difference of 6 Da (with a 0.5-Da window) for lysine and arginine, one miscleavage allowed, and a 0.8-min retention time window.
Results
Programming and software interface
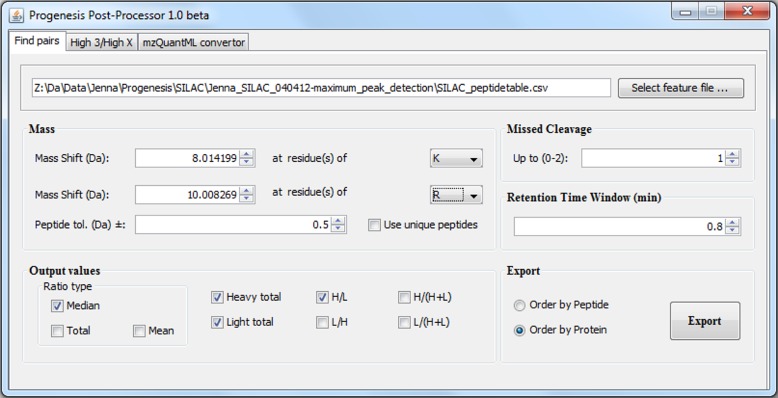
The software interface (Fig. 1) is coded in Java and has two major parts: (1) Progenesis file reader classes; and (2) the mzQuantML application programming interface (API), called jmzQuantML (http://code.google.com/p/jmzquantml/). Progenesis output files, which are in .csv (comma delimited) format, report observed values and attributes from detected features, peptides, and proteins. The Progenesis file reader classes provide the functions to read the feature list and protein list files; to retrieve specific values for selected peptide, protein, or other attributes; and calculate summary attributes (e.g., replicate number and identified peptide number), and load these into Java objects that can be accessed by other methods. The mzQuantML API is generated using JAXB, a Java Architecture for XML Binding, which provides automated mapping between elements in the XML document and Java objects. In this context, jmzQuantML is used for loading objects from the Progenesis file reader and writing out valid mzQuantML files (marshalling).
FIG. 1.
Screenshot of Progenesis Post-Processor used for SILAC analysis.
SILAC analysis with the Progenesis Post-Processor
A module has been created in the Progenesis Post-Processor for the analysis of SILAC-generated datasets. The module performs the following steps: (1) imports the quantified features from Progenesis in all replicates; (2) orders all results by retention time of the master run assigned by Progenesis (assuming that all replicates have been correctly aligned); (3) searches for peptide pairs in the two-dimensional space (matches are accepted as correct if the following criteria are met: the light and heavy feature both have at least one identification across all replicates of the same peptide sequence; the heavy feature has the correct mass shift [±the user-entered tolerance], and is within a given tolerance in the retention time axis); (4) the software calculates protein-level ratios, allowing the user control over the method used for calculation (discussed below); and (5) exports results for individual peptide pairs and proteins into .csv files for the user to process further. Ratios for proteins are created by one of three methods, offered as choices to the user: median peptide ratio, mean peptide ratio, or ratio of total peptide abundance. Software packages such as MaxQuant favor the median peptide ratio for calculating a protein ratio (Cox and Mann, 2008), although we are not aware of any systematic evaluation. By providing the user some control over this aspect, such comparisons can be made. The user can also select whether to use unique peptides only for quantitation (i.e., those that cannot be matched to other proteins). Users may wish to experiment with this parameter on a case-by-case basis, depending on the similarity between different proteins within their search database. We have experimented with relaxing the criteria for making peptide pair identification, for example not requiring that an identification has been made to both the light and heavy peptide (based on a database search); however, we believe this increases the number of incorrect ratios in the final result, without a great increase in sensitivity (data not shown). The outcome is that users can select a relatively relaxed tolerance for peptide pair matching (say 0.5 Da), without adversely affecting data quality (even if almost all matches are accurate to<5 ppm). Missed cleavages are currently only handled for lysine and arginine (supporting protocols using trypsin, ArgC, or LysC). If the user selects that n missed cleavages are allowed, the module searches for the peptide pair with mass shifts of (1.n) * mass shift specified, according to the number of lysine or arginine amino acids observed in the peptide sequence (since the peptide sequence is known prior to matching). As such, if the user specifies to allow no missed cleavages (say for a+8 Da on K protocol), the pair matching module will not identify any peptide pairs for which the sequence contains two lysines, since it will not search for the heavy peptide at+16 Da. Such results are therefore automatically excluded from the calculation of protein ratios.
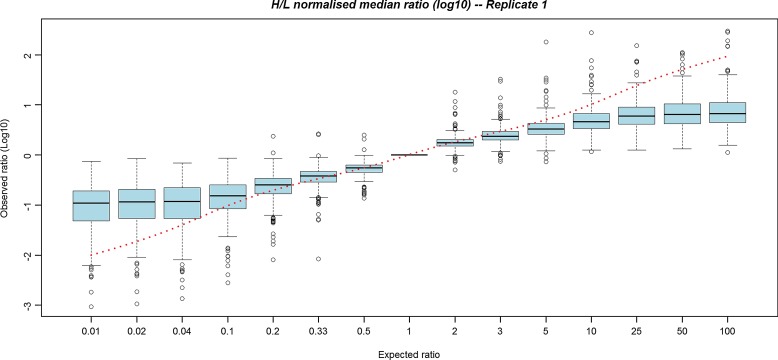
As detailed in the methods section, we have generated an example dataset for testing the performance of the software for which we have control over the expected ratios. The datasets were generated on the Waters SYNAPT HDMSe instrument, for which, to our knowledge, there is currently no software provision for this type of analysis. Samples were generated with expected ratios of 1:100, 1:50, 1:25, 1:10, 1:5, 1:3, 1:2, 1:1, 2:1, 3:1, 5:1, 10:1, 25:1, 50:1, and 100:1, by mixing peptides from parallel grown cultures in either unlabeled or labeled medium. To remove any variation due to the samples being grown in parallel plus incomplete heavy labeling, we have applied post-processing to our results, using the ratios obtained from the 1:1 mix to apply a consistent normalization factor to all other samples. The results from a single run are provided as box plots in Figure 2, showing the protein ratio observed plotted versus the expected protein ratios (Supplementary Figs. S1 and S2; see online supplementary material at http://www.liebertonline.com). Protein ratios were calculated by taking the median peptide ratio, using all peptide pairs matched to the protein as input. Two further replicates are provided as supplementary information, since all show the same general pattern. The results are also summarized in Table 1. In each replicate, it can be observed that broadly accurate ratios are observed for conditions for which there are up to fivefold changes in peptide abundance (i.e., the expected and observed values are close, particularly for ratios 1:2 and 1:3). Higher ratios appear to be flattened, such that a maximum of 1:10 ratios appear observable, even for the spike at 1:100. At this stage it is not simple to determine if this is a facet of this specific benchmarking dataset, or more broadly is due to the use of Progenesis LC-MS in this way, although it should be noted that similar effects have generally been observed for analyses with iTRAQ reagents (Ow et al., 2009). There have been few benchmarking studies on SILAC for which there is a known “ground truth,” due to the difficulty of assembling peptides in known abundances in cell culture. We feel there is value at this stage in making the Post-Processor available, to enable other groups to benchmark the performance of Progenesis LC-MS for SILAC, under a wider range of test conditions.
FIG. 2.
A box plot of the first replicate of the SILAC data set. The x-axis displays the expected ratios of proteins in each run (calculated from the median peptide ratio, using all peptides matched to a given protein), and the y-axis displays the observed log10 ratios. The dotted line displays the expected values.
Table 1.
The Expected Ratios for Proteins Detected in the Three Replicate Runs, Along with the Calculated Median and Mean Ratios, and the Standard Deviation of the Values Observed
| Replicate 1 | ||||||||||||||
| Expected | 0.01 | 0.02 | 0.04 | 0.1 | 0.2 | 0.33 | 0.5 | 2 | 3 | 5 | 10 | 25 | 50 | 100 |
| Median | 0.106 | 0.111 | 0.113 | 0.151 | 0.249 | 0.378 | 0.558 | 1.747 | 2.371 | 3.291 | 4.580 | 5.991 | 6.462 | 6.685 |
| Std dev | 0.133 | 0.139 | 0.138 | 0.149 | 0.214 | 0.263 | 0.233 | 1.551 | 3.078 | 12.388 | 19.550 | 13.538 | 14.616 | 33.015 |
| Mean | 0.136 | 0.143 | 0.151 | 0.183 | 0.272 | 0.401 | 0.563 | 2.007 | 2.928 | 4.848 | 7.606 | 8.834 | 10.061 | 13.722 |
| Replicate 2 | ||||||||||||||
| Expected | 0.01 | 0.02 | 0.04 | 0.1 | 0.2 | 0.33 | 0.5 | 2 | 3 | 5 | 10 | 25 | 50 | 100 |
| Median | 0.095 | 0.103 | 0.110 | 0.154 | 0.248 | 0.382 | 0.562 | 1.778 | 2.497 | 3.658 | 5.101 | 6.169 | 6.782 | 6.739 |
| Std dev | 0.124 | 0.110 | 0.139 | 0.140 | 0.176 | 0.168 | 0.101 | 0.436 | 1.049 | 2.477 | 7.616 | 9.238 | 14.255 | 19.823 |
| Mean | 0.123 | 0.129 | 0.140 | 0.180 | 0.269 | 0.393 | 0.566 | 1.834 | 2.698 | 4.127 | 6.509 | 8.653 | 10.108 | 11.503 |
| Replicate 3 | ||||||||||||||
| Expected | 0.01 | 0.02 | 0.04 | 0.1 | 0.2 | 0.33 | 0.5 | 2 | 3 | 5 | 10 | 25 | 50 | 100 |
| Median | 0.100 | 0.108 | 0.121 | 0.157 | 0.242 | 0.377 | 0.562 | 1.749 | 2.463 | 3.475 | 4.797 | 6.005 | 6.502 | 6.724 |
| Std dev | 0.113 | 0.109 | 0.111 | 0.113 | 0.119 | 0.114 | 0.087 | 0.360 | 0.904 | 1.916 | 8.918 | 9.564 | 11.772 | 17.495 |
| Mean | 0.127 | 0.131 | 0.146 | 0.179 | 0.260 | 0.384 | 0.568 | 1.789 | 2.597 | 3.900 | 6.475 | 8.343 | 9.313 | 10.815 |
A total of 222 proteins were quantified in all three replicates, using two peptides or greater for quantification.
Top3 results for label-free benchmarking
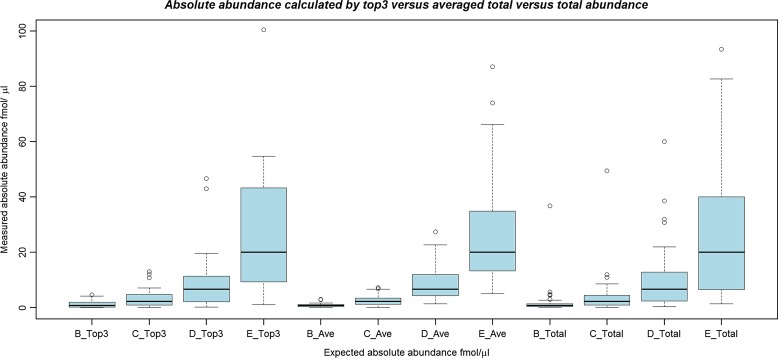
The top3 module was developed to extract the highest three intensity peptides in each replicate and use these to compute a pseudo-absolute value of abundance. At present no additional threshold is applied to exclude peptides from the list, although in future releases we will provide options for excluding modified peptides or peptides resulting from missed cleavages. The software also calculates two other data types: the average of the total abundance, calculated by summing the abundance from all peptides divided by the number of peptides contributing, and the average abundance of the three most abundant peptides (top3). The user can then calculate estimated absolute abundance values via knowledge of proteins that were spiked in known abundance. In the CPTAC dataset, 48 proteins were added in known absolute abundances so all should exhibit the same calculated absolute abundance. For the B–E conditions (0.74, 2.2, 6.7, and 20 fmol/μL spike), we analyzed the data initially with Progenesis, and then ran the post-processor to extract top3 and average total abundance. We have also compared these data types versus the total abundance calculated directly by Progenesis LC-MS. In each condition, 41 out of the 48 UPS proteins were detected, and we used the median estimate of absolute abundance (across the 41 proteins) to convert top3 or total abundance values into fmol/μL estimates. The results in the form of a box plot are shown in Figure 3 and a summary table (Table 2). The results demonstrate that all three methods of calculation produce relatively sensible values for absolute abundance (the expected and observed mean values are fairly close), although the average of the total appears to produce the most reliable estimates of absolute abundance (difference from the mean and standard deviation is lowest in all four comparisons).
FIG. 3.
A box plot displaying the observed values of absolute abundance (fmol/μL), calculated by the top3 protocol (“_Top3”), average of the total abundance (“_Ave”), and total abundance (“_Total”), in conditions B–E for 41 UPS proteins. Expected values and summary statistics are displayed in Table 2.
Table 2.
Summary Results for CPTAC Analysis by Top3
| Protein (n=41) | B_Top3 | B_Ave | B_Total | C_Top3 | C_Ave | C_Total | D_Top3 | D_Ave | D_Total | E_Top3 | E_Ave | E_Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected (fmol/μL) | 0.740 | 0.740 | 0.740 | 2.200 | 2.200 | 2.200 | 6.700 | 6.700 | 6.700 | 20.000 | 20.000 | 20.000 |
| Mean (fmol/μL) | 1.197 | 0.857 | 2.099 | 3.267 | 2.600 | 4.133 | 8.805 | 8.730 | 10.479 | 28.499 | 25.935 | 27.609 |
| Absolute difference from mean | 0.457 | 0.117 | 1.359 | 1.067 | 0.400 | 1.933 | 2.105 | 2.030 | 3.779 | 8.499 | 5.935 | 7.609 |
| Standard deviation | 1.255 | 0.745 | 5.741 | 3.247 | 1.849 | 7.829 | 9.777 | 6.329 | 11.988 | 28.235 | 19.015 | 26.586 |
Average total abundance and total abundance measurement, displaying the expected absolute abundance (fmol/μL), the mean observed abundance (in 41 out of 48 UPS proteins quantified with ≥3 peptides), absolute difference of the observed from the expected value, and the standard deviation.
It should be noted that the original protocol (Silva et al., 2006) was intended for use on the Waters SYNAPT MSE instruments. These data were generated on a Thermo Orbitrap, which may explain the difference, but clearly this is an interesting topic for further investigations, which will be facilitated by this tool. We also believe that this tool may be useful for processing datasets originating from Waters Instruments, since Progenesis LC-MS is compatible with Waters RAW files, and PLGS is limited in the number of replicates that can analyzed at once.
Export to mzQuantML
We have built an exporter to write out a single mzQuantML file, capturing peptide and protein values, from the Progenesis output files (accepting feature, peptide, and protein files). As part of the formal standardization process for mzQuantML, several files generated by the software have been used to demonstrate that genuine label-free experiments can be encoded in the standard, thus fulfilling an important role in the standardization process (http://code.google.com/p/mzquantml/). A number of open-source software packages are currently implementing mzQuantML input, such as Proteosuite (http://www.proteosuite.org/), and OpenMS (Sturm et al., 2008), allowing for re-analysis and visualization of datasets generated by other software.
Conclusions
We have generated a software application for post-processing results files generated by the Progenesis LC-MS software package. It employs two additional protocols beyond the native support of Progenesis, for SILAC and top3 quantitation, on which we have performed preliminary benchmarking to demonstrate that results are sensible and the software is functional. We expect that the software will continue to develop and will offer additional quantitation protocols as they are requested. We have also used the software to generate example files in the developing mzQuantML data standard from the PSI, to facilitate the standardization of this format.
Supplementary Material
Acknowledgments
We would also like to gratefully acknowledge current funding sources that have supported this work from BBSRC (BB/I00095X/1 to A.R.J. and BB/G009112/1 to R.J.B.), and EU FP7 grant ProteomeXchange (grant no. 260558).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Carroll K.M. Simpson D.M. Eyers C.E., et al. Absolute quantification of a metabolic pathway in yeast: deployment of a complete QconCAT approach. Mol Cell Proteomics. 2011 Sep;10(12):19. doi: 10.1074/mcp.M111.007633. M111.007633. Epub 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F. Lawless C. Hubbard S.J. Hermjakob H. Jones A.R. A critical appraisal of techniques, software packages and standards for quantitative proteomic analysis. OMICS. 2012;16 doi: 10.1089/omi.2012.0022. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S.P. Rist B. Gerber S.A. Turecek F. Gelb M.H. Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotech. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Jones A.R. Eisenacher M. Mayer G., et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Molecular & Cellular Proteomics. 2012 2012 Feb 27; doi: 10.1074/mcp. first published on. M111.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L. Chambers M. Sturm M., et al. mzML—a community standard for mass spectrometry data. Mol Cell Proteomics. 2011 Aug;10(1):17. doi: 10.1074/mcp.R110.000133. R110.000133. Epub 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.-E. Blagoev B. Kratchmarova I., et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ow S.Y. Salim M. Noirel J. Evans C. Rehman I. Wright P.C. iTRAQ underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- Paulovich A.G. Billheimer D. Ham A.-J.L., et al. Interlaboratory study characterizing a yeast performance standard for benchmarking LC-MS Platform performance. Mol Cell Proteomics. 2010 2009 Oct 26;9(2):242–254. doi: 10.1074/mcp.M900222-MCP200. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P.L. Huang Y.N. Marchese J.N., et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Silva J.C. Gorenstein M.V. Li G.-Z. Vissers J.P.C. Geromanos S.J. Absolute quantification of proteins by LCMSE. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Sturm M. Bertsch A. Gropl C., et al. OpenMS—An open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.