Abstract
The multi-subunit replication factor C (RFC) complex loads circular proliferating cell nuclear antigen (PCNA) clamps onto DNA where they serve as mobile tethers for polymerases and coordinate the functions of many other DNA metabolic proteins. The clamp loading reaction is complex, involving multiple components (RFC, PCNA, DNA, and ATP) and events (minimally: PCNA opening/closing, DNA binding/release, and ATP binding/hydrolysis) that yield a topologically linked clamp·DNA product in less than a second. Here, we report pre-steady-state measurements of several steps in the reaction catalyzed by Saccharomyces cerevisiae RFC and present a comprehensive kinetic model based on global analysis of the data. Highlights of the reaction mechanism are that ATP binding to RFC initiates slow activation of the clamp loader, enabling it to open PCNA (at ~2 s−1) and bind primer–template DNA (ptDNA). Rapid binding of ptDNA leads to formation of the RFC·ATP·PCNAopen·ptDNA complex, which catalyzes a burst of ATP hydrolysis. Another slow step in the reaction follows ATP hydrolysis and is associated with PCNA closure around ptDNA (8 s−1). Dissociation of PCNA·ptDNA from RFC leads to catalytic turnover. We propose that these early and late rate-determining events are intramolecular conformational changes in RFC and PCNA that control clamp opening and closure, and that ATP binding and hydrolysis switch RFC between conformations with high and low affinities, respectively, for open PCNA and ptDNA, and thus bookend the clamp loading reaction.
Keywords: DNA replication, RFC clamp loader, PCNA clamp, ATPase mechanism, pre-steady-state kinetics
Introduction
Efficient DNA replication requires the coordinated actions of DNA polymerases and several accessory proteins. Central among accessory proteins at the replication fork are the circular clamp, whose primary function is to tether polymerases to primer–template DNA (ptDNA) during synthesis, and the clamp loader, whose function is to load clamps onto DNA for use by polymerases and other DNA metabolic proteins.1,2 In addition to their critical role in enhancing replicative DNA polymerase processivity, circular clamps help orchestrate numerous protein functions at target sites on DNA, including Okazaki fragment processing and ligation, translesion DNA synthesis, DNA repair, and chromatin remodeling, among others.3,4 Not surprisingly, clamp and clamp loader proteins are highly conserved through evolution, and their core structure–function properties appear to be shared by organisms ranging from bacteria to humans.1 In eukaryotes, the proliferating cell nuclear antigen (PCNA) clamp serves to increase the processivity of DNA polymerases δ and ε.5 PCNA is a trimer of three identical subunits arranged in the form of a ring with a large enough diameter to enclose duplex DNA.6,7 Replication factor C (RFC) serves to load PCNA clamps onto DNA in a reaction driven by its ATPase activity.8 RFC is a multi-protein complex comprising one large subunit, RFC-A (95 kDa), and four smaller subunits, RFC-B to RFC-E (36–40 kDa), arranged in the form of a claw that can bind PCNA at its base and ptDNA within its central chamber and forge a topological link between the two macromolecules (Fig. 1); nomenclature note: RFC-A, RFC-B, RFC-C, RFC-D, and RFC-E are RFC-1, RFC-4, RFC-3, RFC-2, and RFC-5, respectively.9-11 RFC subunits belong to the AAA+ family of proteins, containing Walker A and B, and SRC (arginine finger) motifs, that utilize ATP binding and hydrolysis to perform mechanical work.12,13
Fig. 1.
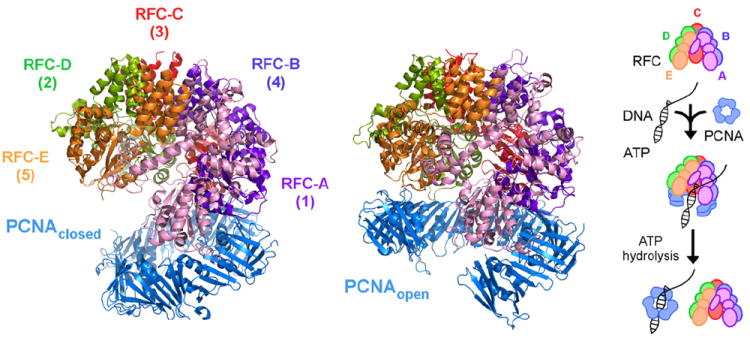
Model of the RFC·ATPγS·PCNA complex and PCNA loading reaction. Crystal structure of RFC bound to ATPγS and a closed PCNA clamp10 and a computationally derived model of RFC bound to ATPγS and open PCNA.42 The MD model represents current thinking about PCNA opening in a right-handed spiral conformation to allow entry of DNA during RFC-catalyzed clamp loading on ptDNA (minimal pathway depicted on the right). Note: RFC-A, RFC-B, RFC-C, RFC-D, and RFC-E correspond to RFC-1, RFC-4, RFC-3, RFC-2, and RFC-5, respectively.
In order to load PCNA onto DNA, RFC must at minimum (a) bind the clamp, (b) open the clamp at an inter-subunit interface/stabilize the clamp in open form, (c) bind the target DNA site (e.g., primer–template junction) and position it within the open clamp, and (d) facilitate closure of the clamp around DNA and release the clamp·DNA product. Progression of these events is driven by distinct steps in the ATPase cycle, and details of the reaction mechanism are under active investigation in several model organisms. Biochemical studies of clamp loaders from T4 bacteriophage,14,15 Escherichia coli,16,17 Pyrococcus furiosus,18 Archaeoglobus fulgidus,19 Saccharomyces cerevisiae,10,20-22 and humans,23,24 among others, have identified common mechanistic features of the ATPase-coupled clamp loading reaction. Mainly, ATP binding is required for formation of a clamp loader·clamp·ptDNA complex, and ATP hydrolysis is required for dissociation of the complex to release the clamp·ptDNA product.22,25-27 More detailed information about the reaction mechanism is available from transient kinetic studies of E. coli and bacteriophage T4 clamp loaders relative to the eukaryotic ones;16,28 for example, recent reports indicate that E. coli γ complex undergoes slow, ATP-induced isomerization that favors binding to β clamp before ptDNA29 and appears to bind and open the clamp rather than simply bind it in open form.30
Kinetic studies of the S. cerevisiae RFC clamp loader have also begun to parse individual steps in the PCNA loading pathway, advancing our understanding of a eukaryotic clamp loading mechanism.20,21,31 A model mechanism based on transient kinetic analysis of RFC ATPase activity suggests that the clamp loader undergoes a slow conformational change (activation) on binding ATP, which enables it to both open PCNA and bind ptDNA.20 The presence of PCNA speeds up RFC activation, indicating that this early step establishes a preferred order of events in which PCNA binding and/or opening occurs before binding of ptDNA. The ATPase data also indicated another slow step, later in the reaction following ATP hydrolysis. It was proposed that this step is associated with product release, including the PCNA·ptDNA complex.20 Although the ATPase kinetics helped identify the occurrence and general location of rate-limiting steps in the clamp loading pathway, they could not reveal the nature of the transactions in these key steps. Another recent study of S. cerevisiae RFC-catalyzed PCNA loading examined interactions between Cy5-labeled PCNA and Cy3-labeled forked DNA by single-molecule and ensemble Förster resonance energy transfer (FRET) methods and proposed intermediate steps in the reaction; in this case also, the nature of these steps and their role in the kinetic mechanism remained unclear.31
In the current study, we specifically addressed the question of what occurs during rate-limiting steps in the PCNA loading reaction, by monitoring the reaction from the perspective of the different reactants—PCNA (opening and closure), ptDNA (binding and release) and ATP (ATP hydrolysis and phosphate release)—and performing global analysis of all the kinetic data for a more comprehensive view of the reaction mechanism. An associated goal was to link between individual steps in the ATPase catalytic cycle with the interactions and conformational transitions of the proteins and DNA, in order to elucidate the mechanochemical coupling process by which clamp loaders load clamps onto DNA.
Results
Measured opening of the PCNA clamp in preparation for assembly on ptDNA
ATP-bound RFC can bind and open the PCNA clamp, as has been demonstrated previously by different methods such as labeling of the exposed inter-subunit interface25,32 or decrease in FRET between a tryptophan–5-[2(acetyl)aminoethyl-]aminonaphthalene-1-sulfonate (AEDANS) donor–acceptor pair positioned across the interface when the clamp is opened (PCNA-WCAEDANS: F185W, K107C).20,21 We utilized the latter method to monitor transient opening and closure of PCNA during the first clamp loading cycle catalyzed by RFC. In stopped-flow experiments, a solution of RFC and PCNA-WCAEDANS was mixed rapidly with a solution of ATP, and the decrease in FRET signal as a consequence of PCNA opening was monitored over time. The signal was converted to FRET efficiency using data from parallel experiments with tryptophan-free PCNA-FCAEDANS, including correction for a small level of inter-protein FRET due to RFC tryptophan residues (see Materials and Methods).33 The total change in FRET efficiency from 0.95 to ~0.7 as PCNA-WCAEDANS converts from closed to open form is comparable to previously reported values (0.95 and 0.66, respectively).21 The kinetic trace shown in Fig. 2a was fit empirically to a 2-exponential function and yielded kopen(1) = 3.4 s−1 and kopen(2) =0.8 s−1 (net rate, ~1.6 s−1). The apparent slow rate of PCNA opening raised two key questions: (a) Do the kinetics reflect slow PCNA opening or a preceding rate-limiting step in the pathway, and (b) does PCNA opening limit subsequent events in the clamp loading pathway? These questions were addressed as described below, and an additional question about the apparent biphasic PCNA opening kinetics was addressed during global data analysis and development of the model mechanism.
Fig. 2.
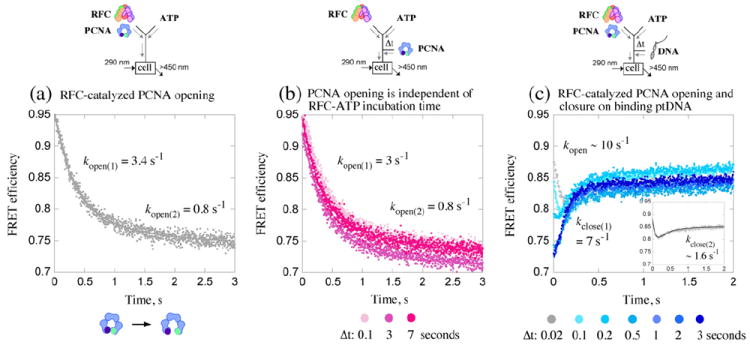
Kinetics of PCNA opening and closure around DNA. Opening/closing of PCNA clamp is measured by change in FRET between tryptophan (donor; λEX=290 nm) and AEDANS (acceptor; λEM>450 nm) at the PCNA-WCAEDANS inter-subunit interface. (a) Rapid mixing of RFC and PCNA with ATP results in FRET efficiency decrease as PCNA opens; 2-exponential fit of the data yields kopen(1)=3.4 s−1 and kopen(2)=0.8 s−1. (b) Pre-incubation of RFC with ATP (Δt=0.02–10 s; 0.02, 3, 7 s shown) prior to addition of PCNA does not alter the apparent PCNA opening rates. (c) Pre-incubation of RFC and PCNA with ATP (Δt=0.02–3 s) followed by addition of ptDNA results in FRET efficiency increase as PCNA closes (around DNA). At shorter Δt (e.g., 0.02 s; gray trace), only a fraction of PCNA has been opened, and both clamp opening (FRET decrease) and closing (biphasic FRET increase) are detected after ptDNA addition (see inset); 3-exponential fit of the data yields kopen=10 s−1, kclose(1)=7 s−1, and kclose(2)=1.6 s−1. At longer Δt (e.g., 3 s; dark-blue trace), all PCNA is open, and only clamp closing is detected at kclose(1)=7 s−1. Final reactant concentrations: 0.6 μM RFC, 0.25 μM PCNA, 0.25 μM ptDNA (when present), and 0.5 mM ATP. Rate constants obtained from the analytical fits were utilized as initial estimates for global data analysis.
For the first question, sequential-mixing experiments were performed in which RFC was pre-incubated with ATP for varying times (Δt= 0.02–10 s) and then mixed with PCNA-WCAEDANS, and the change in FRET was monitored over time. The purpose of incubating RFC with ATP was to allow completion of any slow steps in the reaction before observing PCNA opening. However, as shown in Fig. 2b, even after extended pre-incubation of RFC and ATP, PCNA opening occurs at the same rates as in Fig. 2a (3 s−1 and 0.8 s−1 on average; data shown for Δt=0.1, 3, and 7 s); note: ATP hydrolysis is minimal during pre-incubation (kcat =0.05 s−1 in the absence of DNA).20 The net PCNA opening rate of ~1.6 s−1 is comparable to the 2.2 s−1 rate reported previously.21 Also consistent with the previous report, this slow rate does not change with RFC concentration (Supplementary Fig. S1), further supporting the hypothesis that it reflects clamp opening. The actual event may involve, for example, isomerization of RFC·ATP to capture open PCNA and/or formation and isomerization of the RFC·ATP·PCNAclosed complex to the RFC·ATP·PCNAopen complex.
In order to answer the second question, we performed experiments to measure PCNA closure, which occurs later in the reaction after ptDNA binding and ATP hydrolysis.20 Sequential-mixing experiments were performed in which RFC and PCNA-WCAEDANS were pre-incubated with ATP for varying times (Δt=0.02–3 s) to allow PCNA opening and then mixed with excess ptDNA, and the change in FRET was monitored over time. At short pre-incubation times, both decrease (PCNA opening) and subsequent increase (PCNA closing) in FRET efficiency were detected, indicating an ordered series of events—PCNA binding/opening, followed by ptDNA binding, and then PCNA closure—in the reaction (Fig. 2c). For example, the Δt=0.02 s trace fit empirically to a 3-exponential function indicates FRET decrease at kopen ~10 s−1 and biphasic FRET increase at kclose(1) ~ 6 s−1 and kclose(2)~ 1.6 s−1, indicating a mixture of species in the reaction (Fig. 2c, inset, Δt=0.02 s). Interestingly, PCNA opening apparently occurs faster when ptDNA is added to the reaction. With longer Δt, more PCNA is opened during the pre-incubation period, and correspondingly, more of the PCNA closing phase is observed following addition of ptDNA. At Δt≥ 2 s, all PCNA is open, and only PCNA closure is observed at kclose(1)=7.3 s−1 and kclose(2)=0.4 s−1 (the second phase is reduced at longer Δt). The FRET efficiency levels off at ~0.85 as the reaction enters steady state. The apparent biphasic nature of PCNA closing was addressed during global data analysis. The PCNA opening/-closing kinetics suggest that formation of an RFC·ATP·PCNAopen intermediate is obligatory and rate determining for subsequent events in the pathway. This hypothesis was examined further by analysis of RFC interactions with ptDNA and its ATPase activity.
Assembly of the RFC·ATP·PCNAopen·ptDNA intermediate is followed by slow release of the PCNA·ptDNA product
The PCNA opening/closing data indicated that ptDNA can bind rapidly to the RFC·ATP·PCNA complex and perhaps accelerate PCNA opening, and subsequently, the clamp closes (presumably around ptDNA). In order to test this hypothesis, we directly measured transient interactions between the proteins and ptDNA. Binding of ptDNA to RFC (±PCNA) was reported by increase in fluorescence of 5-(and 6-)carboxytetramethylrhodamine (TAMRA) dye conjugated to the 3′ primer end of the 40/65-nt ptDNA substrate (Supplementary Fig. S2; Kd=1 nM). Sequential mixing experiments were performed in which RFC and PCNA were preincubated with ATP for varying times (Δt=0.02–3 s) and then mixed with ptDNATAMRA, and the fluorescence signal was monitored over time. The relative change in TAMRA fluorescence intensity over time shows biphasic kinetics (Fig. 3a), with an initial increase followed by decrease in signal. In the first phase the rate and amplitude of ptDNA binding increase with pre-incubation time and reach maximum at Δt=2 s, supporting our hypothesis that slow formation of the RFC·ATP·PCNA intermediate precedes rapid binding of ptDNA. Higher concentrations of RFC and PCNA result in larger fraction of bound ptDNA; however, a long preincubation time is still required for maximum binding (Supplementary Fig. S2). We speculated that the second phase of decreasing TAMRA fluorescence reflects dissociation of ptDNA from RFC following ATP hydrolysis (presumably in the form of the PCNA·ptDNA complex), and then the signal levels off in the steady-state phase of the reaction. The trace at Δt=2 s was fit empirically to a 2-exponential function and yielded kup=12.2 s−1 and kdown=4.8 s−1 for the two phases. The rates provide an estimate of the bimolecular ptDNA binding constant (kon ~1×108 M−1 s−1) and indicate linkage between PCNA closure (kclose=7 s−1; Fig. 2c) and ptDNA release from RFC (kdown=4.8 s−1).
Fig. 3.
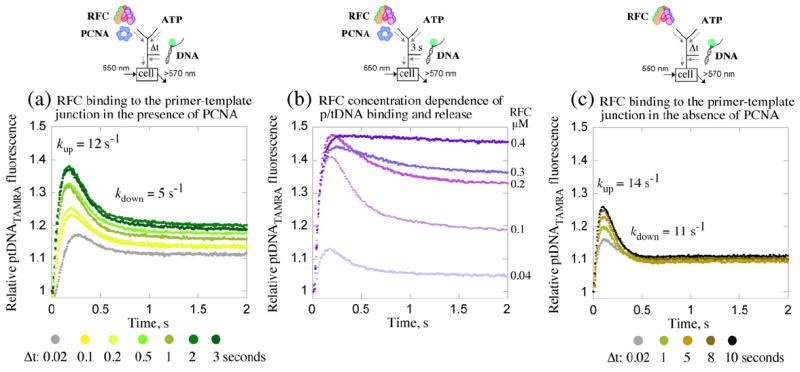
Kinetics of ptDNA binding and release. DNA binding to the RFC·ATP·PCNA complex is measured by change in fluorescence intensity of TAMRA-labeled ptDNA (λEX=550 nm, λEM>570 nm). (a) Pre-incubation of RFC and PCNA with ATP (Δt=0.02–3 s) followed by addition of 3′ primer-labeled ptDNATAMRA results in initial signal increase on ptDNA binding, followed by decrease; the fraction of bound ptDNA increases with Δt. A 2-exponential fit of the Δt=3 s data yields kup=12.2 s−1 (kon ~1×108 M−1 s−1) and kdown=4.8 s−1. (b) As RFC concentration is raised (0.04–0.4 μM; Δt=3 s), more ptDNA remains bound (high fluorescence), indicating that the signal decrease is due to ptDNA release (from RFC·ADP·Pi·PCNA·ptDNA). (c) ATP binding activates RFC to bind ptDNA rapidly even in the absence of PCNA (kup=14 s−1, kdown=11 s−1); however, the fraction of bound ptDNA appears lower, and longer Δt is required for maximum binding than in the presence of PCNA (a). Final reactant concentrations: 0.1 μM RFC (a and c), 0.4 μM PCNA, 0.04 μM ptDNATAMRA, and 0.5 mM ATP. Rate constants obtained from the analytical fits were utilized as initial estimates for global data analysis.
The possibility that the decrease in TAMRA fluorescence reflects an intramolecular event (e.g., a change in RFC·ATP·PCNA·ptDNA conformation) rather than ptDNA release from RFC was addressed by performing the same sequential mixing experiment described above, except at a fixed pre-incubation time (Δt=3 s) andwith varying concentrations of the RFC·ATP·PCNA complex (0.04–0.4 μM). As shown in Fig. 3b, the second phase is lost as the complex concentration rises and is barely detectable at 0.4 μM (ptDNATAMRA=0.04 μM); that is, the fraction of bound ptDNA at steady state increases with RFC·ATP·PCNA concentration. We interpreted these data to mean that ptDNA is released from RFC toward the end of the clamp loading reaction (as signaled by decrease in fluorescence) and that the free ptDNA can be bound again by the excess RFC·ATP·PCNA complex present in solution (note: the experiment in Fig. 3a was performed with 0.1 μM RFC and 0.04 μM ptDNATAMRA).
Another question we tackled using the ptDNATAMRA assay was whether formation of the RFC·ATP·PCNA complex is obligatory for RFC binding to ptDNA. Sequential mixing experiments were performed in the absence of PCNA, wherein RFC was pre-incubated with ATP for varying times (Δt=0.02–10 s; 0.1 μM RFC and 0.04 μM ptDNA) and then mixed with ptDNATAMRA, and the change in fluorescence intensity was monitored over time. The biphasic kinetic traces in Fig. 3c show that RFC binding to ptDNA increases with Δt and reaches maximum at Δt≥8 s. The binding rate constant is similar to that in the presence of PCNA (kon ~1×108 M− 1 s− 1); however, the complex dissociates at a faster rate (kdown=11 s−1), which may account for the lower fraction of bound ptDNA in the absence versus presence of PCNA (Fig. 3a). These data reveal that ATP binding activates RFC for binding to the primer–template junction of ptDNA; however, the process is slower in the absence of PCNA, and the resulting RFC·ATP·ptDNA complex has about 2-fold shorter half-life. A plot of the peak fraction of bound ptDNA versus Δt yields exponential rate constants of ~0.4 s−1 without PCNA and ~3 s−1 with PCNA, indicating that ATP-binding-induced RFC activation occurs ~8-fold slower in the absence of PCNA (plots not shown). The kinetic difference favors RFC binding to PCNA prior to binding ptDNA and thus likely increases the efficiency of the clamp loading reaction.
Rapid ATP hydrolysis and slower phosphate release track ptDNA binding to RFC and release of PCNA·ptDNA from RFC, respectively
In a previous study of RFC ATPase kinetics measured under a variety of conditions,20 we found that the clamp loader hydrolyzes ATP and releases phosphate (Pi) at maximal rate and amplitude in the presence of PCNA and DNA, after pre-incubation of ≥2 s with ATP and PCNA. These data suggested that ptDNA binding to the RFC·ATP·PCNA complex triggers a burst of ATP hydrolysis followed by Pi release and then a slow steady-state phase.20 We measured Pi release kinetics again in this study for comparison with the PCNA opening/closing and ptDNA binding/release kinetics described above. When RFC, PCNA, and ATP are pre-incubated for Δt≥2 s and then mixed with excess ptDNA, ATP is hydrolyzed rapidly, as measured by formation of [32P]ADP from [32P]ATP in rapid chemical quench experiments (hydrolysis rate kADP=20–50 s−1).20 The data shown in Fig. 4 fit to a single exponential +linear function with a kADP of 45 s−1. Pi release follows thereafter, although after a lag phase detectable even at the longest Δt tested (Fig. 4, expanded view); note: at 10 μM concentration, the 7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl)coumarin-labeled phosphate binding protein (MDCC-PBP) sensor, which binds Pi rapidly and with high affinity, is not a limiting factor in the reaction (kon=1.4×107 M−1 s−1; Kd=0.1 μM).34 The lag phase and apparently slow Pi release rate (kPiRelease ~10 s−1) indicate another slow step in the reaction between ATP hydrolysis and Pi release and also suggest linkage between Pi release, PCNA closure (kclose=7 s−1; Fig. 2c), and ptDNA release from RFC (kdown=4.8 s−1; Fig. 3a). Finally, the linear steady-state phase at 1.6 μM s−1 yields a turnover rate, kcat, of 1.1 s−1, assuming three active ATPase sites per RFC (1 and 0.5 μM RFC in [32P] ATPase and Pi release experiments, respectively) (see data analysis below for determination of ATPase stoichiometry). This catalytic rate meets the requirement of loading PCNA every 1–2 s to match the rate of Okazaki fragment synthesis by polymerase δ.35
Fig. 4.
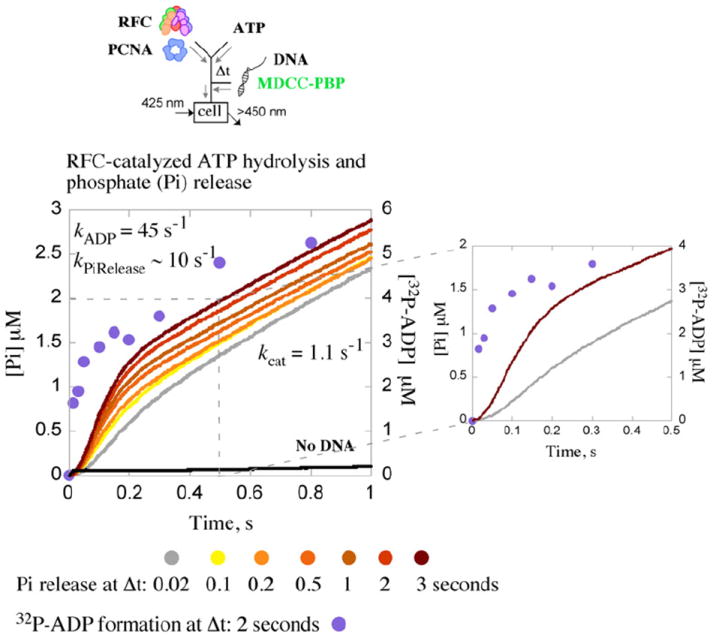
Kinetics of ATP hydrolysis and phosphate (Pi) release. RFC-catalyzed Pi release is measured by change in fluorescence intensity of MDCC-PBP. Pre-incubation of RFC and PCNA with ATP (Δt=0.02–3 s) followed by addition of ptDNA results in a burst of Pi release followed by a linear steady-state phase; the burst amplitude increases with Δt, indicating slow step(s) in the reaction preceding ATP hydrolysis and Pi release. Additionally, a lag prior to the Pi burst phase (even at Δt=3 s), not seen for [α 32P]ADP formation ([α 32P]ADP data shown in purple for Δt=2 s),20 indicates slow step(s) between ATP hydrolysis and Pi release (see expanded view). An exponential+linear fit of the Δt=2 s ADP data yields a burst rate of kADP=45 s− 1 and steady-state kcat=1.2 s−1 (slope/3×[RFC]); similar analysis of the Δt=3 s Pi release data yields kPiRelease=10 s−1 and kcat=1.1 s−1. Final reactant concentrations: 1 μM ([32P]ATPase) or 0.5 μM (Pi release) RFC, 2.5 μM ([32P]ATPase) or 1 μM PCNA (Pi release), 2.5 μM ptDNA, 0.5 mM ATP, and 10 μM MDCC-PBP (Pi release). Rate constants obtained from the analytical fits were utilized as initial estimates for global data analysis.
A comprehensive kinetic model of RFC, PCNA, ptDNA, and ATP transactions in the clamp loading reaction
All the data obtained in this study were globally fit with various models of the clamp loading mechanism in order to determine a single model and set of parameters that could best explain the transient events measured thus far. The goal was to construct a minimally sufficient model—one without excessive simplifying assumptions but also one that is well constrained by the available data. In order to do so, we used the KinTek Explorer program, which can simultaneously fit data from different types of experiments based on numerical integration of the rate equations describing the model.36 The starting point was an earlier kinetic model based on pre-steady-state ATP hydrolysis and Pi release data,20 which was modified to include the events measured in this study. An assumption retained from the earlier version was that all ATPase sites on RFC were considered equivalent with respect to ATP hydrolysis and Pi release; thus, the model treated RFC as a single ATPase unit whose stoichiometry was allowed to float during the fitting process (this assumption was considered reasonable, since the burst of ATP hydrolysis and Pi release is fit well by a single exponential function, and there are no data indicating that RFC subunits hydrolyze ATP at different rates). Scheme 1 shows a kinetic model of the reaction to which all the data fit best. Figure 5a provides a visual summary of the different events following a 3-s pre-incubation of RFC with ATP and PCNA (Δt=2 s for [32P]ADP). The fits for each data set of PCNA opening (Fig. 2b), PCNA opening/closing (Fig. 2c), ptDNA binding/release (Fig. 3a), ATP hydrolysis, and Pi release (Fig. 4) are shown as black lines overlaying the corresponding experimental data in Fig. 5b–f, respectively (only a subset of the data from Figs. 2c, 3a, and 4 at Δt=0.02, 0.2, and 2 s are shown in Fig. 5 for clarity); the same plots are also shown in Supplementary Fig. S3 on a log scale for time.
Scheme 1.
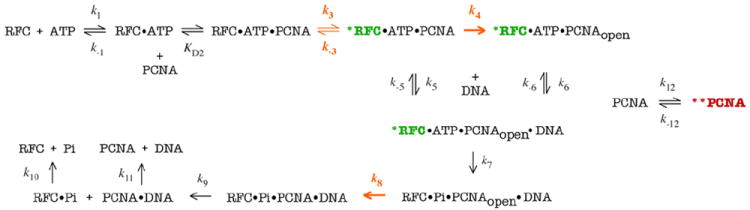
A minimal model of the RFC-catalyzed PCNA loading mechanism. All the kinetic data were fit to this model. Activated RFC is designated as *RFC. Three slow steps in the mechanism (k3, k4 and k8) are highlighted in orange. A separate step shows equilibrium between loading-active (PCNA) and loading-inactive (**PCNA) conformations. Steps where the reverse rate constants are not yet known or expected to be very small are considered irreversible.
Fig. 5.
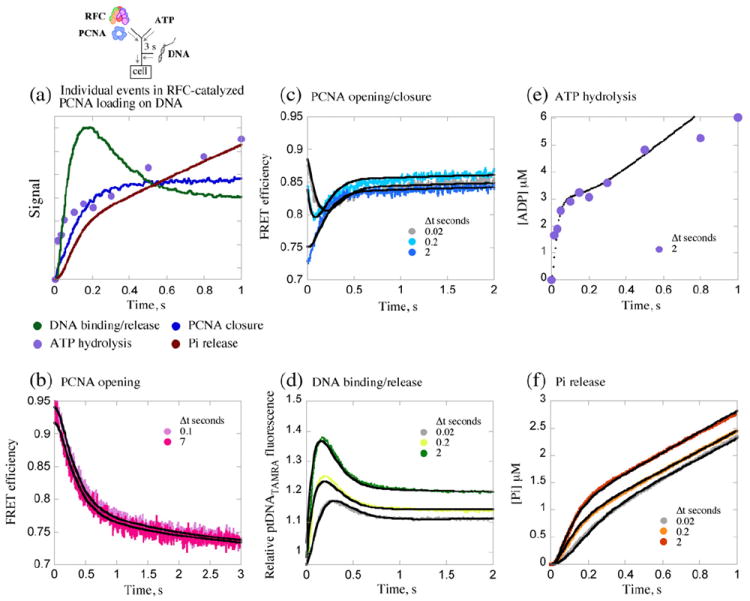
Global analysis of all the kinetic data supports a minimal model of the PCNA loading mechanism. (a) Overlay of data from PCNA opening/closing, DNA binding/release, ATP hydrolysis, and Pi release experiments provides a visual of rapid ptDNA binding to the RFC·ATP·PCNA complex (formed during pre-incubation, Δt=3 s), triggering ATP hydrolysis, PCNA closure, Pi release, PCNA·ptDNA dissociation, and catalytic turnover. Data for (b) PCNA opening (Δt=0.1 and 7 s), (c) PCNA closing after ptDNA addition, (d) ptDNA binding/release, (e) ATP hydrolysis (Δt=2 s), and (f) Pi release [only Δt=0.02, 0.2, and 2 s shown in (c), (d), and (f) for clarity] were all fit simultaneously to the model shown in Scheme 1. The black lines are simulations generated by the model based on parameters listed in Table 1.
In addition to standard error estimates derived from nonlinear regression, we performed a confidence contour analysis to assess whether the data are sufficient to define key unknown kinetic parameters in the model (those lacking independent measurements at this time). This analysis assesses the extent to which each parameter can be varied while the other parameters float during data fitting, and the results are scored by the minimal attainable χ2 value; a 10% increase in χ2 was used as the boundary for setting confidence limits for each parameter (Supplementary Fig. S4).36,37 The best-fit parameters are listed in Table 1, with standard errors from nonlinear regression as well as upper and lower confidence limits from confidence contour analysis.
Table 1.
Best-fit parameters for the RFC reaction mechanism
| Parameters | Best-fit values | Event |
|---|---|---|
| k1 | 100 μM-1 s-1 | ATP binding |
| k-1 | 100 s-1 | ATP dissociation |
| k2a | 513 ± 3.5 s-1 b (462 – 590)c | PCNA binding |
| k-2 (KD2) | 51 ± 0.4 s-1 (0.1 μM) | PCNA dissociation |
| k3 | 7.7 ± 0.05 s-1 | RFC activation |
| k-3 | 0.077 ± 0.001 s-1 | |
| k4 | 4.4 ± 0.03 s-1 | PCNA opening |
| k5, k6 | 50 ± 0.4 s-1 | ptDNA binding to RFC·PCNA complexes |
| k-5, k-6 (KD5, KD6) | 0.05 ± 0.001 s-1 (0.001 μM) | |
| k7 | 45 ± 1 s-1 | ATP hydrolysis |
| k8 | 8.3 ± 0.09 s-1 (7.7 – 8.9) | RFC inactivation and PCNA closure |
| k9 | 500 s-1 | PCNA·DNA release |
| k10 | 500 s-1 | Pi release |
| k11 | 2.4 ± 0.03 s-1 (2 – 2.8) | PCNA·DNA dissociation |
| k12 | 12.3 ± 0.1 s-1 (10.5 – 15.5) | PCNA – PCNA** equilibrium |
| k-12 | 0.56 ± 0.05 s-1 | |
| N | 3.1 ± 0.01 (3 – 3.2) | ATPase sites per RFC |
Parameters shown in bold were linked during global fitting and confidence contour analysis.
Standard error values are shown for parameters allowed to float during global fitting of the data by nonlinear regression analysis.
Upper and lower limit values for parameters varied during confidence contour analysis are shown in parentheses.
The model (Scheme 1) begins with RFC binding ATP with a bimolecular rate constant of 100 μM− 1 s− 1 (k1) and dissociation rate of 100 s−1 (k−1) to form the RFC·ATP complex. We assumed fast ATP binding to RFC since the ATPase rates are independent of ATP concentration in our experiments. The dissociation rate was set to be consistent with the measured affinity of RFC for ATPγS (Kd ~1 μM), a slow-hydrolyzing ATP analog.20,38 The next step in the model is RFC binding to PCNA. Previous reports indicate weak interaction between clamp and clamp loader proteins in the absence of ATP that becomes tight in the presence of ATP or ATPγS (Kd=0.001 μM for RFC and PCNA).22,39 We assumed a dissociation constant of 0.1 μM for the initial interaction (Kd2=k−2/k2) and a fast rate of PCNA binding (k2 ~500 μM−1 s−1), since RFC was incubated with PCNA in the sequential-mixing experiments (note: pre-steady-state ATPase kinetics and Δt required for maximum activity are the same whether PCNA is added with RFC or with ATP).20 Kd2 was allowed to float during data fitting but linked with an equilibrium constant of 0.01 for the next step, ATP-activated RFC·PCNA interaction (1/ K3=k−3/k3), to yield a net Kd of 0.001 μM (Kd2×1/K3) for the *RFC·ATP·PCNA complex, as reported22 [note: asterisk (*) denotes ATP-activated RFC]. Data fitting yielded k3=7.7 s−1 and k−3=0.077 s−1 for this step (Table 1). For the next step in the reaction, PCNA opening, the fit yielded a rate constant of 4.4 s−1 (k4), which results in a net rate constant knet of 2.7 s−1 for formation of the *RFC·ATP·PCNAopen complex [knet=k3k4/(k3+k4+k−3)], similar to the measured net rate of 1.6 s−1 (Fig. 2) and the previously reported rate of 2.2 s−1 for PCNA opening.21 The reverse rate constant (k−4) was set to zero, since allowing it to float during data fitting yielded a very low value and the *RFC·ATP·PCNAopen complex is known to be stable until after ATP hydrolysis. Total FRET efficiency was set at 0.95 for closed PCNA and 0.66 for open PCNAduring data fitting.21 The linked parameters k2, k−2, k3, k−3, and k4 were allowed to vary during confidence contour analysis.
The next step in the reaction is ptDNA binding to the *RFC·ATP·PCNAopen complex, which occurs at a fast bimolecular binding constant of 50 μM−1 s−1 (k5) according to the fit, and is comparable to the 100 μM−1 s−1 constant estimated from data in Fig. 3a. The ptDNA dissociation rate (k−5=0.05 s−1) was linked to k5 in order to maintain the measured Kd of 0.001 μM for the interaction (Supplementary Fig. S2). The relative change in fluorescence signal for free: bound ptDNA was set at 1:2.1 during data fitting, based on quantum yields of ptDNATAMRA alone and in the presence of RFC, PCNA, and non-hydrolyzable ATPγS (Supplementary Fig. S2 and data not shown). ptDNA was also allowed to bind the *RFC·ATP·PCNA closed complex, to account for apparently faster PCNA opening observed on addition of ptDNA to the reaction (Fig. 2c, Δt=0.02 s). Sakato et al. on RFC ATPase mutants presents additional evidence of ptDNA-induced PCNA opening.38 The rate constants for this step were the same as k5, k−5 (k6=50 μM−1 s−1 and k−6=0.05 s−1). Assembly of the *RFC·ATP·PCNAopen·ptDNA complex marks the end of the first phase of the reaction, which is distinguished by ATP-induced rate-determining isomerization of RFC and PCNA into conformations that favor interaction with ptDNA.
Binding of ptDNA triggers a burst of ATP hydrolysis by RFC, which occurs at a rate of 45 s−1 (k7) according to the fit, within the experimentally measured range of 20–50 s−1 (Fig. 4).20 Further analysis of ptDNA binding is necessary to determine whether this rate reflects ATP hydrolysis or an associated rate-determining event, such as ptDNA-binding-induced change in RFC and/or PCNA conformation. This possibility is supported by the observation that the ptDNA binding rate apparently reaches a maximum of ~17 s−1 at high RFC concentrations (Supplementary Fig. S2), and it may also account for the slight lag observed in ptDNA binding, even at long Δt, that is not matched exactly by the current minimal model (Supplementary Fig. S3). After ATP hydrolysis, a slow step in the reaction appears to limit Pi dissociation from RFC, as indicated by the lag in the Pi release (Fig. 4). The apparent rates of PCNA closure (Fig. 2c) and ptDNA release from RFC (Fig. 3a) are similar to Pi release (Fig. 4); therefore, we considered each of these as possible rate-determining events in the reaction. By varying their order in the model and allowing the parameters to float in different combinations during global fitting, we found that all data are best fit with the slow step occurring at a rate of 8.3 s−1 (k8) and that any of these three events (or an associated event not measured explicitly yet) may well be the rate-limiting one (k8 was allowed to vary during confidence contour analysis). In the model in Fig.5, PCNA closure is shown as the slowest of the three (k8) followed by dissociation of PCNA·ptDNA (k9) and Pi (k10); the latter two steps can occur in any order and are set at a fast rate of 500 s−1 in the model. As noted above, all three events can be considered interchangeable for now as long as at least one of them determines the 8.3 s−1 rate constant (note: given the uncertainty in the model in this phase and the lack of kinetic data on reverse rates, we assumed that these steps are irreversible). While the exact nature of the slow step cannot be ascertained at this time, it likely involves isomerization of RFC and PCNA after ATP hydrolysis to allow PCNA closure around ptDNA and to lower RFC affinity for PCNA·ptDNA and Pi products. Notably, one of the RFC subunit ATPase mutants described in Sakato et al. releases ptDNA alone at the same fast rate as wild-type RFC but exhibits slower PCNA closing and PCNA·ptDNA complex dissociation rates.38 Based on this finding, it is likely that the rate-determining step involves PCNA closure. This step distinguishes the second phase of the clamp loading reaction, which ends with disassembly of the RFC·ADP·Pi·PCNA·ptDNA complex. It should be noted that the kinetics of ADP dissociation from RFC have not been measured explicitly yet; hence, the timing and mechanistic significance of this event remain unknown, and it has not been included in the model.
Once the products are released, the catalytic cycle can be considered complete. An additional (irreversible) step involving PCNA·ptDNA dissociation was added to the model in order to account for the steady-state reaction conditions in experiments that contain low concentrations of PCNA and ptDNA. The data fitting yielded a rate of 2.4 s−1 for this step (k11), which may be slow enough to influence the observed steady-state rate (the reverse rate k−11 was fixed at zero). Previous studies indicate that clamps may dissociate slowly even from short linear ptDNA, such as the 40/65-nt substrate in our experiments, because of multiple contacts with the double-stranded DNA backbone as well as single-stranded DNA (ssDNA) bases at the primer–template junction.40,41 Consistent with this interpretation, increasing PCNA and ptDNA concentration in the simulation results in a slight rise in the steady-state rate (the difference is within error of corresponding experimental measurements; data not shown). A final step was added to the model in order to account for the second slow phase detected in the PCNA opening/closing data (Fig. 2b and c). The biphasic kinetics may be indicative of a two-step process for PCNA opening and closure or perhaps alternate conformations of PCNA in the reaction. In our minimal model, the clamp partitions between two forms, one that binds RFC (PCNA) and one that does not (**PCNA), and the data fitting yields forward and reverse rate constants of 12.3 s−1 (k12) and 0.6 s−1 (k−12) for the equilibrium (k11 as well as linked k12 and k−12 were allowed to vary during confidence contour analysis). While inclusion of this step in the model is rather speculative at this time, there is prior evidence from molecular dynamics (MD) simulations that clamps can adopt different conformations, one or more of which may be bound preferentially by the clamp loader.42,43
Finally, the global data fitting also yielded a stoichiometry of 3.1 ATP molecules hydrolyzed per RFC in the burst phase. A scaling parameter representing the number ATPase sites on RFC was allowed to float during data fitting and confidence contour analysis (a best-fit value of 3 was also obtained from the earlier model based on analysis of ATP hydrolysis and Pi release data alone). 20 Notably, a recent model for the E. coli γ complex clamp loader mechanism proposes that three ATP molecules are hydrolyzed in one catalytic turnover,29 and a new crystal structure of the bacteriophage T4 clamp loader also shows three active ATPase sites.44 Thus, hydrolysis of three ATP molecules may be necessary and sufficient to drive clamp loading across the evolutionary spectrum.
Discussion
To load a circular clamp onto DNA—a critical event in several DNA metabolic processes—a clamp loader has to manipulate the two macromolecules into a topologically linked product. The requisite energy is supplied by ATP binding and hydrolysis catalyzed by multiple clamp loader subunits. The question of how the chemical reaction is coupled to the mechanical work is being addressed by structural and kinetic studies of severalmodel clamp loaders. It is known that the reaction occurs in two main stages: (a) Assembly—on binding ATP, the loader forms a complex containing an open clamp and ptDNA, and (b) Disassembly—on ATP hydrolysis, the loader releases the clamp·ptDNA product. It is also clear that ptDNA binding stimulates ATP hydrolysis and transition from one stage of the reaction to the next. Nevertheless, the question of how the reaction progresses through each stage has yet to be resolved—particularly in the case of eukaryotic clamp loaders. To this end, we have used a transient kinetic approach to detect formation and decay of intermediate species in the PCNA loading reaction catalyzed by S. cerevisiae RFC.
In a recent pre-steady-state kinetic study of RFC ATPase activity we discovered a slow step in the reaction after ATP binding to clamp loader. The data revealed that three ATPase-active subunits of RFC catalyze a rapid burst of ATP hydrolysis on binding ptDNA, but only after prolonged incubation with ATP (±PCNA). A minimal kinetic model based on global fitting of the ATP hydrolysis and phosphate (Pi) release data yielded a rate of ~5 s−1 for this step, followed by rapid ptDNA binding, and then ATP hydrolysis at a rate of ~50 s−1.20 We proposed that a rate-determining event, in which RFC converts from “ATPase-inactive” to “ATPase-active” conformation, is an early checkpoint in the pathway toward ordered assembly of a complex poised to load PCNA. Another slow step was identified after ATP hydrolysis, prior to or related to Pi dissociation, at a rate of ~6 s−1. We speculated that this event involving PCNA, ptDNA, ADP, and/or Pi is a late checkpoint in the pathway toward ordered formation and release of topologically linked PCNA·ptDNA. Our goal here was to measure the reaction kinetics from the perspective of the clamp and ptDNA, and complement the ATPase data for a more comprehensive view of the clamp loading mechanism. To this end, we used fluorescence-based in-solution assays to monitor PCNA opening and closure as well as ptDNA binding and release in real time. The ATPase kinetics were measured concurrently in order to clarify the mechanochemical basis of clamp loading.
Identity and timing of key events in the clamp loading reaction
We used an assay developed by the Benkovic group to measure PCNA opening and closure by FRET between W185 and C107AEDANS across the inter-subunit interface. Consistent with their report,21 the measured distance between the fluorophores increases from 13 Å for PCNA alone to 34 Å for PCNA in the presence of RFC and ATP. A model of the RFC·ATPγS·PCNAopen complex,43 derived from MD simulations of an RFC·ATPγS·PCNAclosed crystal structure,10 yields an average distance of 39 Å between the two residues (Fig. 1). Given the similarity between the experimental and theoretical values for open PCNA, we consider the MD-based structure a good model for the RFC·ATP·PCNAopen complex measured in our kinetic analysis (Fig. 2).
Crystal structures of E. coli γ complex and S. cerevisiae RFC provide important clues about structural transitions leading to the open-clamp complex.10,45,46 The first γ complex structure revealed the general clamp loader architecture of five subunits with C-terminal domains arranged in a circular collar and N-terminal AAA+ ATPase modules arranged in the shape of a claw. In this case, the three ATP-binding sites at the δ′–γ and γ–γ subunit interfaces were nucleotide free, and the complex appeared to be in an asymmetric, inactive conformation. 45 The next structure was of an RFC·ATPγS·PCNAclosed complex in which the ATPase activity of each RFC subunit was suppressed by replacing the catalytic “arginine finger” with glutamine. ATPγS was bound to the composite ATP-binding sites at all subunit interfaces (despite the lack of conserved catalytic residues in RFC-E), and the associated inter-subunit interactions helped organize the clamp loader into a spiral conformation matching the double helix (Fig. 1).10 We propose that, in the slow activation process initiated by ATP binding, the clamp loader subunits begin to adopt a more symmetric arrangement around a central helical axis, aligning the ATPase modules to form catalytically competent active sites and to contact the duplex portion of ptDNA (Fig. 6, step I).
Fig. 6.
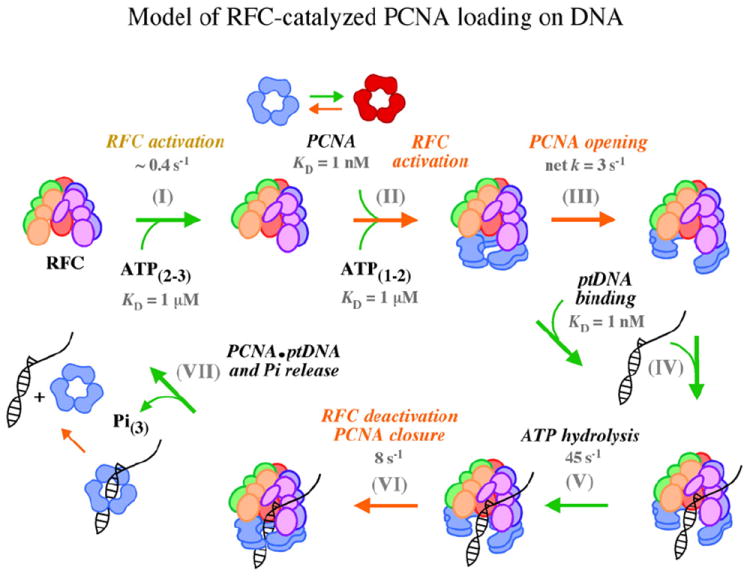
A model pathway for RFC-catalyzed PCNA loading on ptDNA. The schematic depicts key steps in the clamp loading reaction determined thus far. (I) Binding of two to three ATP molecules to RFC is followed by (II) RFC interaction with PCNA, additional binding of one to two ATP molecules,20 and slow activation steps involving conformational changes in RFC and PCNA that (III) lead to a stable open-clamp complex. (IV) ptDNA binds rapidly and with high affinity to activated RFC·ATP·PCNA complexes, (V) triggering ATP hydrolysis. (VI) Next, another slow step involving conformational changes in RFC and PCNA leads to closure of the clamp around ptDNA, (VII) release of products (Pi, PCNA·ptDNA), and catalytic turnover. Under the conditions of our in vitro experiments, slow dissociation of PCNA·ptDNA may contribute to the turnover rate (slow steps are indicated by orange arrows). The proposed partitioning of PCNA between loading-active and loading-inactive forms is also shown.
ATP binding alone does not appear to be sufficient for optimal activation of RFC. The clamp loader binds only two to three ATP molecules in the absence of PCNA and an additional one to two molecules in its presence.20,38,47 Furthermore, our ptDNA binding (Fig. 3) and ATPase kinetic data20 provide clear evidence that RFC activation is accelerated by PCNA. In the crystal structure,10 the clamp is closed and in contact with the bases of RFC-A, RFC-B, and RFC-C subunits only, while RFC-D and RFC-E are suspended above the plane of the clamp (Fig. 1). In the MD-derived model,43 the open clamp is in an out-of-plane right-handed spiral conformation and in contact with all RFC subunits, including RFC-D and RFC-E, which are in a matching spiral conformation (Fig. 1). We propose that as RFC binds ATP and begins the transition to active form, its interaction with PCNA drives cooperative structural changes in both proteins that result in additional ATP binding and stabilization of the complex in a spiral, open-clamp conformation (Fig. 6, steps II and III). Recent MD analysis of PCNA indicates that it is a flexible, mechanically compliant protein that can transiently twist open in left- or right-handed spirals. Indeed, the calculated energetic cost of deforming PCNA into the wide-open conformation detected by FRET between W185 and C107AEDANS is very low.42 In the absence of RFC, however, an open PCNA clamp is estimated to reclose within nanoseconds, and the closed form is predominant. In the presence of RFC, interactions between the clamp loader base and the entire circumference of the clamp could trap and secure a transient spiral conformation.43 In this view, the RFC·ATPγS·PCNAclosed crystal structure10 represents an intermediate en route to an open-clamp complex that can bind ptDNA rapidly. The MD results are consistent with a mechanism in which ATP binding favors an RFC conformation that binds PCNA with high affinity, and capture of open PCNA by RFC with in this complex leads to mutual stabilization of the proteins in an active conformation for the next stage of the reaction (Fig. 6, step IV). A recent study of E. coli γ complex indicates that this clamp loader binds and opens β clamp in a two-step process as well.30
Another crystal structure of E. coli γ complex bound to ADP·BeF3 and ptDNA was solved recently,46 and it provides clues about structural transitions leading to a ptDNA-bound complex on the brink of ATP hydrolysis. In the RFC·ATPγS·PCNAclosed structure discussed earlier, only RFC-A and RFC-C ATP-binding sites appeared near catalytic competency, whereas the nucleotides in RFC-B and RFC-D sites were loosely coordinated.10 In contrast, the ATPase modules are in a highly symmetric helical arrangement in the ptDNA-bound γ complex structure, and all three sites are catalytically competent with the arginine finger positioned to stimulate ATP hydrolysis (coordinating BeF3 in this case). Thus, it appears that binding of ptDNA in the central chamber drives further adjustment of inter-subunit contacts, bringing all catalytic sites on line for ATP hydrolysis. Consistent with these structural data, ptDNA binding to the activated RFC·ATP·PCNA complex (or activated RFC·ATP complex)20 is followed by a burst of ATP hydrolysis (Fig. 4; Fig. 6, step V).
Key mechanistic features of this stage of the reaction are highlighted here. First, complete coordination of ATP results in a clamp loader conformation that better complements the clamp surface.46 The kinetics of PCNA opening/closure suggest that interaction of ptDNA with the RFC·ATP·PCNA complex leads to faster PCNA opening. Perhaps initial contacts between RFC, PCNA, and ptDNA promote further conformational changes that result in all three macromolecules adopting optimal complementary structures. Second, all ATPase sites in the γ complex·ADP·BeF3·ptDNA structure are in essentially the same active conformation, suggesting that ATP hydrolysis occurs in a concerted fashion. This interpretation is consistent with our proposed model in which three ATP molecules are hydrolyzed at the same apparent rate by the RFC·ATP·PCNAopen·ptDNA complex. The bacteriophage T4 gp44/62 clamp loader also hydrolyzes all bound ATP at the same rate in the presence of gp45 clamp and DNA.28 Third, convergence between the number of ATP molecules hydrolyzed rapidly by γ complex, which has three ATP-binding sites,29 and RFC, which has five ATP-binding (four ATPase active) sites, implies that hydrolysis of three ATP molecules in a catalytic turnover is necessary and possibly sufficient for loading a clamp onto DNA and may be an evolutionarily conserved feature of the mechanism. Earlier studies have shown that only RFC-B, RFC-C, and RFC-D ATPase activity is required for clamp loading.32,48 In the case of T4 gp44/62, rapid hydrolysis of four ATP molecules has been reported;28 however, a new crystal structure indicates three active ATPase sites in this clamp loader as well.44 It remains possible that there is a fraction of inactive RFC complex in our preparation or a fourth ATP molecule is hydrolyzed at a slow rate that is indistinguishable from kcat. Sakato et al. investigates the contribution of individual RFC subunits to the PCNA loading mechanism.38
The kinetic data also indicate a slow step in the reaction immediately following ATP hydrolysis—at or prior to PCNA closure, and/or ptDNA and Pi dissociation. PCNA·ptDNA, ADP, and Pi product release from RFC may occur simultaneously or in a preferred sequence, but currently, there is no experimental evidence to resolve which event occurs first and/or is rate determining. As noted earlier, the energetic barrier for closure of free PCNA appears to be minimal;43 therefore, once ptDNA is positioned in the open clamp, reversal of the clamp loader to an inactive conformation with low affinity for the clamp and ptDNA may be sufficient to end the reaction. Disengagement of PCNA from RFC and rapid re-closure of the interface would trap ptDNA within the clamp and lead to release of topologically linked product. We speculate that once ATP is hydrolyzed, RFC undergoes slow inactivation to relax back into an asymmetric conformation, as suggested by the nucleotide-free γ complex structure.45 PCNA closes rapidly on losing contact with RFC-D and RFC-E subunits (Fig. 6, step VI), and PCNA·ptDNA and Pi are released (Fig. 6, step VII).
Several mechanistic questions remain about the second stage of the reaction that require further experimentation. First, whether change in RFC conformation is truly a rate-determining event at the end of the catalytic turnover, as proposed here; this question can be addressed by fluorescence-based assays that monitor RFC isomerization during the reaction. Second, whether ADP release has any role in ending the reaction; the kinetics of ADP release following ATP hydrolysis have not been measured for any clamp loader yet, and the question remains open. Third, whether the ATPase activity of each RFC subunit has the same or different role in ending the reaction; this question is addressed in Sakato et al.38 Fourth, whether interactions between DNA and cationic residues inside the clamp help trigger clamp closure and release, as was speculated upon discovery of contacts between E. coli β clamp and ptDNA.40 According to a recent report, mutation of 12 cationic residues inside PCNA causes barely 2-fold reduction in RFC ATPase activity, suggesting that interactions between PCNA and DNA do not influence the loading mechanism;49 however, since the study measured steady-state ATPase rates (kcat), it is not known whether the interactions affect particular steps in the reaction. Our initial pre-steady-state analysis suggests that mutation of select cationic residues in PCNA can disrupt clamp closure and DNA release (Y.Z. and M.M.H., unpublished data). Finally, a recent report from the Benkovic group raised a provocative question about whether RFC functions catalytically, since their data suggest stoichiometric PCNA loading on forked ptDNA.31 However, an earlier study from the Burgers group had shown that RFC releases PCNA·ptDNA after ATP hydrolysis, indicating catalytic function.22 The DNA binding kinetics reported here also indicate that RFC releases ptDNA toward the end of the reaction. We have found that RFC can bind ssDNA with high affinity50 (Supplementary Fig. S2), suggesting that some fraction of RFC may associate with the template portion of ptDNA (a forked DNA substrate provides more ssDNA binding sites).31 However, ssDNA does not trigger a burst of ATP hydrolysis by RFC (Supplementary Fig. S5), indicating nonspecific interaction, and the RFC·ssDNA complex dissociates at least 10-fold faster than RFC·ptDNA.50 We have proposed that fleeting contact with ssDNA is part of a scanning mechanism employed by RFC to find primed sites for loading PCNA.50 Potential effects of single-strand binding protein (replication protein A) on the kinetics of ptDNA binding/release by RFC are yet to be tested. Regarding the proposal that RFC functions non-catalytically,31 we speculate that the presence of mixed species in the reaction, including varying fractions of unactivated and activated RFC bound to PCNA, forked DNA, and ATP/ADP/Pi (and combinations thereof), may have influenced the kinetics; furthermore, the experiments were performed with a truncated version of RFC (RFC-A ΔN), which can load PCNA onto DNA but has significantly reduced activity (lower stoichiometry and rate) relative to full-length wild-type RFC, according to pre-steady-state ATPase data.20
Links between the RFC ATPase reaction and the work of loading PCNA onto DNA
Clamp loader structural dynamics and ATPase activity are geared toward transforming chemical energy into mechanical work. In terms of ligand binding and release-induced allosteric effects, the following changes in protein conformation during the reaction appear significant: (1) Nucleotide-free RFC exists in a relaxed conformation with relatively weak inter-subunit interactions between the AAA+ ATPase modules. ATP binding causes the first allosteric transition that results in stronger intersubunit interactions and ordering around a central helical axis.10 (2) This structural rearrangement causes RFC-A, RFC-B, and RFC-C subunits to present a favorable surface for electrostatic interactions with PCNA. (3) Contact between PCNA and RFC is accompanied by another transition, in which twisting of antiparallel β-strands at a PCNA intersubunit interface42,43 and corresponding adjustment of RFC stabilize the complex into a spiral with open PCNA. RFC in complex with PCNA can bind additional ATP and both proteins present a positively charged surface to match the helical pitch of double-stranded DNA. (4) Electrostatic interactions between RFC and DNA drive another transition, in which all the interfacial nucleotide-binding sites tighten coordination of ATP and catalytic residues are set for hydrolysis.46 (5) ATP hydrolysis initiates the next cascade of conformational changes that alter the network of contacts between RFC subunits, between RFC and ptDNA, and between RFC and PCNA, allowing clamp closure around ptDNA. (7) Release of ATP hydrolysis products resets RFC to a nucleotide-free conformation with lower affinity for PCNA and ptDNA. Allosteric transitions coupled to ligand binding/release may go in both directions along the reaction coordinate depending on the associated changes in free energy. However, the decrease in free energy associated with ATP hydrolysis can alter the free-energy landscape for RFC isomerization in a directional manner toward PCNA loading. The sequence of conformational changes noted above, highlighted by ATP-binding-led stabilization of the RFC·PCNAopen complex and ATP-hydrolysis-led destabilization of the RFC·PCNAopen·ptDNA complex, provides a basic mechanistic explanation of how ATPase activity drives clamp loading onto DNA.
Materials and Methods
Proteins and DNA
Wild-type, full-length S. cerevisiae RFC was expressed in BL21(DE3) E. coli cells (Agilent Technologies, Santa Clara, CA) using a dual-plasmid system(pLANT2/RIL-RFC-A/E and pET11a-RFC-B/C/D) and purified as described previously.51 Wild-type PCNA, (pET11a-PCNA), PCNA-WC (pET22b-PCNA-F185W/K107C/C22S/C30S/C62S/ C81S; a gift from Stephen Benkovic, Penn State University), and PCNA-FC (pET22b-PCNA-K107C/C22S/C30S/ C62S/C81S) were expressed in BL21(DE3) cells, purified and labeled with N-(iodoacetyl)-N′-(5-sulfo-1-naphthyl) ethylenediamine (Invitrogen, Carlsbad, CA) as described, 21,52 with minor modifications. PCNA was purified from E. coli cell lysate by ammonium sulfate fractionation followed by DE52 DEAE cellulose (Whatman, Piscataway, NJ) and Q Sepharose FF (GE Healthcare, Piscataway, NJ) ion-exchange chromatography. After labeling, PCNAAEDANS was separated fromfree dye using Centricon 10 (Millipore, Billerica, MA) and P-6 gel-filtration (Bio-Rad Laboratories, Hercules, CA). E. coli PBP was purified and labeled with MDCC (Invitrogen) as described previously.34 Protein concentrationswere determined by Coomassie Plus assay (Pierce, Rockford, IL).
ptDNA (3′ junction) was prepared by annealing 40-nt primer (5′-ATT TCC TTC AGC AGA TAG GAA CCA TAC TGA TTC ACA TGG C-3′) and 65-nt template (5′-TAG TTA GAA CCT AAG CAT ATT AGT AGC CAT GTG AAT CAG TAT GGT TCC TAT CTG CTG AAG GAA AT-3′). The DNAs were purchased from Integrated DNA Technologies (Coralville, IA) and purified by urea gel electrophoresis. TAMRA (Invitrogen)-labeled primer–template was prepared by labeling the 3′ primer end modified with an amino linker, as described previously.53
PCNA opening and closing kinetics
PCNA opening/closing was measured on a stopped-flow instrument (KinTek Corp., Austin, TX). Single-mixing (RFC and PCNA mixed with ATP) and double-mixing (RFC and PCNA pre-incubated with ATP for Δt=0.02–3 s and then mixed with ptDNA or RFC pre-incubated with ATP for Δt=0.02–10 s and then mixed with PCNA) experiments were performed at 25 °C. Changes in PCNA-WCAEDANS or PCNA-FCAEDANS (lacking donor W185) fluorescence were measured over time (λEX=290 nm or 336 nm, λEM>450 nm) as described previously.20 In all experiments, the final reactant concentrations were 0.6 μM RFC, 0.25 μM PCNA, 0.25 μM ptDNA (when present), and 0.5 mM ATP in buffer A [30 mM Hepes-NaOH (pH 7.5), 10 mM MgCl2, 100 mM NaCl, 2 mM DTT]. Three or more kinetic traces (1000 data points each) were averaged, and the signal was converted to FRET efficiency, as follows, and plotted versus time.
ET is FRET efficiency, and IAD and IA are fluorescence intensities of AEDANS acceptor in the presence (PCNA-WCAEDANS) or absence (PCNA-FCAEDANS) of the tryptophan donor in PCNA at λEX=290 nm. εA (1800M−1 cm−1) and εD (4100 M−1 cm−1) are the extinction coefficients of AEDANS and tryptophan at 290 nm.21 A small contribution by RFC tryptophan residues to the FRET signal (interprotein FRET) was first corrected as described previously.33 Briefly, normalized fluorescence data were calculated for PCNA-WCAEDANS [FAD (290)] and PCNA-FCAEDANS [FA (290) and FA (336)] by dividing background-subtracted kinetic traces measured in the presence of RFC by those measured in the absence of RFC. FAD (290) is related to IAD + X, FA (290) is related to IA + X, and FA (336) is related to IA, where X is the amount of inter-protein FRET. FAD (290), FA (290), and FA (336) weremultiplied by 3.12, 1, and 1, respectively (relative fluorescence intensities of PCNA-WCAEDANS at λEX=290 nm, PCNA-FCAEDANS at λEX=290, and PCNA-FCAEDANS at λEX=336 nm, measured on both a fluorometer and the stopped flow) to obtain corresponding IAD+X, IA+X, and IA values. By subtracting IA from IA + X, we obtained X and, consequently, IAD. These corrected IAD and IA values were used to calculate ET. The data were fit to single or double exponential functions for initial estimation of rate constants.
ptDNA binding kinetics
DNA binding was measured by double-mixing stopped-flow experiments in which RFC and PCNA (when present) were pre-incubated with ATP for various times (Δt=0.02–10 s) and then mixed with ptDNATAMRA in buffer A. TAMRA fluorescence was measured over time (λEX=550 nm, λEM>570 nm) as described previously.20 Final reactant concentrations were 0.1 μM RFC, 0.4 μM PCNA, 0.04 μM ptDNATAMRA, and 0.5 mM ATP in buffer A containing 0.05 mg/ml bovine serum albumin. RFC concentration dependence was measured by varying RFC from 0.04 to 0.4 μM at Δt=3 s. Three or more kinetic traces were averaged, and the change in fluorescence intensity (relative to the initial measurement) was plotted versus time. The data were fit to a double exponential function for initial estimation of rate constants.
ATP hydrolysis and phosphate (Pi) release kinetics
ATP hydrolysis was measured under pre-steady-state conditions by double-mixing experiments on a quench-flow instrument (KinTek Corp.), as described previously.20 RFC and PCNA were pre-incubated with [α 32P]ATP for Δt=2 s and then mixed with ptDNA in buffer A. At varying times, the reaction was quenched with 4 M formic acid,28,54 and the products were resolved by TLC (0.5 M formic acid and 0.5 M LiCl) and quantified. Final reactant concentrations were 1 μM RFC, 2.5 μM PCNA, 2.5 μM ptDNA, and 500 mM ATP. [α 32P]ADP concentration was plotted versus time, and the data were fit to an exponential + linear function for initial estimation of rate constants.
Pi release was measured under pre-steady-state conditions by double-mixing stopped-flow experiments inwhich RFC and PCNA were pre-incubated with ATP for various times (Δt=0.02–3 s) and then mixed with ptDNA and MDCC-PBP in buffer A containing a Pi contaminant mopping system of 0.1 unit/ml purine nucleoside phosphorylase (PNPase; Sigma-Aldrich, St. Louis, MO) and 0.2 mM 7-methylguanosine (R. I. Chemical Inc., Orange, CA). Changes in MDCC-PBP fluorescence were measured over time (λEX=425 nm, λEM>450 nm) as described previously.20 Final reactant concentrations were 0.5 μM RFC, 1 μM PCNA, 2.5 μM ptDNA, 0.5 mM ATP, and 10 μM MDCC-PBP. Three or more kinetic traces were averaged, and Pi concentration, determined from calibration curves generated by parallel experiments with standard Pi solution (Sigma-Aldrich), was plotted versus time after subtracting a small amount of Pi formed during Δt in the absence of ptDNA. The data were fit to an exponential + linear function for initial estimation of rate constants.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R15 GM094047-01 (M.M.H.). We thank Stephen J. Benkovic for the PCNA-WC clone and Smita S. Patel and Michael O’Donnell for helpful discussions.
Abbreviations used
- PCNA
proliferating cell nuclear antigen
- RFC
replication factor C
- ptDNA
primer–template DNA
- Pi
inorganic phosphate
- TAMRA
5-(and 6-)carboxytetramethylrhodamine
- AEDANS
5-[2(acetyl)aminoethyl-]aminonaphthalene-1-sulfonate
- MDCC-PBP
7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin-labeled phosphate binding protein
- FRET
Förster resonance energy transfer
- ssDNA
single-stranded DNA
- MD
molecular dynamics
Footnotes
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2011.12.018
References
- 1.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Indiani C, O’Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 3.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 5.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 7.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 8.Yoder BL, Burgers PM. Saccharomyces cerevisiae replication factor C.I. Purification and characterization of its ATPase activity. J Biol Chem. 1991;266:22689–22697. [PubMed] [Google Scholar]
- 9.Gerik KJ, Gary SL, Burgers PM. Overproduction and affinity purification of Saccharomyces cerevisiae replication factor C. J Biol Chem. 1997;272:1256–1262. doi: 10.1074/jbc.272.2.1256. [DOI] [PubMed] [Google Scholar]
- 10.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 11.Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, et al. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]
- 12.Neuwald AF. Evolutionary clues to eukaryotic DNA clamp-loading mechanisms: analysis of the functional constraints imposed on replication factor C AAA+ ATPases. Nucleic Acids Res. 2005;33:3614–3628. doi: 10.1093/nar/gki674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Smiley RD, Zhuang Z, Benkovic SJ, Hammes GG. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: multiple pathways of holoenzyme formation. Biochemistry. 2006;45:7990–7997. doi: 10.1021/bi0603322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pietroni P, von Hippel PH. Multiple ATP binding is required to stabilize the “activated” (clamp open) clamp loader of the T4 DNA replication complex. J Biol Chem. 2008;283:28338–28353. doi: 10.1074/jbc.M804371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom LB. Dynamics of loading the Escherichia coli DNA polymerase processivity clamp. Crit Rev Biochem Mol Biol. 2006;41:179–208. doi: 10.1080/10409230600648751. [DOI] [PubMed] [Google Scholar]
- 17.O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci USA. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seybert A, Wigley DB. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. EMBO J. 2004;23:1360–1371. doi: 10.1038/sj.emboj.7600130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Levin MK, Sakato M, Zhou Y, Hingorani MM. Mechanism of ATP-driven PCNA clamp loading by S. cerevisiae RFC. J Mol Biol. 2009;388:431–442. doi: 10.1016/j.jmb.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci USA. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- 23.Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O’Donnell M, Hurwitz J. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci USA. 1998;95:11607–11612. doi: 10.1073/pnas.95.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Gibbs E, Kelman Z, O’Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hingorani MM, O’Donnell M. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem. 1998;273:24550–24563. doi: 10.1074/jbc.273.38.24550. [DOI] [PubMed] [Google Scholar]
- 26.Williams CR, Snyder AK, Kuzmic P, O’Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the gamma complex. J Biol Chem. 2004;279:4376–4385. doi: 10.1074/jbc.M310429200. [DOI] [PubMed] [Google Scholar]
- 27.Anderson SG, Thompson JA, Paschall CO, O’Donnell M, Bloom LB. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli gamma complex. Biochemistry. 2009;48:8516–8527. doi: 10.1021/bi900912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trakselis MA, Berdis AJ, Benkovic SJ. Examination of the role of the clamp-loader and ATP hydrolysis in the formation of the bacteriophage T4 polymerase holoenzyme. J Mol Biol. 2003;326:435–451. doi: 10.1016/s0022-2836(02)01330-x. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JA, Paschall CO, O’Donnell M, Bloom LB. A slow ATP-induced conformational change limits the rate of DNA binding but not the rate of beta clamp binding by the Escherichia coli gamma complex clamp loader. J Biol Chem. 2009;284:32147–32157. doi: 10.1074/jbc.M109.045997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschall CO, Thompson JA, Marzahn MR, Chiraniya A, Hayner JN, O’Donnell M, et al. The E. coli clamp loader can actively pry open the betasliding clamp. J Biol Chem. 2011;286:42704–42714. doi: 10.1074/jbc.M111.268169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar R, Nashine VC, Mishra PP, Benkovic SJ, Lee TH. Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies. Proc Natl Acad Sci USA. 2010;107:19736–19741. doi: 10.1073/pnas.1014139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281:35531–35543. doi: 10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- 33.Alley SC, Abel-Santos E, Benkovic SJ. Tracking sliding clamp opening and closing during bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry. 2000;39:3076–3090. doi: 10.1021/bi992377r. [DOI] [PubMed] [Google Scholar]
- 34.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, Webb MR. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 35.Langston LD, O’Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 37.Johnson KA, Simpson ZB, Blom T. FitSpace explorer: an algorithm to evaluate multi-dimensional parameter space in fitting kinetic data. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 38.Sakato M, O’Donnell M, Hingorani MM. A central swivel point in the RFC clamp loader controls PCNA opening and loading on DNA. J Mol Biol. 2012;416:163–175. doi: 10.1016/j.jmb.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder AK, Williams CR, Johnson A, O’Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: II. Uncoupling the beta and DNA binding activities of the gamma complex. J Biol Chem. 2004;279:4386–4393. doi: 10.1074/jbc.M310430200. [DOI] [PubMed] [Google Scholar]
- 40.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurence TA, Kwon Y, Johnson A, Hollars CW, O’Donnell M, Camarero JA, Barsky D. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J Biol Chem. 2008;283:22895–22906. doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adelman JL, Chodera JD, Kuo IF, Miller TF, III, Barsky D. The mechanical properties of PCNA: implications for the loading and function of a DNA sliding clamp. Biophys J. 2010;98:3062–3069. doi: 10.1016/j.bpj.2010.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tainer JA, McCammon JA, Ivanov I. Recognition of the ring-opened state of proliferating cell nuclear antigen by replication factor C promotes eukaryotic clamp-loading. J Am Chem Soc. 2010;132:7372–7378. doi: 10.1021/ja100365x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. Crystal structures show how the sliding clamp that facilitates processive replication is loaded onto DNA. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 46.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, et al. The mechanism of ATP-dependent primer–template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt SL, Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem. 2001;276:34784–34791. doi: 10.1074/jbc.M011633200. [DOI] [PubMed] [Google Scholar]
- 49.McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA–DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hingorani MM, Coman MM. On the specificity of interaction between the Saccharomyces cerevisiae clamp loader replication factor C and primed DNA templates during DNA replication. J Biol Chem. 2002;277:47213–47224. doi: 10.1074/jbc.M206764200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finkelstein J, Antony E, Hingorani MM, O’Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 52.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, Coman MM, Sakato M, O’Donnell M, Hingorani MM. Conserved residues in the delta subunit help the E. coli clamp loader, gamma complex, target primer–template DNA for clamp assembly. Nucleic Acids Res. 2008;36:3274–3286. doi: 10.1093/nar/gkn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2–Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


