Abstract
RNA viruses are excellent experimental models for studying evolution under the theoretical framework of population genetics. For a proper justification of this thesis we have introduced some properties of RNA viruses that are relevant for studying evolution. On the other hand, population genetics is a reductionistic theory of evolution. It does not consider or make simplistic assumptions on the transformation laws within and between genotypic and phenotypic spaces. However, such laws are minimized in the case of RNA viruses because the phenotypic space maps onto the genotypic space in a much more linear way than on higher DNA-based organisms. Under experimental conditions, we have tested the role of deleterious and beneficial mutations in the degree of adaptation of vesicular stomatitis virus (VSV), a nonsegmented virus of negative strand. We also have studied how effective population size, initial genetic variability in populations, and environmental heterogeneity shapes the impact of mutations in the evolution of vesicular stomatitis virus. Finally, in an integrative attempt, we discuss pros and cons of the quasispecies theory compared with classic population genetics models for haploid organisms to explain the evolution of RNA viruses.
RNA Viruses: Biological and Population Properties
Despite their great functional and structural diversity, all RNA viruses share the following properties (1): (i) Cell–virus junction is mediated by means of specific membrane receptors. (ii) A viral particle penetrates the cell, loses its capsid, and releases its nucleic acids within the cell. (iii) The replication of the viral genome is regulated by the expression of viral genes (i.e., RNA replicase is encoded by the virus genome). (iv) The component parts of the viruses are assembled and released as virions out of the cell. In addition, these properties are complemented with four others that are relevant to understanding the evolution of RNA viruses. (v) The number of viral particles in a given infected organism may be as high as 1012 (1). Such population sizes, several orders of magnitude larger than any population size for DNA-based organisms, are related to viral generation time. (vi) In fact, a single infectious particle can produce, on average, 100,000 copies in 10 h (1). If the replication machinery is working optimally, a new RNA genome is produced every 0.4 s. (vii) Genome sizes range between 3 and 30 kb. Accordingly, the number of genes per genomes is also very small. (viii) Finally, RNA viruses show extremely high mutation rates (2). Because of the lack of proofreading by their replicases, RNA viruses show the highest mutation rates among living beings (2), on the order of one mutation per genome and replication round.
The above-mentioned properties of large population size, high replication rate, and short generation time are responsible, in general, for the extremely high genetic variability of RNA viral populations. Recombination and segmentation also may play an important role in generating new genetic variability (1). In any case, the extent of genetic variability per generation time of any RNA virus is usually much higher than that corresponding to any DNA-based organism, providing an excellent opportunity for studying ongoing evolution in accessible terms for human observers.
RNA Viruses Meet the Population Genetics Theory of Evolution: Theoretical Background
According to population genetics, evolution is the change in the genetic properties of populations. Changes considered to be evolutionarily relevant are those inherited via the genetic material. Population genetics, in a more formal sense, is the study of those variables that are responsible for changes in the frequency of alleles in populations. In essence, the theory is reductionistic because it makes simplified assumptions on the transformation laws, especially those related to development, that are operating within and between genotypic and phenotypic spaces (3). Such simplifications allow the estimation of allele frequencies of a given generation as a function (probabilistic and/or deterministic) of frequencies from previous generations, as well as a set of state variables, mostly including mutation, selection, migration, and random drift. The vast majority of these transformation laws are not considered by population genetics or, if considered, they are incorporated as linear transformations, i.e., the genes of the genotypic space map linearly on the phenotypic space. When applied, for instance, to a phenotypic trait such as fitness, linearity means that a change in a gene promotes a certain fitness change, and that the genotype is an array of independent units contributing to the fitness in an additive way. Mendelian laws are the only transformation laws formally incorporated to the core of the population genetics theory (3).
It seems likely that populations of organisms governed by simple transformation laws (specially epigenetic) will meet the theoretical predictions of population genetics much better than those organisms governed by unknown, but probably very complex, transformation laws. Therefore, RNA viruses should better meet the theoretical predictions of population genetics. As the number of their genes is small, the type and number of epistatic interactions among their products should be of minor relevance compared with organisms with larger genomes (4). Thus, epistasis is expected to be of minor importance in simple-genome RNA viruses (4).
The environment of an RNA virus has several components, all of which could have different effects on its adaptive process. The closest environmental component is formed by cytoplasmic components of the infected cell, but intercellular spaces within tissues, tissues within individual hosts, and the ecological environment where host species are living are other components that modulate the adaptive response of RNA viruses. The theoretical framework to understand the dynamics of haploid organisms can be found in a series of classic papers that appeared many years ago (5–20). These studies, together with more recent statistical procedures for testing the presence of positive Darwinian selection or neutrality at the nucleotide level (21–23), constitute the main body of population genetics to account for the dynamics of RNA viral populations.
Experimental Virus Model and Fitness Assays
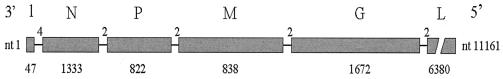
Vesicular stomatitis virus (VSV) is the prototype of the well-defined Rhabdoviridae family. It has a wide host range of vertebrates and arthropods. This virus is identified by its elongated bullet-shaped form of approximately 180 × 70 nm in size, with a nucleocapsid covered by a lipid-rich envelope. The RNA of the virion is complementary in its sequence to the mRNA for the viral proteins. The Rhabdoviridae are the simplest of the so-called minus-strand viruses. The VSV genome contains approximately 11.2 kb and transcribes into five mRNAs coding for five proteins (Fig. 1). All of the studies described here are in vitro experiments with VSV, in which viruses were grown in different types of cell cultures depending on the experiments, although BHK cells (from baby hamster kidney) were the usual ones.
Figure 1.
Schematic representation of the VSV genome, with its nontranscribed leader (l), and five consecutively transcribed mRNAs (gray blocks). Each letter refers to the proteins. From left to right: N, nucleocapsid; P, phosphoprotein; M, matrix; G, glycoprotein; L, viral replicase. Numbers below each block and above lines are lengths in nts of the genes and nontranscribed sequence, respectively.
The study of fitness effects of state variables and/or experimental regimes on viral populations is a three-step process. First, before any experimental treatment, relative fitness assays of two VSV competing clones (stock clones) were carried out as described in the third step. The first competing clone was a surrogate wild type, and the second was one of the following four different resistant to monoclonal antibodies mutant (MARM) clones. MARM C is an approximately neutral variant (with fitness 1.02 ± 0.03 relative to wild type) that contains an Asp259 → Ala substitution in the surface glycoprotein (G, Fig. 1). This amino acid substitution allows the mutant to replicate under I1 mAb concentration levels that neutralize the wild-type clone (24). MARM R clone was isolated after repeated plaque-to-plaque transfers of MARM C and showed a lower fitness than the parental virus (0.87 ± 0.05). The I1 MAb phenotype of MARM X is conferred by an Asp257 → Val substitution in the G protein. It has a much higher fitness (2.52 ± 0.16) relative to the wild type, acquired after 61 consecutive transfers of large virus populations on BHK cells. Finally, MARM F is an extremely low fitness clone (0.00015 ± 0.00001) obtained after 20 plaque-to-plaque transfers from MARM X. MARM C and X originally were isolated from the wild-type virus by picking spontaneous I1 mAb-resistant clones (24). Therefore, they were isogenic with wild type with the only exception of the above mutations responsible for the resistance.
The second step corresponds with the experiment itself (see next four sections), normally carried out with one of the four MARM clones, and in which viral populations experienced different demographic regimes and environmental conditions.
Third, fitness of the evolved viral populations was evaluated by competition assays with the ancestral wild-type clone in the following form. The evolved MARM population was mixed with a known amount of the wild-type clone. A differential quantitation of MARM clone, compared with the total virus, was done by parallel plating of the virus with and without I1 mAb. These virus mixtures then were used to initiate replicate serial competition passages. After each competition passage, the resulting virus mixture was 104-fold diluted and used to initiate the next competition transfer by infection of a fresh monolayer. The number of competition passages varied between two and a maximum of five, depending on the speed with which one competitor displaced the other. The antilogarithm of the slope of the regression ln[pt/(1 − pt)] = ln[p0/(1 − p0)] + tlnW̄ is taken as an estimate of the mean fitness of the corresponding MARM population relative to the wild type, where pt and 1 − pt are the proportions at passage number t of MARM and wild type, respectively (25–28).
The Dynamics of Deleterious Mutations in Finite Populations
When finite populations with high mutation rates are considered, a significant proportion of the mutants should be deleterious. If populations are asexual and small in size, mutation-free individuals become rare and can be lost by random genetic drift. In that case a kind of irreversible ratchet mechanism gradually will decrease the mean fitness of the populations (19). Chao (29) provided the first experimental evidence for the action of Muller's ratchet in RNA viruses. As can be observed in Table 1, there is a common pattern of fitness decline, but the magnitude of decline strongly depended on the virus studied. For instance, in the case of VSV (25, 26, 30, 31) we performed genetic bottleneck passages (plaque-to-plaque transfers) and quantified the relative fitness of bottlenecked clones by allowing direct competition in mixed infections as described above. We documented variable fitness drops after 20 or more plaque-to-plaque transfers of VSV. The relevance of these findings for evolutionary biology is clear: whenever bottlenecks occur, fitness decreases.
Table 1.
Percentage of fitness decline, with respect to the corresponding initial viral clone, in experiments with different RNA viruses subjected to a different number and continuous bottleneck transfers
| Virus | No. of bottleneck passages | % of fitness
decrease
|
Reference | |
|---|---|---|---|---|
| Average | Range | |||
| φ6 | 40 | 22 | 12–33 | 29 |
| VSV | 20 | 18 | 0.1→99 | 25, 26 30, 31 |
| FMDV | 30 | 60 | 14→99 | 32 |
| HIV-1 | 15 | 94 | 89–99 | 33 |
| MS2 | 20 | 17 | 16–18 | 34 |
FMDV, foot-and-mouth disease virus.
In relation to the accumulation of deleterious mutations we carried out two additional types of studies. First, we explored which is the model that better accounts for the distribution of deleterious mutational effects. Second, we addressed the question of how large the effective population size should be to overcome the Muller's ratchet effect.
After a series of independent experiments of plaque-to-plaque passages of approximately 120 generations for each of the three VSV mutant clones (MARM C, MARM R, and MARM X, with 24, 16, and nine independent mutation-accumulation lines, respectively), relative mean fitness was estimated and the nature of its distribution was studied (35). Because fitness effects were not normally distributed, we fitted the observed distribution to alternative models. Table 2 shows the basic statistics of the fitting of three different models. The first model tested was the negative exponential (36). The only parameter of this model, α, is the inverse of the expected fitness (1/W̄). As can be observed in Table 2, although significant to each mutant data set, R2 values are not good. The model shows the property that mutations with small effects are more common than mutations with larger effects. The second model tested was the gamma distribution (37). This model has two parameters, α and β, related with fitness as: W̄ = α/β. This model fits significantly better to the data than the negative exponential (35). However, it still fails to explain those cases with large fitness effects for MARM X and MARM C mutants (Table 2; see ref. 35 for more details). For a better description of larger deleterious effects, we considered a third model, which is a linear combination of a uniform and a gamma distribution. In this model a fraction p of the mutants are drawn from a uniform distribution and 1 − p from a gamma distribution (35, 38). MARM X and MARM C fit better than the gamma model. The fit for MARM R is not better than that obtained with the gamma model, but this could be caused by the low number of replicates for this clone.
Table 2.
Fitting the deleterious fitness effects distribution of MARM X, C, and R to three theoretical models
| MARM | Mean fitness | Model | Predicted fitness | Parameters
|
P* | R2 | ||
|---|---|---|---|---|---|---|---|---|
| α | β | p | ||||||
| X | 0.43 ± 0.28 | Exponential | 0.49 | 2.05 ± 0.24 | — | — | 0.74 | |
| Gamma | 0.40 | 48.23 ± 17.86 | 19.39 ± 7.19 | — | 0.0001 | 0.92 | ||
| Compound | 0.44 | 400.05 ± 245.91 | 158.32 ± 96.89 | 0.41 ± 0.08 | 0.0031 | 0.96 | ||
| C | 0.74 ± 0.26 | Exponential | 0.93 | 1.08 ± 0.12 | — | — | 0.60 | |
| Gamma | 0.82 | 38.28 ± 11.22 | 31.23 ± 9.28 | — | <0.0001 | 0.89 | ||
| Compound | 0.72 | 168.78 ± 25.25 | 147.97 ± 22.33 | 0.42 ± 0.02 | <0.0001 | 0.99 | ||
| R | 0.70 ± 0.28 | Exponential | 0.78 | 1.28 ± 0.20 | — | — | 0.73 | |
| Gamma | 0.67 | 5.90 ± 1.68 | 3.97 ± 1.07 | — | 0.0021 | 0.94 | ||
| Compound | 0.66 | 6.72 ± 3.96 | 5.25 ± 3.64 | 0.44 ± 0.37 | 0.3510 | 0.95 | ||
See text for more details.
P, significance level for the partial F-test used for testing the reduction in the error sum of squares for models with an increasing number of parameters.
The distribution of deleterious effects, on the other hand, strongly depended on which one of the three VSV mutant clones was used in the experiment. Table 3 shows E(ΔW), the mean fitness reduction per generation, for the three MARM clones. They are statistically different from zero and proportional to the initial fitness (27). Applying Mukai–Bateman's method of a constant distribution of mutational fitness effects (35), we obtained (see Table 3) the estimates of U, the deleterious mutation rate per genome and generation, and E(s), the expected selection coefficient against deleterious mutations. The average mean fitness reduction for the entire data set was −0.26% per generation, which is compatible with the idea that fitness was reduced by the accumulation of many mutations (U ≈1) of small effect (−0.1% each). In other words, the huge decline in fitness reported above was the consequence of the accumulation of many mutations of small effect. The effect of deleterious mutations on long-term survival of viral population should be greater if we consider large numbers of mutations of small average effect compared with few changes of large effect because, in the latter case, selection should be more efficient in purifying deleterious load.
Table 3.
Mean estimates, with standard error, of E (ΔW), the mean fitness change per generation, as well as U, the deleterious mutation rate and, E(s), its average effect under a constant model of effect distribution
| MARM | E(ΔW) | U | E(s) |
|---|---|---|---|
| X | −0.00311 ± 0.00020 | 1.5753 ± 0.5676 | −0.0022 ± 0.0008 |
| C | −0.00210 ± 0.00004 | 0.8839 ± 0.0541 | −0.0024 ± 0.0001 |
| R | −0.00252 ± 0.00004 | 1.1610 ± 0.0519 | −0.0022 ± 0.0001 |
The population size necessary to stop Muller's ratchet not only depended on the demographic experimental regimes of the viral populations, but also on the genetic composition and the fitness of the initial populations. In a series of bottleneck experiments performed with MARM X, MARM C, MARM U, and MARM N populations (for biological properties of the last two mutants not mentioned in this review see ref. 39), initial fitness and fitness after 20 2-to-2, 5-to-5, and 30-to-30 clone passages was estimated. Table 4 gives a summary of the results obtained. As can be observed, after 20 2-to-2 clone passages the initial low-fitness MARM N clones showed slight fitness changes. When the virus transmission rate was raised to five, fitness gain was observed in the six experiments performed. The neutral MARM C and U mutants, with passage transmission rates of five, gave similar mean fitness results to the MARM N mutant at a transmission rate of two (i.e., no significant change in fitness). Finally, the high fitness MARM X clone showed no fitness changes with a transmission rate of 30, but fitness decrease at a transmission rate of five clones (39). The new fitness values were similar after 20 more passages with 5-to-5 plaque transfers in MARM X, MARM U, and MARM N clones (40). The results obtained can be explained by the sampling of infectious particles of lower, equal, or greater fitness than the average fitness showed by the original nonbottlenecked population (see Table 4). Mutants of equal or greater fitness should be sampled more frequently from populations with low fitness, because highly fit populations have lower probability of undergoing further mutational improvement. One discrepancy with this explanation was observed when similar experiments were done with the extremely debilitated MARM F clone (28). We did not expect any fitness recovery after 20 single plaque-to-plaque (1-to-1) passages, but we observed significant fitness recoveries, ranging from 0.11 to 0.94 (28). A partial explanation for this may be that competition is taking place between variants within a single plaque. In the context of a highly mutable population, a beneficial mutation that confers faster replication may occur during the growth of a single plaque. Then, the probability of picking this fitter clone from the plaque is very high because of its higher frequency.
Table 4.
Mean fitness, with standard error (based on six different experiments), after different population dynamics for different MARM clonal populations
| MARM | Fitness before passage | Passage dynamics | Mean fitness after passage |
|---|---|---|---|
| X | 3.05 ± 0.03 | 5-to-5 | 1.70 ± 0.03*** |
| 30-to-30 | 3.00 ± 0.40ns | ||
| U | 1.00 ± 0.20 | 5-to-5 | 1.30 ± 0.20ns |
| C | 0.91 ± 0.03 | 5-to-5 | 1.20 ± 0.20ns |
| N | 0.38 ± 0.01 | 2-to-2 | 0.38 ± 0.01ns |
| 5-to-5 | 0.55 ± 0.05** |
ns, not significant; **, P < 0.01; ***, P < 0.001.
Adaptation: Competition in Constant, Changing, and Subdivided Environments
Until now, we have shown experimental evidence for the accumulation of many deleterious mutations during the evolution of RNA viral populations, especially those subjected to strong bottlenecking, a common feature of natural virus populations. But from time to time, the appearance of mutations with beneficial effect on fitness cannot be ruled out. This, and the next two sections, will provide experimental evidence for the presence of such mutations and its effect on the long-term evolution of RNA viruses. All of the topics treated can be reduced to the following two: (i) is there evolution of fitness in competing populations under different environmental situations, and (ii) is there any limitation to the rate of viral adaptation?
In population biology, the competitive exclusion principle (41) states that in the absence of niche differentiation, one competing species always will eliminate or exclude the other. The very high error frequency during RNA genome replication and, hence, its rapid evolution, may render the prolonged coexistence of two (or more) genetically distinct viral populations unlikely. Different genomes that may have different replication abilities and/or encapsidation rates are constantly being generated. Then, the appearance of one or more mutations in one of the competing populations may confer enough selective advantage to its carrier as to facilitate its fixation. However, Clarke et al. (42) showed that two viral competing populations of approximately initial equal fitness could coexist through numerous generations during prolonged replication in a constant seasonal environment. A mechanism that may explain the long preservation of both populations is frequency-dependent selection. Evidence of that also has been obtained (43). The biological scenario where frequency-dependent selection might be present is as follows: low-frequency genotype faces a new and sparsely populated biological niche, whereas the niche of the most common genotype may approach saturation. This scenario requires certain niche heterogeneity. For the cell–culture environment we can imagine factors like age, density stage of the cellular cycle, location, etc. It is then possible that each viral genotype is using this smooth variability in a constant environment in different ways.
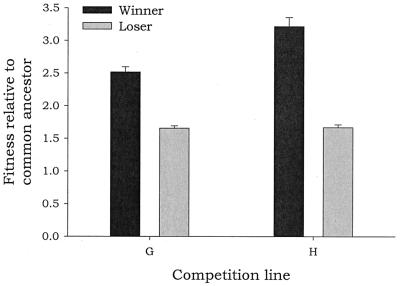
To explain the relevance of competition in evolution, Van Valen (44) proposed the Red Queen's hypothesis according to which each species is competing in a zero-sum game against others; each game is a dynamic equilibrium between competing species where, although both are increasingly adapted, no species can ever win. This is exactly what we observed (42). Fig. 2 shows the relative fitness estimates of both winners and losers of two competition series (H and G) compared with the fitness of the initial competing populations. As can be observed, both winners and losers show a higher fitness than the initial ones.
Figure 2.
Relative fitness to the ancestral populations of winners and losers in long-term competition experiments (G and H). Initial populations were neutral. Modified from ref. 42.
The process of adaptation cannot continue forever for a constant environment. Once the virus becomes adapted to the new environment, no further improvement is expected unless the environment changes. However, what type of trajectory is followed by fitness during the adaptation of a viral population? In a study of the fitness recovery of the MARM F clone under different demographic regimes (28) we observed that, for the case of large and consecutive population expansions, a maximum fitness value close to 1 was reached after few passages in four independent experiments. Moreover, the rate of fitness increases slowed down. Although the issue is still controversial, a hyperbolic function on a logarithmic scale [lnW̄t = lnW̄0 + at/(b + t), where t is the passage number, a is the asymptotic log-fitness value, and b is the passage number at which the log fitness is equal to half the maximum value, seems to provide a good fit to the observed trajectories (Table 5). It catches some relevant properties of the fitness trajectories. The model has two main properties: mean fitness evolves rapidly during the first passages and asymptotically approaches a maximum value. The reason for the deceleration in the rate of adaptation has to do with the availability of beneficial mutations and the magnitude associated with each available mutation. At the beginning of the process, when the viral population is far from the optimum, any beneficial mutation will increase fitness by driving it upward in a fitness landscape (45). However, as the population approaches the fitness peak, only fine-tuning mutations, less abundant than the former, will be needed. Once the virus becomes adapted to its new environment, no further improvement is expected unless the environment changes.
Table 5.
Estimated parameters for the hyperbolic model of fitness, with standard errors, in four independent experiments of fitness recovery of MARM F clone during 40 large and consecutive population expansions
| Replicate | ln W̄0 | a | b | R2 | W̄ |
|---|---|---|---|---|---|
| I | −9.3 ± 0.7 | 9.3 ± 0.8 | 0.4 ± 0.2 | 0.95 | 1.1 ± 0.0 |
| II | −7.9 ± 0.6 | 7.9 ± 0.7 | 0.3 ± 0.1 | 0.95 | 1.2 ± 0.1 |
| III | −9.0 ± 0.6 | 9.3 ± 0.8 | 0.8 ± 0.3 | 0.99 | 1.2 ± 0.2 |
| IV | −10.0 ± 0.3 | 10.2 ± 0.4 | 1.2 ± 0.2 | 1.00 | 1.0 ± 0.1 |
Last column corresponds to fitness estimates at passage 40. For more details see text and ref. 28.
The issue regarding fitness changes also should be considered when viruses shift among environments. Three types of in vitro experiments have been done in relation to environmental changes. First, experiments of adaptation of VSV to new hosts (46, 47) has been done. VSV clones, previously adapted to BHK cells, gained fitness after few passages on alternative new host cells. It is controversial, however, if VSV also concurrently increased fitness on ancestral BHK cells. This question needs to be addressed under appropriate and more replicated experiments. Second, the fitness changes associated with adaptation to fluctuating environments also were explored. Experiments have been done with VSV and Eastern equine encephalitis virus (47, 48), where viruses were grown in two new cell types, changing daily. Both viruses were selected with increased fitness in both novel environments. Adaptation seems, however, to be reversible. VSV clones adapted to persistently replicate in LL5 insect cells showed low fitness on BHK cells and in mouse brain (49), but attenuated virus soon recovered fitness and virulence after a few passages back in BHK. Third, in vitro experiments were performed with antiviral agents added to the media. When cells were treated with α-interferon, the size of VSV viral populations experienced a dramatic reduction (99.9%). Those viruses that developed α-interferon resistance quickly increased their fitness in the presence of the interferon (50). However, fitness decreased when interferon was removed from the media.
Another question that could be of great importance to viral populations is its eventual differentiation into subpopulations (for instance different organs or tissues within an individual), and then subsequent migration among subpopulations. We have experimentally modeled this situation by means of an in vitro system to simulate migration of VSV among isolated homogeneous host cell populations (see ref. 51 for a detailed description of the experimental protocol). The results clearly demonstrated a positive correlation between migration rate and the magnitude of the mean fitness across subpopulations and, with less support, a decrease in the fitness difference among subpopulations with the magnitude of the migration. The results, in full agreement with population genetics theory, can be explained by the spread of beneficial mutations, originated in single isolated populations, through the entire population.
Clonal Interference Imposes a Limit on the Rate of Virus Adaptation
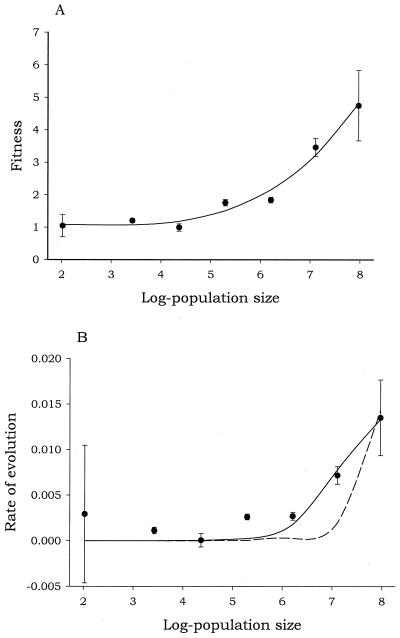
Viral populations adapt through the appearance and fixation of beneficial mutations. In large asexual populations such as of VSV, where mutation rate and population size are the key state variables, beneficial mutations may arise frequently enough that two or more are coexisting. When this happens, among other properties of the clonal interference model (52), we expect, for increasing population sizes, that (i) the fitness effect associated with a fixed beneficial mutation will be larger, and (ii) the rate of adaptation will tend toward a limit. These two predictions were demonstrated to hold in experimental populations of VSV (53). As already described, we carried out experiments with two competing populations of initially similar fitness (i.e., MARM C and wild type). Experiments were done at seven (five times replicated) different effective population sizes ranging from 100 to 108 viral particles. Once one of two variants became fixed, we measured its fitness relative to its nonevolved counterpart. The major results are summarized in Fig. 3. The larger the population size, the bigger the effect associated with a beneficial mutation that becomes fixed in the population (Fig. 3A). In addition (Fig. 3B), the rate of adaptation slows down with increasing effective population size as a consequence of longer times required for fixation of beneficial mutations: in other words, the winning clone must outblock more alternative less-beneficial genotypes.
Figure 3.
(A) Influence of effective population size on the fitness effect of beneficial mutations that are fixed. The model of the fitted line is described in ref. 53. (B) Effect of the effective population size on the rate of evolution. See text for more details. The dashed line corresponds to the fit of a liner model to the data. The solid line is the fit to a hyperbolic model. Both curves appear to be exponential because of the logarithmic scale of x axis.
From data shown in Fig. 3 it is possible to estimate the rate at which beneficial mutations are generated, as well as its average effect on fitness (52). The estimated value for the beneficial mutation rate (53) was 6.4 × 10−8 beneficial mutations per genome and generation. The effective population sizes studied, all higher than 109, warranted the possibility of appearance of a beneficial mutation, giving support to the assumption that a single beneficial mutation in each lineage is responsible for the fitness increase (Fig. 3). Comparing our estimate with that of the total mutation rate in VSV (2–3.5 substitutions per genome and generation), we can infer that one of 2 × 108 mutations produced in VSV can be considered as beneficial. On the other hand, the maximum-likelihood estimate of the mean selective advantage of all beneficial mutations produced in the population (see ref. 53 for details) was 31% per day. Most mutations have small deleterious effect on fitness, whereas the few beneficial ones that appear in the population will have a large effect on it.
Nucleotide Diversity and Fitness Recovery in the Evolution of a Highly Debilitated VSV Experimental Population: The Sampling Problem
As mentioned above, we have studied fitness recovery in the evolution of a highly sick VSV viral population clone (MARM F) obtained from 20 consecutive plaque-to-plaque passages (28). The MARM F clone was diluted and plated on a monolayer of BHK cells, and four well-isolated plaques were collected. Fitness of each subclone was estimated and the relative mean fitness was 0.00015 relative to wild type (28). After a few large population passages (ref. 28, regime E), the four subclones recovered fitness several orders of magnitude, and by passage 40 it reached a mean value of 1.1 ± 0.0, not significantly different from the wild type. Then, we estimated fitness recovery in one of the subclones at three more times (after 28 h, 2 days, and 5 days of large consecutive passages) (Table 6). In addition, two VSV genome regions, representing ≈10% of the VSV genome were sequenced. One of them, 514 nt long, includes part of the G gene, and the other one, 510 nt long, comprises part of the P and M genes (see Fig. 1), including a 56-nt noncoding region of the P gene. This noncoding region contains a highly conserved sequence of 11 nt (3′-AUACUUUUUUU-5′) in the vesiculovirus genus, which is involved in the synthesis of the poly(A) tail during transcription process (54). The poly(U) tract of this sequence suffers frequent insertions and deletions that could involve a fitness reduction. Thus, in VSV, removal of a single U residue abolished the synthesis of the monocistronic upstream transcript completely (54). A less dramatic effect showed the addition of U residues to the VSV signal. We sequenced 20 clones of each region from isolates taken at six time points of the experiment: 0 h (the original subclone), 28 h, 2 days, 5 days, 8 days, and after 40 days of daily mass expansion (28, 55). Some of the results obtained are shown in Table 6. As can be observed, the number of synonymous and nonsynonymous polymorphic sites was low for both regions at any time. These numbers, as well as nucleotide diversity, were expected to increase from 0 h to 40 days. They were derived from a single plaque with an extremely low initial fitness. Such tendency was observed only when comparing the sample taken at day 40 with any of the other five moments for both regions (average between sampled corrected distances of 0.0024 ± 0.0003 for region G, and 0.0037 ± 0.0004 for regions M–P). However, the low numbers of mutations detected in this study forced us to consider two key points related with the estimation of genetic variability of RNA viral populations and its relationship with fitness change. First, how correct are the available estimates of the mutation rate for VSV of 10−3 to 10−4 substitutions per nucleotide and per replication round (2)? If that figure is valid, then how large should both nucleotide and population samples be to detect a significant proportion of its genetic variability? It is expected that the fitness recovery of an expanding population of a highly debilitated clone should be characterized at the molecular level by repeated nonsynonymous substitution fixation, a pattern that has not been observed in the regions studied. If we consider that the estimated number of beneficial mutations in VSV expanding populations is low, then it might be possible that the two regions selected have not been the appropriate ones. This statement points toward a much higher nucleotide sampling. However, as can be seen in Table 6, there is a reduction of the frequency of VSV molecules showing indels in the U-stretch of the intergenic region. As mentioned above, indels in this region have an effect on the levels of transcription of the adjacent genes and can be responsible for the fitness reduction of the MARM F clone. A recent study (56) on the evolution of highly debilitated foot-and-mouth disease virus clones under continuous population expansions give support to this idea. Escarmís et al. (56) observed that the original debilitated clone had six-point mutations spread over the genome in addition to an elongated internal polyadenylate tract immediately preceding the second initiation AUG codon (32). The point mutations were replaced and the polyadenylate tract disappeared after a large number of passages (56).
Table 6.
Mean fitness, with standard error, number of polymorphic, nonsynonymous (NS) and synonymous (S) sites as well as nucleotide diversity, π (estimated as number of substitutions per site), with standard error, of regions G (510 nt) and M-P (514 nt)
| Time | Fitness | G region
|
M-P
region
|
Indels | ||||
|---|---|---|---|---|---|---|---|---|
| NS | S | π (× 103) | NS | S | π (× 103) | |||
| 0 h | 0.00041 ± 0.00004 | 0 | 0 | 0.00 ± 0.00 | 0 | 0 | 0.00 ± 0.00 | 0(14), +1(4), +2(1), −1(1) |
| 28 h | 0.22 ± 0.08 | 0 | 0 | 0.00 ± 0.00 | 0 | 1 | 0.20 ± 0.32 | 0(17), +1(3) |
| 2 d | 0.22 ± 0.07 | 0 | 0 | 0.00 ± 0.00 | 2 | 0 | 0.39 ± 0.24 | 0(14), +1(4), +2(1), +13(1) |
| 5 d | — | 0 | 0 | 0.00 ± 0.00 | 2 | 0 | 0.39 ± 0.24 | 0(19), +2(1) |
| 8 d | 0.40 ± 0.10 | 1 | 0 | 0.20 ± 0.30 | 0 | 0 | 0.00 ± 0.00 | 0(19), +6(1) |
| 40 d | 1.10 ± 0.00 | 3 | 2 | 2.12 ± 0.32 | 1 | 1 | 0.39 ± 0.24 | 0(19), +2(1) |
In the studied M-P noncoding region of the VSV genome there is a highly conserved stretch of seven Us (see text). Indel distribution corresponds to the number (between parenthesis) of zero, added (+), or deleted (−) Us found in a 20-clone sample. The values have been obtained in six time points of an experiment of fitness recovery of a highly debilitated MARM clone.
Quasispecies and Population Genetics Theories of the Evolution of RNA Viruses
There is abundant theoretical literature related to the dynamics of populations in which mutation is a frequent event. This is the case of Eigen and Schuster's notion of quasispecies (57) where the target of selection is no longer a single fittest genotype but rather a cloud of mutants distributed around a most frequent one quoted as master sequence. Following those authors, population genetics theory is a good descriptor for those populations where mutations are rare events and purifying selection is the main evolutionary force generating a homogeneous population, in addition to possible neutral variation fixed by random drift (58). They also suggested that the quaspecies theory is able to handle all situations, from small to large mutation rates. If true, then we have two different theories with different explanatory power, formally being the quasispecies theory of a wider application range than population genetics, in a way similar to the statement that general relativity is more general than Newton's mechanics. However, the theoretical models of population genetics are not compromised at all by the assumption of small mutation rates, as it can be appreciated when studying any formal presentation of the theory, and even much less when considering the evolution of haploid organisms (5–20). Consequently, in terms of mutation rates we have at least two competing theories of similar application range.
One important issue of the quasispecies theory is the notion that the target of selection is not just the fastest growing (i.e., the fittest) replicator, but a broad spectrum of mutants produced by erroneous copying of the original sequence. Nowak (59) and Eigen (60) defined the quasispecies in precise mathematical terms as the “dominant eigenvector which belongs to the largest eigenvalue of the replication matrix.” This replication matrix contains the replication rates (i.e., selective values) of each mutant class, as well as mutation probabilities. However, because neutral mutants are not considered and population size is infinite, genetic drift formally seems not to operate in the quasispecies framework. The dynamics of a population of infinite size under continuous selection pressure will be determined by selective advantage of the fastest replicating variant. However, the frequency of a given mutant also depends on the probability with which it is generated by mutation from other closely related variants and their frequencies. The consequence of this scenario (59) is that the individual sequence no longer serves as the unit (or target) of selection. On the contrary, the entire quasispecies should be this target.
The thesis of the present work is that both population genetics and quasispecies theories, when applied to the evolution of RNA viral populations, might have different explanatory power and/or application range. Historically, they represent two research traditions, and both have their own theoretical tools to explain the evolutionary dynamics of highly mutating populations of infinite size in which genetic drift is not present and mutants have always different selection coefficients. However, population genetics theory (i) does not require selective imposition of the fittest genotype, as the proponents of quasispecies theory seem to suggest, and (ii) formally contemplates the action of random drift and eventual fixation of some neutral mutants.
Most of the evidence giving support to the quasispecies theory comes from RNA viruses because they show high mutation rates and reach very high population numbers in a short time. In fact, virus populations normally consist of a widely dispersed mutant distribution rather than a homogeneous one formed by a single, most-fit, wild-type sequence (60). However, the realization of high levels of genetic variability in viral populations does not constitute per se an observation giving exclusive support to the quasispecies theory or to the theory of population genetics. A pattern of extremely high genetic variability also can be derived from a model exclusively based on mutation and selection on single replicons, a result derivable from the theoretical background on haploid organisms (5–20).
One central issue that could discriminate between both theories is related to the unit of selection of highly mutating replicons. According to the quasispecies theory, as previously stated, the unit is not the single replicon but a set of related mutant sequences, whose degree of kinship can be expressed by a genetic distance. Such a set of closely related sequences forms an evolving cloud that has a higher evolutionary (i.e., adaptive) plasticity than an evolving population exclusively based on independent replicons. Population genetics is not a theory based exclusively on the idea that the unit of selection is the individual. Group or kin selection models are relevant components of the population genetics theory. Consider, for instance, the evolution of virulence in parasite organisms. Explanatory hypotheses have been advanced that are not only based on tradeoff at the individual level between virulence and transmission rate (for a review see ref. 61), but also on group selection (62).
There is an agreement between quasispecies theory and group selection models, because the cloud or distribution of closely related mutants fulfills the three conditions that the latter imposed to consider a group as the target of selection. First, we are dealing with replicative entities whose population structure promotes fast genetic divergence (63). Second, the members of the quasispecies are intimately related (64). Third, the whole distribution of mutants should be considered as an individual instead of a group (64). In summary, objections and explanatory power of both theories can be exchanged. Once stated the formal equivalence between quasispecies theory and group selection models, the major problem to be solved is to gain experimental evidence in favor of a supra individual unit of selection. However, few experimental or field observations have been reported, and the issue is still highly controversial.
Acknowledgments
We thank Prof. Amparo Latorre for critical reading and suggestions to improve the manuscript. This work has been supported by Grant PM97-0060-C02-02 from the Spanish Dirección General de Enseñanza Superior. A.B. and R.M. also have been recipients of fellowships from the Ministerio de Educación y Ciencia.
Abbreviations
- VSV
vesicular stomatitis virus
- BHK
baby hamster kidney
- MARM
monoclonal antibodies mutant
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 2.Drake J W, Holland J J. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewontin R C. The Genetic Basis of Evolutionary Change. New York: Columbia Univ. Press; 1974. [Google Scholar]
- 4.Elena S F. J Mol Evol. 1999;49:703–707. doi: 10.1007/pl00000082. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein J. Theor Pop Biol. 1971;2:391–403. doi: 10.1016/0040-5809(71)90028-1. [DOI] [PubMed] [Google Scholar]
- 6.Cook R D, Nassar R F. Biometrics. 1972;28:373–384. [PubMed] [Google Scholar]
- 7.Cannings C. Adv Appl Prob. 1974;7:4–5. [Google Scholar]
- 8.Cannings C. Adv Appl Prob. 1975;7:264–282. [Google Scholar]
- 9.Drobník J, Dlouhá J. J Theor Biol. 1966;11:418–435. doi: 10.1016/0022-5193(66)90102-0. [DOI] [PubMed] [Google Scholar]
- 10.Trajstman A C. Biometrics. 1973;29:701–711. [PubMed] [Google Scholar]
- 11.Moran P A P. Camb Philos. 1957;54:463–467. [Google Scholar]
- 12.Karlin S, McGregor J. Theor Pop Biol. 1971;2:60–70. doi: 10.1016/0040-5809(71)90005-0. [DOI] [PubMed] [Google Scholar]
- 13.Emigh T H. Genetics. 1979;92:323–337. doi: 10.1093/genetics/92.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emigh T H. Genetics. 1979;92:339–351. doi: 10.1093/genetics/92.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie J H. Genet Res. 1973;21:115–120. [Google Scholar]
- 16.Gladstein K. Theor Pop Biol. 1976;10:383–394. doi: 10.1016/0040-5809(76)90025-3. [DOI] [PubMed] [Google Scholar]
- 17.Robson D S. Genetics. 1957;42:487–498. doi: 10.1093/genetics/42.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strobeck C. Am Nat. 1979;113:439–444. [Google Scholar]
- 19.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 20.Haigh J. Theor Pop Biol. 1978;14:251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- 21.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 22.Gillespie J H. The Causes of Molecular Evolution. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 23.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 24.VandePol S B, Lefrancois L, Holland J J. Virology. 1986;148:312–325. doi: 10.1016/0042-6822(86)90328-4. [DOI] [PubMed] [Google Scholar]
- 25.Duarte E A, Clarke D K, Moya A, Domingo E, Holland J J. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke D, Duarte E, Moya A, Elena S F, Domingo E, Holland J J. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elena S F, González-Candelas F, Novella I S, Duarte E A, Clarke D K, Domingo E, Holland J J, Moya A. Genetics. 1996;142:673–679. doi: 10.1093/genetics/142.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elena S F, Dávila M, Novella I S, Holland J J, Domingo E, Moya A. Evolution. 1998;52:309–314. doi: 10.1111/j.1558-5646.1998.tb01633.x. [DOI] [PubMed] [Google Scholar]
- 29.Chao L. Nature (London) 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 30.Duarte E A, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. J Virol. 1993;67:3620–3623. doi: 10.1128/jvi.67.6.3620-3623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duarte E A, Novella I S, Ledesma S, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. J Virol. 1994;68:4295–4301. doi: 10.1128/jvi.68.7.4295-4301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escarmís C, Dávila M, Charpentier N, Bracho A, Moya A, Domingo E. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 33.Yuste E, Sánchez-Palomino S, Casado C, Domingo E, López-Galíndez C. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De la Peña M, Elena S F, Moya A. Evolution. 2000;54:686–691. doi: 10.1554/0014-3820(2000)054[0686:eodmao]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 35.Elena S F, Moya A. J Evol Biol. 1999;12:1078–1088. [Google Scholar]
- 36.Mukai T, Chigusa S I, Mettler L E, Crow J F. Genetics. 1972;72:335–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keightley P D. Genetics. 1994;138:1315–1322. doi: 10.1093/genetics/138.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elena S F, Ekunwe L, Hajela N, Oden S A, Lenski R E. Genetica. 1998;102/103:349–358. [PubMed] [Google Scholar]
- 39.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Mol Gen Genet. 1996;252:733–738. doi: 10.1007/BF02173980. [DOI] [PubMed] [Google Scholar]
- 41.Hardin G. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 42.Clarke D K, Duarte E A, Elena S F, Moya A, Domingo E, Holland J J. Proc Natl Acad Sci USA. 1994;91:4821–4824. doi: 10.1073/pnas.91.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elena S F, Miralles R, Moya A. Evolution. 1997;51:984–987. doi: 10.1111/j.1558-5646.1997.tb03679.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Valen L. Evol Theory. 1973;1:1–30. [Google Scholar]
- 45.Orr H A. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- 46.Holland J J, de la Torre J C, Clarke D K, Duarte E A. J Virol. 1991;65:2960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novella I S, Hershey C L, Escarmis C, Domingo E, Holland J J. J Mol Biol. 1999;287:459–465. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- 48.Weaver S C, Brault A C, Kang W, Holland J J. J Virol. 1999;73:4316–4326. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novella I S, Clarke D K, Quer J, Duarte E A, Lee C H, Weaver S C, Elena S F, Moya A, Domingo E, Holland J J. J Virol. 1995;69:6805–6809. doi: 10.1128/jvi.69.11.6805-6809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novella I S, Cilnis M, Elena S F, Kohn J, Moya A, Domingo E, Holland J J. J Virol. 1996;70:6414–6417. doi: 10.1128/jvi.70.9.6414-6417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miralles R, Moya A, Elena S F. J Gen Virol. 1999;80:2051–2059. doi: 10.1099/0022-1317-80-8-2051. [DOI] [PubMed] [Google Scholar]
- 52.Gerrish P J, Lenski R E. Genetica. 1998;102/103:127–144. [PubMed] [Google Scholar]
- 53.Miralles R, Gerrish P J, Moya A, Elena S F. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- 54.Conzelmann K K. Annu Rev Genet. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 55.Bracho A, Moya A, Barrio E. J Gen Virol. 1998;79:2921–2928. doi: 10.1099/0022-1317-79-12-2921. [DOI] [PubMed] [Google Scholar]
- 56.Escarmís C, Dávila M, Domingo E. J Mol Biol. 1999;285:495–505. doi: 10.1006/jmbi.1998.2366. [DOI] [PubMed] [Google Scholar]
- 57.Eigen M, Schuster P. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 58.Nowak M, Schuster P. J Theor Biol. 1989;137:375–395. doi: 10.1016/s0022-5193(89)80036-0. [DOI] [PubMed] [Google Scholar]
- 59.Nowak M. Trends Ecol Evol. 1992;7:118–121. doi: 10.1016/0169-5347(92)90145-2. [DOI] [PubMed] [Google Scholar]
- 60.Eigen M. Trends Microbiol. 1996;4:216–218. doi: 10.1016/0966-842X(96)20011-3. [DOI] [PubMed] [Google Scholar]
- 61.Frank S. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 62.Miralles R, Moya A, Elena S F. Genet Res. 1997;69:165–172. doi: 10.1017/s0016672397002735. [DOI] [PubMed] [Google Scholar]
- 63.Wade M J. Proc Natl Acad Sci USA. 1976;73:4604–4607. doi: 10.1073/pnas.73.12.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hull D L. Annu Rev Ecol Syst. 1980;11:311–332. [Google Scholar]