SUMMARY
The indolocarbazole biosynthetic enzymes StaC, InkE, RebC, and AtmC mediate the degree of oxidation of chromopyrrolic acid on route to the natural products staurosporine, K252a, rebeccamycin, and AT2433-A1, respectively. Here we show that StaC and InkE, which mediate a net 4-electron oxidation, bind FAD with a micromolar Kd, whereas RebC and AtmC, which mediate a net 8-electron oxidation, bind FAD with a nanomolar Kd, while displaying the same FAD redox properties. We further create RebC-10x, a RebC protein with ten StaC-like amino acid substitutions outside of previously characterized FAD binding motifs and the complementary StaC-10x. We find that these mutations mediate both FAD affinity and product specificity, with RebC-10x displaying higher StaC activity than StaC itself. X-ray structures of this StaC catalyst identify the substrate of StaC as 7-carboxy-K252c and suggest a unique mechanism for this FAD-dependent enzyme.
INTRODUCTION
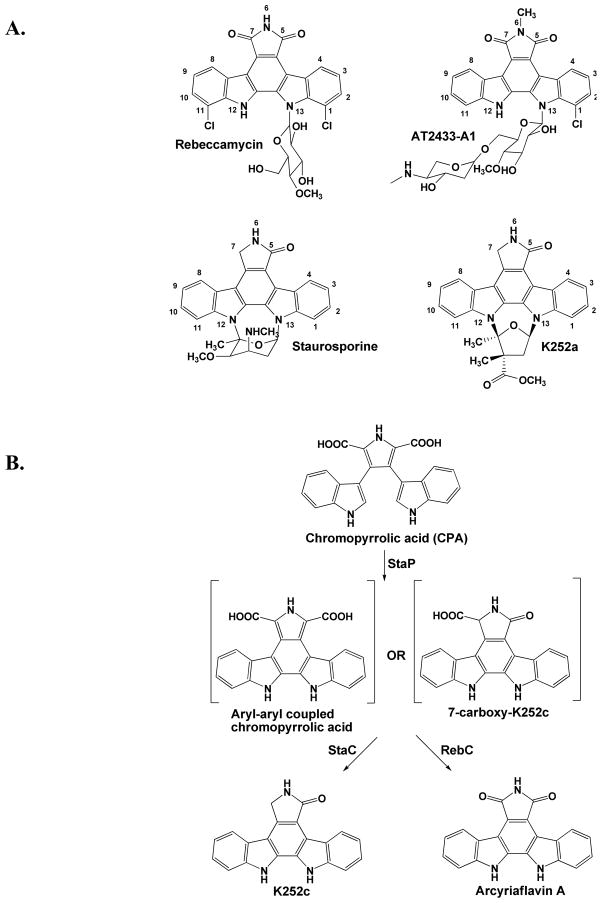
Indolocarbazoles, a subset of the L-tryptophan derived bisindole class of alkaloid natural products, include a variety of molecules of pharmaceutical interest (Drennan and Ryan, 2009). Isolated from Streptomyces and other soil- and marine-dwelling actinomycete bacteria (Jensen et al., 2007; Sánchez et al., 2006), staurosporine (Figure 1A) has no assigned native function but has proven to be a potent protein kinase inhibitor (Ruegg and Burgess, 1989) with an analog (7-hydroxy-staurosporine, also known as UCN-01) in clinical trials as an anti-cancer agent (Edelman et al., 2007; Jimeno et al., 2008; Welch et al., 2007). Rebeccamycin (Figure 1A), isolated from Lechevalieria aerocolonigenes, shares a very similar aglycone scaffold with staurosporine and an analog of this compound, becatecarin, is also in clinical trails afforded by its antitumor activity (Dowlati et al., 2009; Nock et al., 2011). However, unlike staurosporine, rebeccamycin analogs inhibit DNA replication by stabilizing DNA-topoisomerase I complexes (Bailly et al., 1997), showing the therapeutic diversity of indolocarbazole compounds. To exploit the full medicinal potential of indolocarbazoles and to generate a more diverse array of pharmaceutically active compounds, a better understanding of their biosynthetic pathways and the key branch points is desirable.
Figure 1.
Indolocarbazole natural products and reaction schemes. (A) Chemical structures of rebeccamycin, AT2433-A1, staurosporine, and K252a with standard numbering. (B) Reaction scheme for StaP/StaC mediated production of K252c from CPA, and reaction scheme for StaP/RebC mediated production of arcyriaflavin A from CPA.
The biosynthetic pathways of staurosporine, rebeccamycin, and other indolocarbazoles involve the oxidation and subsequent coupling of two tryptophan molecules to generate chromopyrrolic acid (CPA) (Figure 1B). At this step, the pathways diverge to generate the diverse aglycone scaffolds characteristic of indolocarbazoles. In the staurosporine pathway, CPA is converted to K252c in a net 4-electron oxidation, and then further tailored, whereas CPA is converted to arcyriaflavin A via a net 8-electron oxidation in the rebeccamycin biosynthetic pathway (Figure 1). Both oxidations involve two enzymes, the first of which, StaP or RebP, are cytochrome P450 enzymes that catalyze aryl-aryl coupling reactions to generate a reactive intermediate and can be used interchangeably between the two pathways (Sánchez et al., 2005). The second enzyme of each pathway, StaC or RebC, is thought to intercept this intermediate and enable its conversion to the corresponding aglycone (Howard-Jones and Walsh, 2006). For StaC, the reaction yields an asymmetric molecule with a carbonyl group at the C-5 position and a fully reduced C-7 carbon (numbering in Figure 1A), while the product of RebC is a symmetric molecule, with carbonyl groups at both the C-5 and C-7 positions. Without either StaC or RebC in the reaction mix, a variety of products are generated with varying oxidation states at the C-7 position; however, if either enzyme is included, a single product is observed (Howard-Jones and Walsh, 2006).
StaC and RebC share 65% sequence identity and both enzymes contain three motifs typical of flavin hydroxylase proteins (Figure S1) (Eppink et al., 1997; Howard-Jones and Walsh, 2006; Ryan et al., 2007). Surprisingly though, only RebC co-purifies with FAD in E. coli recombinant expression systems. Nonetheless, StaC is capable of converting FAD to FADH2 using NAD(P)H (Howard-Jones and Walsh, 2006). Two protein homologues of RebC and StaC, InkE and AtmC (Figure S1), are involved in separate indolocarbazole biosynthetic pathways. Both contain the same three motifs and the enzymes share 56% sequence identity with one another. InkE is involved in the biosynthesis of K252a (Figure 1A) (Kim et al., 2007), a molecule that has a carbonyl group at the C-5 position but a fully reduced C-7 carbon. The aglycone of K252a is likely generated through a largely similar pathway as the staurosporine aglycone, with InkE (like StaC) finalizing a net 4-electron oxidation of CPA (Figure 1B). By contrast, AtmC is involved in the biosynthesis of AT2433-A1 (Figure 1A) (Gao et al., 2006), a molecule that has carbonyl groups at the both the C-5 and C-7 positions. The aglycone of AT2433-A1 is likely generated through a highly similar pathway as the rebeccamycin aglycone, with AtmC (like RebC) mediating a net 8-electron oxidation of CPA (Figure 1B).
Here we investigate the role of FAD and the enzyme mechanism of RebC- and StaC-like biosynthetic enzymes. Using isothermal titration calorimetry (ITC) to determine FAD dissociation constants for StaC, RebC, InkE, and AtmC, we find a correlation between FAD affinity and the reaction catalyzed, with tighter FAD affinity linked with RebC/AtmC-like activity and weaker with StaC/InkE-like activity. To investigate whether mutations that alter FAD affinity also alter the type of reaction catalyzed, we use the structure of RebC (Ryan et al., 2007) in combination with comparative sequence analysis of the enzyme family (RebC, StaC, AtmC, and InkE) to generate a RebC protein with ten amino acid substitutions, called RebC-10x, and the complementary StaC-10x protein (Table 1). Excitingly, we find that RebC-10x shows a decrease in FAD affinity (although not to StaC levels) and exhibits strong StaC-like activity, while StaC-10x shows an increase in FAD affinity (although not up to RebC levels) and is a weak RebC-like catalyst. To probe the molecular basis for these enzyme activity conversions, FAD redox potentials are measured and the structure of RebC-10x is determined in native, substrate-bound and product-bound states. These data, along with recent site-directed mutagenesis studies of singly and doubly mutated RebC and StaC proteins (Asamizu et al., 2011; Groom et al., 2011), suggest the structural basis for the differential catalytic activities of these proteins.
Table 1.
Residues interchanged to generate the RebC-10x and StaC-10x constructs (see Figure S1 for a complete sequence alignment).
| RebC | StaC | Function |
|---|---|---|
| Glu36 | Asp37 | FAD binding (adenine) |
| Gln37 | Ala38 | FAD binding (adenine) |
| Thr38 | Gly39 | FAD binding (adenine) |
| Gln117 | Ala118 | FAD binding (phosphate) |
| Arg46 | Lys47 | FAD binding (isoalloxazine) |
| Gly48 | Ser49 | FAD binding (isoalloxazine) |
| Phe216 | Val221 | Substrate binding pocket |
| Ala231 | Ser236 | Substrate binding pocket |
| Arg239 | Asn244 | FAD (isoalloxazine) & substrate binding |
| Thr241 | Val246 | Substrate binding pocket |
RESULTS
The FAD binding affinity correlates with the reaction catalyzed for wild-type enzymes
StaC, RebC, AtmC, and InkE were purified recombinantly without addition of exogenous riboflavin to the media. As reported earlier, StaC purifies without bound FAD, whereas RebC purifies with bound FAD (Howard-Jones and Walsh, 2006). Using a new cell line for protein expression (Rosetta™ (DE3) 2 pLysS cells from Novagen), we were able to increase the amount of FAD co-purified with RebC to ~68%, from ~33% reported previously, when RebC was purified from BL21(DE3) cells (Howard-Jones and Walsh, 2006). Furthermore, we find that AtmC, which catalyzes a RebC-like reaction, purifies with bound FAD, while InkE, which catalyzes a reaction similar to StaC, purifies without bound FAD.
To determine the FAD dissociation constants for these four proteins, we generated the deflavinated (apo) forms and performed isothermal titration calorimetry experiments (ITC). The removal of the FAD cofactor was unnecessary in the case of StaC and InkE, as each of these proteins purifies without bound FAD. For RebC and AtmC, deflavination was accomplished by washing the protein bound to the metal affinity column with a buffer containing 2 M KBr and 2 M urea (Hefti et al., 2003). We find that the dissociation constants (Kd’s) of StaC, RebC, InkE, and AtmC for FAD are 15 ± 2 μM, 20 ± 12 nM, 59 ± 48 μM, and 73 ± 28 nM, respectively (Table 2 and Figures S2A-S2F). While RebC and AtmC, both of which facilitate a net 8-electron oxidation of CPA (Gao et al., 2006; Howard-Jones and Walsh, 2006), have FAD Kd’ s in the low nanomolar range, StaC and InkE, both of which facilitate a net 4-electron oxidation of CPA (Howard-Jones and Walsh, 2006; Kim et al., 2007), have FAD Kd’s in the micromolar range (Table 2). These data suggest that, among this group of enzymes, there is a correlation between, on one hand, tight (nanomolar) binding of FAD and mediation of a net 8-electron oxidation of CPA and, on the other hand, substantially weaker (micromolar) binding of FAD and mediation of a net 4-electron oxidation of CPA.
Table 2.
Dissociation constants for FAD of StaC- and RebC-like proteins, as determined by isothermal titration calorimetry (curves shown in Figure S2).
| Protein | # of ITC experiments | Dissociation Constant for FAD |
|---|---|---|
| StaC | 3 | 15,000 ± 2,000 nM |
| InkE | 4 | 59,000 ± 48,000 nM |
| RebC | 5 | 20 ± 12 nM |
| AtmC | 5 | 73 ± 28 nM |
| RebC-10x | 3 | 600 ± 55 nM |
| StaC-10x-FAD1 | 2 | 560 ± 120 nM |
The FAD bound to StaC-10x could not be removed from the protein (see text).
StaC-10x and RebC-10x have similar, intermediate affinities for FAD
Because of the close sequence similarity of RebC, StaC, AtmC, and InkE, and because the structure of RebC is available (Ryan et al., 2007), the identity of residues likely to mediate the differences in enzymatic activity and FAD affinity between the two groups of enzymes could be readily predicted (Figure S1). With few exceptions, sequence alignment reveals key residues that have one identity in both RebC and AtmC and another identity in both StaC and InkE (Figure S1). Using the RebC crystal structure to filter out residues distant from the expected FAD-binding and active sites, ten RebC residues and ten StaC residues were chosen for complementary mutation to the amino acid found in the homologous enzyme, resulting in our RebC-10x and StaC-10x constructs (Table 1).
To assess their FAD binding affinity, these new constructs were expressed and purified for ITC experiments. Like RebC and AtmC, both RebC-10x and StaC-10x co-purify with FAD. RebC-10x was deflavinated in the same manner as RebC and AtmC, and its Kd for FAD was determined to be 600 ± 55 nM (Figure S2E, Table 2). Unlike RebC-10x, however, it was not possible to produce apo-StaC-10x (See Methods). Therefore as isolated StaC-10x, which co-purifies with 33% FAD occupancy, was used in ITC experiments to yield an approximate Kd of FAD for StaC-10x of 560 ± 120 nM (Figure S2F, Table 2). To understand the relationship of this value to the true Kd, we determined the Kd of FAD for as-purified RebC without removal of FAD (RebC co-purifies with ~68% FAD in our expression system). The Kd of RebC for FAD determined in this manner is 31 nM, a value within the range of values determined using fully deflavinated protein (20 ± 12 nM). Therefore, although the Kd calculated for StaC-10x for FAD is imprecise, we assume that the value is on the order of magnitude of the true Kd value. Thus, by targeting ten amino acids that may be involved in binding FAD, we have altered both StaC’s and RebC’s affinity for FAD, suggesting successful identification of residues important for modulating FAD affinity.
RebC-10x and StaC-10x have almost identical redox potentials to RebC
To determine whether the amino acid replacements had an effect on the FAD midpoint potential, reduction potentials of RebC-10x and StaC-10x were determined and compared to the previously reported reduction potential of RebC, −179 mV (Ryan et al., 2007). Using the xanthine/xanthine oxidase reduction assay (Massey, 1991; Palfey et al., 1994), the reduction potential of FAD-bound RebC-10x was found to be −178 mV ± 5 (Figure S3A), identical within experimental error to that of FAD-bound RebC. Interestingly, the reduction potential of the StaC-10x FAD is also the same −178 ± 2 mV (Figure S3B). These results show that the mutant proteins have adopted or retained a flavin cofactor redox environment similar to that found in RebC.
The 10x mutant proteins have altered activities
Activity assays using a previously described method (Howard-Jones and Walsh, 2006) were performed to investigate the chemical reactivities of both 10x variants. Because the substrate(s) of StaC and RebC have never been isolated, this assay uses the upstream enzyme StaP and its known substrate CPA together with an electron source provided by spinach ferredoxin, flavodoxin NADP+ reductase, and NADPH to generate the reactive intermediate that is the putative substrate(s) for RebC and StaC. The reaction products are then separated and analyzed via HPLC. In the presence of StaP alone, the reaction yields multiple products, oxidized to different degrees, including K252c (a 4-electron oxidation product), 7-hydroxy-K252c (a 6-electron oxidation and the major product), and arcyriaflavin A (an 8-electron oxidation product). The addition of either RebC or StaC to the assay directs the reaction to a single major product, arcyriaflavin A for RebC and K252c for StaC (Howard-Jones and Walsh, 2006).
Interestingly, when RebC-10x is added to the assay, no production of the RebC product arcyriaflavin A is observed. Instead, RebC-10x addition results in production of the StaC product K252c, with a rate that is ~3-fold faster than the rate of the StaC-catalyzed reaction (based on authentic samples and standard curves) (Table 3 and Figure S4). StaC-10x behaves like RebC to the extent that it produces arcyriaflavin A as its major product and has lost nearly all of its activity for the normal StaC product K252c. However, it is not a strong RebC catalyst, generating arcyriaflavin A at rates that are ~8-fold lower than that of wild-type RebC (Table 3 and Figure S4).
Table 3.
Relative rates of arcyriaflavin A and K252c production (traces shown in Figure S4).
| Proteins assayed | Rate of arcyriaflavin A production (relative to StaP alone) | Rate of K252c production (relative to StaP alone) |
|---|---|---|
| StaP / StaC | 1.1 ± 0.3 | 7 ± 1 |
| StaP / RebC | 14 | 1.0 |
| StaP / RebC-10x | 0.79 ± 0.07 | 20 ± 2 |
| StaP / StaC-10x | 1.7 ± 0.4 | 1.3 ± 0.2 |
Structures of RebC-10x display the key characteristics of a flavin hydroxylase and explain the molecular basis for decreased FAD affinity
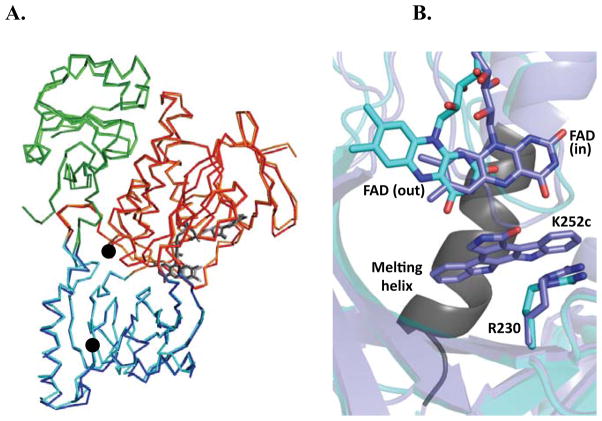
Crystal structures spanning three different states of the RebC-10x reaction (native, substrate-, and product-bound) have been determined in an effort to provide a molecular explanation for the decrease in FAD affinity and gain in StaC-like function. A single crystal structure of K252c-soaked RebC-10x with two molecules in the asymmetric unit has afforded a view of both native and product-bound RebC-10x states at 2.33 Å resolution, while soaking a RebC-10x crystal with StaP substrate CPA (see below) has provided a substrate-bound structure at 2.76 Å resolution (see Table 4 for data collection and refinement statistics). Structures were solved by rigid body refinement against the wild-type RebC structure, with mutations confirmed by Fo–Fc difference density. As expected, RebC-10x adopts the general fold of flavin-dependent hydroxylases (Montersino et al., 2011) with FAD-, substrate-binding and thioredoxin-like domains (Figure 2A). The root mean squared deviation (RMSD) of mainchain atoms between RebC-10x and RebC structures are less than 1 Å, indicating no major conformational changes occur as a result of the ten mutations (Figure 2A). In addition, the mutations do not appear to cause local instability, as B-factors of the mutated sites and their surroundings are very similar when compared to the structure of wild type RebC (Figure S5). Other RebC features retained in RebC-10x, such as the ‘mobile flavin’ and the ‘melting helix,’ are revealed by the structures generated through the soaking of RebC-10x with StaC product, K252c. The ‘out’ position of the mobile flavin is observed in the molecule in the asymmetric unit that contains no bound K252c. This FAD orientation is associated with NADPH reduction due to the afforded solvent accessibility (Figure 2B) (Enroth et al., 1998; Gatti et al., 1994; Ryan et al., 2007). The ‘in’ position or ‘catalytic orientation’ of the mobile flavin is found in the molecule that has density for K252c. Comparison of these two structures also shows that like RebC, residues 354–363 (termed the ‘melting helix’) are disordered when the active site is empty and become ordered when the active site is full (Figure 2). This ‘melting helix’ region is believed to be the entry point for substrate (Ryan et al., 2007).
Table 4.
Data collection and refinement statistics.
| RebC-10x K252c soak (K252c and native structures) | RebC-10X CPA soak (7-carboxy-K252c structure) | |
|---|---|---|
| Data Collection | ||
| Wavelength (Å) | 0.97910 | 0.97918 |
| Space Group | P21 | P21 |
| Cell Dimensions | ||
| a, b, c (Å); β (°) | 64.3, 78.6, 125.7; 99.9° | 63.2, 77.7, 123.1; 98.8° |
| Resolution (Å)1 | 50 – 2.33 (2.42 – 2.33) | 50 – 2.76 (2.86 – 2.76) |
| Rsym (%)1 | 8.2 (52.0) | 5.4 (48.9) |
| <I> / σ(<I>)1 | 19.7 (3.6) | 19.6 (2.2) |
| Completeness (%)1 | 99.4 (98.5) | 94.9 (97.4) |
| Redundancy1 | 5.2 (4.9) | 3.5 (3.5) |
| Total reflections | 256,655 | 101,523 |
| Refinement | ||
| Resolution (Å) | 41.4 – 2.33 | 50 – 2.76 |
| Reflections | 51,610 | 28,820 |
| Rwork / Rfree2 | 0.214 / 0.261 | 0.206 / 0.255 |
| No. of non-hydrogen atoms | ||
| Protein | 7,872 | 7,812 |
| FAD / Indolocarbazole | 106 / 24 | − / 54 |
| Water | 304 | 62 |
| Average B-factors (Å2) | ||
| Protein | 38.1 | 61.7 |
| FAD / Indolocarbazole | 40.2 / 31.1 | − / 69.8 |
| Water | 35.3 | 52.3 |
| Rms deviations | ||
| Bond lengths (Å) | 0.006 | 0.009 |
| Bond angles (°) | 1.0 | 1.6 |
| Ramachandran statistics | ||
| Most favored region (%) | 88.5 | 85.7 |
| Additionally allowed (%) | 10.9 | 14.0 |
| Generously allowed (%) | 0.5 | 0 |
| Disallowed (no. of residues) | 1 | 2 |
Highest resolution shell is shown in parentheses.
R-factor = Σ(|Fobs|-k|Fcalc|)/Σ |Fobs| and R-free is the R value for a test set of reflections consisting of 5% of the diffraction data not used in refinement. The same test set of reflections used in the refinement of the RebC structures was used here.
Figure 2.
Overall structure of RebC-10x aligned with wild type RebC. (A) RebC (lighter colors) and RebC-10x (darker colors) are aligned, showing the FAD binding domain (red), substrate binding domain (blue), thioredoxin-like domain (green), and FAD (grey sticks). RMSD of mainchain atoms is 0.46 Å. See Fig. S5 for comparison of (B) RebC-10x shows retention of the ‘mobile flavin’, where flavin binds in the ‘out’ position of native RebC-10x (cyan) and in the ‘in’ position when product is bound (slate), and the ‘melting helix’, where residues 354–363 (black) become disordered in absence of a bound indolocarbazole ligand (melting helix termini are marked with black circles in (A)). Arg230 is labeled to help relate this view of the active site with those shown in Fig. 3 and S6.
Differences between RebC and RebC-10x are also observed. As expected, multiple protein-FAD interactions are lost when the RebC-10x structure is compared to RebC. These include the loss of van der Waals interactions between Q37 and T38 and the FAD adenosine moiety when these residues are replaced by A37 and G38, resulting in a more solvent accessible cofactor (Figure S6A); a hydrogen bond loss from E36 to the FAD ribose when E36 is replaced with shorter D36 (Figure S6A); a salt-bridge loss from R46 to the FAD α-phosphate when R46 is replaced with shorter K46 (Figure S6D); and another hydrogen bond loss from R239 of RebC to FAD O4 in the ‘out’ position, when R239 is replaced with N239 in RebC-10x (Figure S6E).
CPA-soaked RebC-10x structure reveals putative StaC substrate
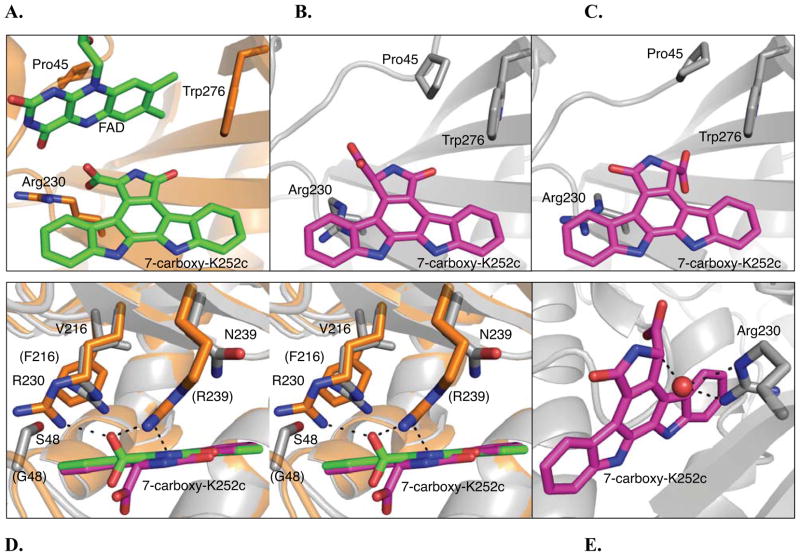
A structure of RebC-10x bound to its putative substrate was obtained by incubating RebC-10x crystals with StaP substrate, CPA, for one week. Previously, the putative substrate for RebC was ‘trapped’ by allowing CPA to spontaneously degrade in the presence of RebC crystals. The crystallized RebC protein selectively bound and stabilized its substrate (7-carboxy-K252c) in its active site, allowing visualization (Ryan et al., 2007). Using this same approach with the StaC catalyst RebC-10x, indolocarbazole density was discovered in the active site of RebC-10x (Figure S7). Since the identity of the StaC (and thus RebC-10x) substrate is not known, a number of indolocarbazoles were modeled into the electron density, including aryl-aryl coupled CPA, arcyriaflavin A, K252c, and the enol tautomer of 7-carboxy-K252c, the molecule identified in the previous RebC CPA soak (Figure 1B). Refinement of these molecules results in significant positive and negative difference density peaks (Figure S7A-D), indicating that they do not accurately describe the electron density. However, when the S-keto tautomer of 7-carboxy-K252c is modeled, little or no residual Fo–Fc difference density appears (Figure S7E) and the refined structure is the best fit to omit density maps (Figure S7G-H). Thus, our data support the presence of the S-keto tautomer of 7-carboxy-K252c in the RebC-10x active site, and suggest that this molecule is likely the substrate for StaC.
Interestingly, while RebC binds 7-carboxy-K252c with its carboxyl group pointed toward R230 and its carbonyl in a more hydrophobic pocket (Figure 3A), RebC-10x binds the S-keto tautomer of 7-carboxy-K252c in two orientations that differ by 180° (Figure 3B-C). One molecule in the asymmetric unit, molecule A, binds substrate in the same orientation as RebC (Figure 3B), while in molecule B, the substrate sits in an alternate orientation, with the carbonyl pointing toward R230 and the carboxyl group near the hydrophobic pocket (Figure 3C). Notably, FAD is missing in these structures. During the week-long CPA soak used to generate this crystal structure, FAD, which binds more weakly to RebC-10x than RebC, dissociates, and a loop containing residues 45 – 48 shifts to occupy its site, repositioning P45 such that it now interacts with W276 (Figure 3A-C). The absence of FAD yields a larger active site, which can accommodate both orientations of the 7-carboxy-K252c. Modeling FAD in the ‘in’ conformation into the active site predicts a tight fit with substrate in either orientation, suggesting that FAD may be in an ‘out’ conformation prior to decarboxylation or that FAD moving ‘in’ might prompt decarboxylation. Importantly, regardless of FAD position, both 7-carboxy-K252c molecules are bound to RebC-10x in the S-keto tautomer form, while the enol tautomer of 7-carboxy-K252c was found in RebC. With active sites of StaC-catalyst RebC-10x so similar in sequence to RebC, it is interesting to consider the molecular basis for this preferential binding.
Figure 3.
RebC and RebC-10x substrate binding pocket. (A) RebC substrate binding pocket (PDB ID 2R0G) with protein backbone shown in orange and cofactor, FAD, and substrate, enol 7-carboxy-K252c, carbons in green and in stick representation. The sidechains, Arg230, Pro45, and Trp276 (representing a hydrophobic region of the binding pocket) are also shown in stick representation. (B) The S-keto form of 7-carboxy-K252c as found in molecule A of the RebC-10x CPA soaked crystal. Protein carbons are colored grey and substrate shown in stick representation and carbons colored magenta. The carboxyl group of substrate is proximal to Arg230, as in RebC. (C) The alternate orientation of substrate, as found in molecule B of the RebC-10x CPA soaked crystal. The carboxyl group of substrate is proximal to Trp276; coloring as in (B). (D) A stereo representation of an overlay of the RebC and RebC-10x (molecule A) active sites, shown from above. Substrate and protein colors the same as (A) – (C); RebC sidechains are labeled in parentheses. (E) A water molecule (red sphere) present in RebC-10x structure is located 2.6, 3.1 and 3.1 Å from two nitrogens of Arg230 and the C-7 position of 7-carboxy-K252c, respectively. In all stick representations, oxygen is shown in red and nitrogen in blue (see Figure S8A for the water binding site in the alternative orientation of substrate). See also Figure S7.
RebC and RebC-10x active site comparisons
Although mutations were designed to alter the affinity of FAD for the enzyme and most reside in the FAD binding pocket, some residues (like G48S, T241V, F216V, R239N) are also near the substrate binding site and are likely to influence both substrate binding and reactivity (Figure S6C-F). While the loss of FAD causes a repositioning of the FAD binding loop (residues 39 – 46), most of the protein backbone in the substrate-binding site of RebC-10x is identical to that of RebC, allowing direct comparison of substrate-bound RebC10x to substrate-bound RebC. A key difference between the two is the removal of salt bridge and hydrogen bonding interactions between R239 and the C-7 carboxyl group and NH-6 position of the RebC planar substrate (Figure 3D). In RebC-10x, N239 is incapable of interacting with either the bound RebC-10x keto substrate, or the overlaid enol tautomer from the RebC structure (Figure 3D).
Another difference between the two active sites is the positioning of another arginine, R230. The residue, present in both RebC and RebC-10x, sits lower in the RebC-10x active site, apparently due to the combination of the G48S and F216V mutations (Figure 3D). In particular, the substitution of S for G48 sterically blocks the RebC R230 position and V for F216 creates room for this residue to move down. This ‘lower’ position of R230 in RebC-10x disrupts the favorable interaction observed in RebC between the guanidinium group of R230 and the 7-carboxyl of substrate in the enol tautomeric form. In RebC, the distance (2.6 Å) as well as the geometry of the interaction should serve to stabilize this orientation of the 7-carboxyl group and thus stabilize the enol form of the substrate (Figure 3D). In contrast, the position of R230 observed in RebC-10x has suboptimal geometry to maintain this interaction, and we observe instead, a new position of the 7-carboxyl moiety and a keto tautomeric form of substrate. This altered orientation of R230 in RebC-10x no longer interacts with substrate, but rather appears to be involved in creation of a new water binding site not observed in RebC. When water is modeled into observed positive Fo–Fc electron difference density adjacent to R230, the difference electron density disappears and the B-factor for this new water in molecules A and B, 65.0 and 58.6 Å2, are consistent with the average B-factors for protein atoms (62.7 Å2) and 7-carboxy-K252c atoms (66.1 Å2), indicating that water is a good fit for this density. Following refinement, the water is located 3.1 and 3.4 Å from C-7 position of the RebC-10x substrate in chains A and B, respectively (Figure 3E and S7A), a position that suggests a catalytic role (see discussion). This water binding site is conserved in both the native and K252c bound RebC-10x molecules. Overall, the active sites of RebC and RebC-10x are similar; the bound products and substrates can be superimposed. However, small changes in side chain positions results in different tautomers of substrate and changes to bound water positions.
DISCUSSION
While the substrates for RebC and StaC can convert to the respective products spontaneously, the presence of these enzymes directs the reactivity toward a single product. In this sense, RebC and StaC are babysitting enzymes, ensuring that the appropriate reaction goes forward, but just as importantly, preventing unwanted reactions. With such high sequence similarly between RebC and StaC, understanding how these enzymes catalyze their differential reactions has been a point of considerable interest in the natural product field.
For RebC, biochemical and structural analysis helped elucidate the nature of the RebC substrate as 7-carboxy-K252c (Ryan et al., 2007), indicating that conversion to product requires both a decarboxylation and oxidation at C-7. With structural analysis firmly placing RebC in the FAD monooxygenase class of enzymes, it has been proposed that RebC catalyzes both the decarboxylation and oxidation of substrate using FAD-dependent hydroxylase activity (Ballou, 2007; Ryan et al., 2007). In contrast StaC, which purifies in a deflavinated state, does not require oxidation activity, raising the question of whether StaC requires FAD for activity. In this work, we find a clear correlation between FAD-binding affinities and the reaction catalyzed, with tighter (nanomolar) binding associated with enzymes that mediate a net 8-electron oxidation of CPA and substantially weaker (micromolar) binding associated with the mediation of a net 4-electron oxidation of CPA. This trend holds for both the wild-type and mutant proteins. However, we find that RebC-10x, which binds its FAD more tightly than StaC by three orders of magnitude, is actually a better StaC catalyst than StaC itself, indicating that truly weak binding of FAD is not required for StaC-like chemistry. In contrast, StaC-10x, which binds FAD more weakly than RebC by 30-fold, is a poor RebC catalyst. Collectively, these data imply that both StaC and RebC are FAD-dependent enzymes in which the tight binding of FAD is more important to the facilitation of RebC-like chemistry. Further support for the identification of StaC as an FAD-dependent enzyme comes from Onaka and co-workers, who showed that StaC activity is substantially diminished when regions of the GXGXXG FAD binding motif are mutated (Asamizu et al., 2011).
With FAD independence ruled out as the source of StaC’s disparate reactivity, we further considered if variations in substrate use and/or FAD-based mechanisms could be the cause. While it is difficult to envision a FAD monooxygenase mechanism that could convert 7-carboxy-K252c to K252c, Onaka and co-workers argued that such a mechanism could be invoked for converting a substrate such as 5-deoxo-7-carboxy-K252c to form K252c (Asamizu et al., 2011). To investigate the nature of the StaC substrate, we employed the same crystallographic trick that was successful for RebC (Ryan et al., 2007): screening CPA degradation products using enzyme crystals to determine which molecule selectively binds. Interestingly, we again find 7-carboxy-K252c in the active site, implying RebC and StaC do indeed share a common substrate. The fact that 7-carboxy-K252c is found in the structures of both RebC and the StaC-like catalyst RebC-10x is consistent with the interchangeable use of the preceding enzymes (StaP and RebP).
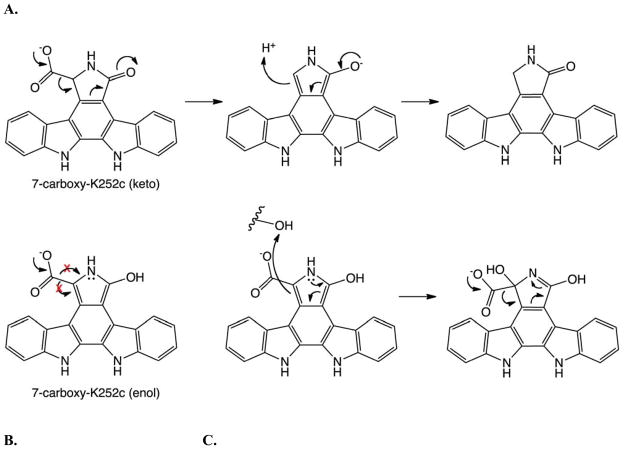
Given that the substrates for RebC and StaC appear to be the same, the source of differential products must lie with the use of a disparate FAD-based mechanism, achieved by the manner in which each enzyme binds its substrate and the amino acid environment near the active site. Due to the high homology between these enzymes, small variations must be all that are required for this shift in mechanism. Here our structures of substrate-bound forms of RebC and the StaC-like catalyst RebC- 10x are most informative. While our data show a common substrate, each enzyme active site is designed to bind a different tautomer of that substrate. A nonplanar keto tautomer of 7-carboxy-K252c is bound to the StaC-like catalyst while RebC binds the planar enol tautomer. Notably, there is a key difference in the chemical reactivity of these two tautomers: in the keto tautomer, the sp3 hybridized C-7 carbon can accept electrons from the spontaneous decarboxylation of 7-carboxy-K252c (Figure 4A). In the enol form, however, spontaneous decarboxylation is not possible due to the sp2 hybridized C-7 carbon (Figure 4B). Thus, RebC binds 7-carboxy-K252c such that decarboxylation would require a change in C-7 hybridization, likely achieved through hydroxylation of C-7 in a FAD-dependent monooxygenase reaction (Ballou, 2007; Ryan et al., 2007) (Figure 4C), while the StaC-like catalyst binds 7-carboxy-K252c primed for decarboxylation without the need for hydroxylation (Figure 4A). Support for an FAD-dependent monooxygenase mechanism for RebC includes the observation that its ‘mobile’ flavin adopts an ‘in’ position in the presence of substrate or when the FAD is reduced, such that the substrate C-7 position is perfectly positioned near the C4a of the isoalloxazine ring for attack on C4a-hydroperoxy-FAD, a common flavin-dependent monooxygenase intermediate (Ryan et al., 2008; Ryan et al., 2007).
Figure 4.
Mechanistic implications of substrate tautomerization. (A) A mechanistic scheme for the decarboxylation of S-keto 7-carboxy-K252c in a StaC-like enzyme, where the reaction is initiated by decarboxylation. (B) Hybridization of the C-7 carbon in the enol tautomer of 7-carboxy-K252c makes decarboxylation as the first step in the mechanism unlikely. (C) A mechanistic scheme for RebC in which oxidation precedes decarboxylation at the C-7 position.
While there is an obvious role for FAD in the RebC mechanistic proposal, its role in a StaC-like catalyst is less clear. As described above, our data and that of the Onaka laboratory support StaC as a FAD-dependent enzyme (Asamizu et al., 2011), and while our redox measurements show that FAD potentials are virtually unchanged among RebC and StaC-like catalysts in the absence of substrate, our structural data do not buttress the idea that StaC is a monooxygenase. Instead, our data suggest that FAD could contribute to the catalysis of decarboxylation through a steric and/or electrostatic mechanism. In an overlay of the substrate-bound structures of RebC and RebC-10x, the carboxyl group of substrate in RebC-10x clashes with the FAD isoalloxazine moiety (Figure S8B-C), indicating that FAD binding and/or movement of the cofactor from the ‘out’ to the ‘in’ position could play a steric role in driving decarboxylation. Alternatively, or in addition, reduced FAD swinging ‘in’ to the active site could serve to facilitate decarboxylation through an electrostatic mechanism. Structural data presented here show that like RebC, the RebC-10x FAD does transition from ‘out’ to ‘in’ positions. In particular, we observe FAD ‘out’ in the native RebC-10x structure and find FAD ‘in’ with the product K252c bound. In contrast to RebC, where the FAD is ‘in’, positioned near substrate following the week-long CPA soak (Ryan et al., 2007), here we find that FAD dissociates from RebC-10x crystals during the same time period. While in agreement with a weaker FAD binding affinity for RebC-10x, FAD dissociation during the soaking experiment is also consistent with the notion of an unfavorably crowded active site following substrate binding.
These two different mechanistic hypotheses for RebC and StaC-like catalysts are also in alignment with the observed FAD binding affinities. For RebC, tight binding of FAD is likely important for the intricate control of the FAD cofactor required for substrate hydroxylation, whereas for StaC-like catalysts, precise control over FAD positioning would not be required in a steric or electrostatic mechanism. Although tighter binding of FAD from micromolar to high nanomolar appears to make for a better StaC catalyst, in vivo, weaker binding of the cofactor could allow for a single FAD to service multiple StaC enzymes. In its babysitter role, StaC protects the 7-carboxy-K252c from unwanted side-reactions while waiting for FAD to bind. While organisms containing the rebeccamycin pathway could also benefit from shared use of FAD by multiple RebC enzymes, in this case, weaker FAD affinity might impair catalysis to too great an extent. Here we find that weakening FAD affinity from low to high nanomolar leads to a poor RebC catalyst.
While the binding of different tautomers of 7-carboxy-K252c by RebC and StaC-like catalysts can explain how two highly homologous FAD-dependent enzymes can use different mechanisms to yield their observed varied products, it is also interesting to consider the number of amino acid changes that are required for this differential binding of substrate. Based on structural analysis, two arginines (R239 and R230) in RebC are responsible for interacting with the 7-carboxyl moiety of 7-carboxy-K252c and appear key to maintaining it in an enol form, requiring RebC-like chemistry for product formation (see Figure 3D and Results). In StaC-like enzymes, the equivalent of R239 is asparagine and R230 adopts an altered position due to G48S and F216V substitutions. As we were preparing this manuscript, two groups reported that conversion of RebC activity to StaC-like activity can be achieved via two simultaneous mutations, F216V and R239N (Asamizu et al., 2011; Groom et al., 2011), with individual mutations of these residues leading to unchanged and dead enzyme, respectively (Asamizu et al., 2011). These mutational data highlight the importance of the residue at position 239 and show that the availability of the ‘lower’ (StaC-like) conformation of R230, afforded by the F216V mutation alone, is not enough to convert a RebC to a StaC. The authors also tried the complementary mutations in StaC and found that this construct, StaC-V221F-N244R, is not an active RebC catalyst (Asamizu et al., 2011). Consistent with this result, our structural data suggest the StaC R230 equivalent, R235, cannot adopt its “RebC-like” orientation unless S48 is converted to G (see Figure 3D).
Collectively, these data provide insight into the key amino acid interactions in the RebC/StaC active sites that afford the product specificity. For RebC activity, R239 and an ‘upper’ position of R230 appear to help stabilize an enol form of 7-carboxy-K252c, requiring the tightly bound FAD to perform monooxygenase activity for product formation. In StaC, the absence of arginine at position 239 and a ‘lower’ position of R230, appear to yield the preferential binding of the keto form of 7-carboxy-K252c. Chemical logic predicts that this keto tautomer would not require FAD monooxygenase activity for conversion, perhaps using the ‘mobile flavin’ as steric or electrostatic driving force for decarboxylation instead. Of course after decarboxylation, protonation of the C-7 carbon is required to form the StaC product, K252c (Figure 4A). Here we find that the ‘lower’ position of R230 creates an ordered water site that is not present in RebC (Figure 3E). This water is in position to act as a general acid to protonate C-7, yielding the StaC product. Interestingly, this water molecule is present regardless of the observed substrate orientation. As such, both orientations of 7-carboxy-K252c shown in Figure 3B, C seem to be catalytically viable. In contrast, only one orientation of substrate is found in the RebC structures and only that orientation has C-7 positioned correctly for hydroxylation.
SIGNIFICANCE
Natural product diversity arises in part from the tailoring of common molecular scaffolds. Enzymatic pathways creating these scaffolds are often duplicated in nature with modifications added to generate product diversity. Frequently these pathways are challenging to study by traditional enzymological approaches due to inherent reactivity of the scaffold molecules, obscuring substrate and product identification. Here we demonstrate that a crystallographic approach can be highly informative in these types of situations. In particular, crystallography has allowed for the identification of substrates for both RebC (Ryan et al., 2007) and StaC (this work). Also, crystallography combined with biochemical analysis has provided insight into how enzymes with homology to the ‘mobile flavin’ class of FAD-dependent monooxygenases have been co-opted to generate RebC-like and StaC-like catalysts. While both RebC and StaC-like enzymes display the so-called ‘melting helix’ modification, with residues of a helix providing an alternative active site entrance for the large bisindole substrates, StaC shows additional departures from traditional ‘mobile flavin’ monooxygenases. In particular, it appears to use a mobile flavin, but not for monooxygenase activity. Instead, we propose the mobile flavin drives a decarboxylation. With as few as two amino acid substitutions in the active site (Asamizu et al., 2011; Groom et al., 2011), RebC and StaC bind distinct isomers of the same substrate to afford different reaction mechanisms. While small changes to the molecular scaffold of a natural product can turn an inert molecule into an anti-tumor drug, small changes to the molecular scaffold of an enzyme can alter the mechanistic role of a required cofactor and promote the formation of a new product.
EXPERIMENTAL PROCEDURES
Generation of expression vectors
The generation of all expression vectors for proteins discussed in this manuscript was carried out as described in the Supplementary Information.
Protein purification
All protein expression vectors were transformed into chemically competent Rosetta™ 2 (DE3) pLysS cells (Novagen). Single colonies were used to inoculate LB media containing 30 mg/L kanamycin and 34 mg/L chloramphenicol. Cultures were grown at 37°C to an OD600 of ~0.45, at which point the temperature was lowered to 21°C. At an OD600 of ~0.65, cultures were induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and grown for ~20 hours at 250 rpm. Cells were pelleted by centrifugation, suspended in 300 mM NaCl, 25 mM Tris-HCl, 5 mM imidazole, pH 8.0, sonicated, and centrifuged at 25,000 rpm to pellet insoluble material. The supernatant was incubated with nickel (II) loaded chelating sepharose fast flow (GE Biosciences) for 40 min at 4°C. Unbound material was removed, and then the column was washed with 25 column volumes of 300 mM NaCl, 25 mM Tris-HCl, 20 mM imidazole, pH 8.0 and then, for AtmC and RebC, washed with 2 M KBr, 2 M urea, 300 mM NaCl, 25 mM Tris-HCl, 20 mM imidazole, pH 8.0 to remove bound flavin, and then re-washed with 300 mM NaCl, 25 mM Tris-HCl, 20 mM imidazole, pH 8.0. Protein was eluted with 300 mM NaCl, 25 mM Tris-HCl, 200 mM imidazole, pH 8.0. Eluted protein was loaded onto a Superdex-200 column pre-equilibrated with 150 mM NaCl, 25 mM HEPES, 10% glycerol, pH 7.5, and 3 mL fractions were collected. Fractions containing purified protein were pooled and used directly in isothermal titration calorimetry experiments. Protein concentration was assayed using absorbance in the linear range at A280, using theoretical extinction coefficients calculated for each protein by ProtParam (Gasteiger et al., 2005). For StaC-10x and RebC-10x, protein concentration was calculated instead using the BioRad Protein Assay (BioRad) calibrated using BSA (New England Biolabs) as a standard.
Kd for FAD of StaC, RebC, InkE, AtmC, StaC-10x, and RebC-10x measured by isothermal titration calorimetry (ITC)
All experiments were carried out using a MicroCal Isothermal Titration Calorimeter in the MIT Biophysical Instrumentation Laboratory. FAD was removed from RebC, AtmC, and RebC-10x as described above. Apo-protein from gel filtration was diluted in gel filtration buffer to the desired concentration. FAD could not be removed from StaC-10x in the same manner. After passing a buffer containing 2M urea and 2M KBr over a metal affinity column with bound StaC-10x to remove FAD, protein could not be eluted from the column using any tested elution buffer. Presumably, the protein aggregated or cross-linked to the column after the loss of FAD, requiring us to use StaC-10x with partial FAD occupancy in this experiments. The use of non-apo StaC-10x complicated the ITC analysis, since a fundamental assumption of Origin Software is that the ligand concentration in the cell is zero prior to the first injection of ligand. This limitation prompted a control experiment with RebC as described in text. The ITC experiments were repeated in at least triplicate, except for the StaC-10x experiment, which was done twice (Figure S2). Equilibrium constants of binding (Kb’s) were determined using the Origin Software and were converted to equilibrium constants of dissociation (Kd’s). The single binding site model was used in determining Kb’s.
Measurement of the redox potential of StaC-10x and RebC-10x
The midpoint potential of purified StaC-10x and RebC-10x were determined using a xanthine oxidase reduction method developed by Massey (Massey, 1991). Experiments were carried out at 25°C under anaerobic conditions in a glovebox using a S. I. Photonics 400 series spectrophotometer, housed in an MBraun Labmaster glove box. The cell solution was buffered by 25 mM HEPES (pH 7.0), and contained 10% glycerol, 150 mM NaCl, 1 μM benzyl viologen, 1 μM methyl viologen, 250 μM xanthine, 15 μM StaC-10x, and 50 μM 1-hydroxyphenazine (1-OHP) as a reference dye (-172 mV). Addition of a catalytic amount of xanthine oxidase (Sigma Aldrich) initiated the reaction, and full spectra were collected every 5 minutes until completion. The reduction of the StaC-10x flavin and 1-hydroxyphenazine were monitored at 480 nm and 370 nm, respectively. For this technique, unlike ITC, the level of flavin incorporation is thought to be irrelevant, as only the flavin cofactor itself is observed spectroscopically. Hence, barring interference of apo protein with the dye molecule (which is extremely unlikely), the presence of apo protein is not thought to influence the redox potential determined.
Activity assays of StaC, RebC, and StaC-10x
All enzyme assays were carried out as described earlier (Howard-Jones and Walsh, 2006) in the presence of 75 mM HEPES pH 7.5, 1 mg/mL BSA, 5 mM NADPH, 20 μM ferredoxin, and 1 μM flavodoxin NADP+ reductase. All assays were carried out in triplicate, except RebC, which was run once.
Crystallization and Structure Determinations
RebC-10x protein was purified as described above. After size exclusion, the protein sample was concentrated to 25 mg/mL (as determined by using the BioRad Protein Assay) using an Amicon Ultra 30K Centrifugal Filter Device in protein buffer (150 mM NaCl, 25 mM HEPES, 10% glycerol, pH 7.5), and flash frozen in liquid nitrogen for storage at −80°C. Crystals of RebC-10x were grown using the hanging-drop diffusion method at room temperature, by incubating 1 μL of protein sample (diluted to 20 mg/mL with protein buffer) with 1 μL of precipitant solution, 100 mM NH4F and 20% PEG 3350 over a 0.5 mL well of precipitant solution. After 3–4 days, rod clusters would appear; these were used to generate microseeds using the Seed Bead kit (Hampton). The seed solution was diluted 100-fold in a 50% protein buffer, 50% precipitant solution and used to serially streak seed 3 drops of 1 μL of 4–6 mg/mL RebC-10x (diluted in protein buffer) and 1 μL of 50 mM NH4F and 10% PEG 3350 which had been pre-equilibrated for three hours. Single crystals appear after 1–2 days. Solutions of CPA and K252c for use in soaking experiments were synthesized as described (Howard-Jones and Walsh, 2006) (Tocris Bioscience), and prepared at 50 and 10 mM in DMSO, respectively. The native/K252c structure was generated from crystals that were soaked in a solution of 25 mM NH4F, 75 mM NaCl, 1 mM K252c, 12.5 mM HEPES (pH 7.5), 5% PEG 3350, and 5% glycerol for three hours at room temperature prior to cryocooling by washing the crystals through a soaking solution with 20% glycerol before submerging in liquid nitrogen. CPA soaked crystals were prepared by incubating single crystals in an identical soaking drop, but substituting K252c for 5 mM CPA. These crystals were incubated at room temperature for one week, then washed in cyroprotectant and cryocooled as described above.
Diffraction data to 2.15 and 2.37 Å resolution for K252c and CPA soaks, respectively, were collected at Beamline 24-IDE at the Advanced Photon Source (Argonne, IL), and processed with HKL2000 (Otwinowski and Minor, 1997) to 2.33 and 2.76 Å resolution, respectively, trimming the resolution such that the I/σ(I) is above 2.0 in the highest resolution bins (Table 4). Both structures were phased using a previously solved RebC – CPA soaked structure (PDB ID 2R0G) stripped of water molecules and ligands as a model for rigid body refinement. Topology and parameter files for FAD, K252c, and S-keto-7-carboxy-K252c were adapted from those used previously (Ryan et al., 2007). Iterative rounds of model building and refinement were done using the programs COOT (Emsley et al., 2010), CNS (Brunger, 2007; Brunger et al., 1998), and PHENIX (Adams et al., 2010). Non-crystallographic symmetry restraints were not used in either structure due to inherit differences of the molecules in the asymmetric unit. No sigma cutoff was included in the refinement and both structures were verified using composite omit electron density maps. The final R factors (work/free/all reflections) of 21.4/26.1/21.6 and 20.6/25.5/21.0 for K252c-soaked and CPA-soaked structures, respectively, are within the typically ranges for structures at or above 2.3 Å resolutions (Badger and Hendle, 2002). In both structures, the 20-residue N-terminal tags containing the His6 affinity sites are disordered. In addition, the RebC-10x K252c soaked structure is missing residues 1–3 and 247–252 in molecule A (K252c-bound structure) and residues 1–2, 353–364, 418–422 in molecule B (native structure) due to disorder. Water molecules were included at positions where +3.5σ Fo-Fc peaks were present. Water molecules in the RebC-10x CPA soaked structure were included only where positive Fo-Fc difference density was present and water binding sites were previously established by higher resolution structures. In this structure, residues 1, 40–41, 248–251, and 418–425 in molecule A and residues 1, 38–40, 247–252, and 418–425 in molecule B are not included in the model. See Table 4 for refinement statistics. The atomic coordinates and structures factors have been deposited in the Protein Data Bank as entries 4EIP and 4EIQ.
Supplementary Material
HIGHLIGHTS.
FAD binding affinities of StaC, RebC, InkE, and AtmC correlate to tailoring function
Crystallographic screening experiment reveals substrate for StaC-like catalyst
The mechanism of product specificity in the RebC / StaC system is clarified
First X-ray structure of a StaC catalyst suggests a unique mechanism from RebC
Acknowledgments
For helpful discussions, we thank Verna Frasca (Origin Software), Deborah Pheasant (ITC), Julie Norville and Peter Carr (experimental conditions for gene synthesis), Danny Yun (three-part ligations), and Elizabeth Nolan, Sarah Mahlstedt, and Elizabeth Sattely (HPLC assays). We also thank Rowena Matthews for the gift of a flavodoxin NADP+-reductase construct, and Carl Balibar for Actinomadura melliaura genomic DNA. This work was supported by the Richard Allan Barry Fund of the Boston Foundation (S.J.E.), NIH grant GM 20011 (C.T.W.), and a Howard Hughes Medical Institute Pre-doctoral Fellowship (K.S.R). C.L.D. is a Howard Hughes Medical Institute Investigator. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
Author contributions: K.S.R. helped design the study, generated StaC-10x and RebC-10x, purified proteins except RebC-10x, carried out ITC experiments, performed initial activity assays, and contributed to writing of the paper. P.J.G. generated RebC-10x, purified RebC-10x, carried out ITC on RebC-10x, performed final activity assays, solved the crystal structures, and co-wrote the paper. M.J.H. determined the redox potential of StaC-10x and RebC-10x. A.R.H-J. generated the InkE, StaC, and RebC expression vectors and designed the previously reported activity assay. S.J.E. and C.T.W. provided mentorship. C.L.D. provided mentorship and co-wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu S, Shiro Y, Igarashi Y, Nagano S, Onaka H. Characterization and Functional Modification of StaC and RebC, Which Are Involved in the Pyrrole Oxidation of Indolocarbazole Biosynthesis. Biosci Biotech Bioch. 2011;75:2184–2193. doi: 10.1271/bbb.110474. [DOI] [PubMed] [Google Scholar]
- Badger J, Hendle J. Reliable quality-control methods for protein crystal structures. Acta Crystallogr D Biol Crystallogr. 2002;58:284–291. doi: 10.1107/s0907444901020133. [DOI] [PubMed] [Google Scholar]
- Bailly C, Riou JF, Colson P, Houssier C, Rodrigues-Pereira E, Prudhomme M. DNA cleavage by topoisomerase I in the presence of indolocarbazole derivatives of rebeccamycin. Biochemistry. 1997;36:3917–3929. doi: 10.1021/bi9624898. [DOI] [PubMed] [Google Scholar]
- Ballou DP. Crystallography gets the jump on the enzymologists. Proc Natl Acad Sci USA. 2007;104:15587–15588. doi: 10.1073/pnas.0707843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Dowlati A, Posey J, Ramanathan RK, Rath L, Fu P, Chak A, Krishnamurthi S, Brell J, Ingalls S, Hoppel CL, et al. Phase II and pharmacokinetic trial of rebeccamycin analog in advanced biliary cancers. Cancer Chemother Pharmacol. 2009;65:73–78. doi: 10.1007/s00280-009-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan CL, Ryan KS. Divergent Pathways in the Biosynthesis of Bisindole Natural Products. Chemistry & Biology. 2009;16:351–364. doi: 10.1016/j.chembiol.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman MJ, Bauer KS, Jr, Wu S, Smith R, Bisacia S, Dancey J. Phase I and pharmacokinetic study of 7-hydroxystaurosporine and carboplatin in advanced solid tumors. Clin Cancer Res. 2007;13:2667–2674. doi: 10.1158/1078-0432.CCR-06-1832. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enroth C, Neujahr H, Schneider G, Lindqvist Y. The crystal structure of phenol hydroxylase in complex with FAD and phenol provides evidence for a concerted conformational change in the enzyme and its cofactor during catalysis. Structure. 1998;6:605–617. doi: 10.1016/s0969-2126(98)00062-8. [DOI] [PubMed] [Google Scholar]
- Eppink MH, Schreuder HA, Van Berkel WJ. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 1997;6:2454–2458. doi: 10.1002/pro.5560061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhang C, Blanchard S, Thorson JS. Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: insights from the AT2433 biosynthetic locus. Chem Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gatti DL, Palfey BA, Lah MS, Entsch B, Massey V, Ballou DP, Ludwig ML. The mobile flavin of 4-OH benzoate hydroxylase. Science. 1994;266:110–114. doi: 10.1126/science.7939628. [DOI] [PubMed] [Google Scholar]
- Groom K, Bhattacharya A, Zechel DL. Rebeccamycin and staurosporine biosynthesis: insight into the mechanisms of the flavin-dependent monooxygenases RebC and StaC. Chembiochem. 2011;12:396–400. doi: 10.1002/cbic.201000580. [DOI] [PubMed] [Google Scholar]
- Hefti MH, Milder FJ, Boeren S, Vervoort J, van Berkel WJ. A His-tag based immobilization method for the preparation and reconstitution of apoflavoproteins. Biochim Biophys Acta. 2003;1619:139–143. doi: 10.1016/s0304-4165(02)00474-9. [DOI] [PubMed] [Google Scholar]
- Howard-Jones AR, Walsh CT. Staurosporine and rebeccamycin aglycones are assembled by the oxidative action of StaP, StaC, and RebC on chromopyrrolic acid. J Am Chem Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno A, Rudek MA, Purcell T, Laheru DA, Messersmith WA, Dancey J, Carducci MA, Baker SD, Hidalgo M, Donehower RC. Phase I and pharmacokinetic study of UCN-01 in combination with irinotecan in patients with solid tumors. Cancer Chemother Pharmacol. 2008;61:423–433. doi: 10.1007/s00280-007-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Park JS, Chae CS, Hyun CG, Choi BW, Shin J, Oh KB. Genetic organization of the biosynthetic gene cluster for the indolocarbazole K-252a in Nonomuraea longicatena JCM 11136. Appl Microbiol Biotechnol. 2007;75:1119–1126. doi: 10.1007/s00253-007-0924-x. [DOI] [PubMed] [Google Scholar]
- Massey VA. Simple method for the determination of redox potentials. In: Curti B, Ronchi S, Zanetti G, editors. Flavins and Flavoproteins. Berlin, Germany: Walter de Gruyter; 1991. pp. 59–66. [Google Scholar]
- Montersino S, Tischler D, Gassner GT, van Berkel WJH. Catalytic and Structural Features of Flavoprotein Hydroxylases and Epoxidases. Adv Synth Catal. 2011;353:2301–2319. [Google Scholar]
- Nock CJ, Brell JM, Bokar JA, Cooney MM, Cooper B, Gibbons J, Krishnamurthi S, Manda S, Savvides P, Remick SC, et al. A phase I study of rebeccamycin analog in combination with oxaliplatin in patients with refractory solid tumors. Invest New Drugs. 2011;29:126–130. doi: 10.1007/s10637-009-9322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palfey BA, Entsch B, Ballou DP, Massey V. Changes in the catalytic properties of p-hydroxybenzoate hydroxylase caused by the mutation Asn300Asp. Biochemistry. 1994;33:1545–1554. doi: 10.1021/bi00172a035. [DOI] [PubMed] [Google Scholar]
- Ruegg UT, Burgess GM. Staurosporine, K-252 and UCN-01: potent but nonspecific inhibitors of protein kinases. Trends Pharmacol Sci. 1989;10:218–220. doi: 10.1016/0165-6147(89)90263-0. [DOI] [PubMed] [Google Scholar]
- Ryan KS, Chakraborty S, Howard-Jones AR, Walsh CT, Ballou DP, Drennan CL. The FAD Cofactor of RebC Shifts to an IN Conformation upon Flavin Reduction. Biochemistry. 2008;47:13506–13513. doi: 10.1021/bi801229w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KS, Howard-Jones AR, Hamill MJ, Elliott SJ, Walsh CT, Drennan CL. Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC. Proc Natl Acad Sci USA. 2007;104:15311–15316. doi: 10.1073/pnas.0707190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Méndez C, Salas JA. Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. Nat Prod Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- Sánchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc Natl Acad Sci USA. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S, Hirte HW, Carey MS, Hotte SJ, Tsao MS, Brown S, Pond GR, Dancey JE, Oza AM. UCN-01 in combination with topotecan in patients with advanced recurrent ovarian cancer: a study of the Princess Margaret Hospital Phase II consortium. Gynecol Oncol. 2007;106:305–310. doi: 10.1016/j.ygyno.2007.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.