Abstract
In this paper we determine the extent to which host-mediated mutations and a known sampling bias affect evolutionary studies of human influenza A. Previous phylogenetic reconstruction of influenza A (H3N2) evolution using the hemagglutinin gene revealed an excess of nonsilent substitutions assigned to the terminal branches of the tree. We investigate two hypotheses to explain this observation. The first hypothesis is that the excess reflects mutations that were either not present or were at low frequency in the viral sample isolated from its human host, and that these mutations increased in frequency during passage of the virus in embryonated eggs. A set of 22 codons known to undergo such “host-mediated” mutations showed a significant excess of mutations assigned to branches attaching sequences from egg-cultured (as opposed to cell-cultured) isolates to the tree. Our second hypothesis is that the remaining excess results from sampling bias. Influenza surveillance is purposefully biased toward sequencing antigenically dissimilar strains in an effort to identify new variants that may signal the need to update the vaccine. This bias produces an excess of mutations assigned to terminal branches simply because an isolate with no close relatives is by definition attached to the tree by a relatively long branch. Simulations show that the magnitude of excess mutations we observed in the hemagglutinin tree is consistent with expectations based on our sampling protocol. Sampling bias does not affect inferences about evolution drawn from phylogenetic analyses. However, if possible, the excess caused by host-mediated mutations should be removed from studies of the evolution of influenza viruses as they replicate in their human hosts.
It is well known that some pathogenic microbes undergo adaptation in response to laboratory culture. Host-mediated (HM) mutations have been particularly well studied in the influenza A virus (1). However, this phenomenon has been documented in many other viruses, such as HIV, Japanese encephalitis virus, hepatitis A, and Sendai virus as well (2–5). Molecular evolution studies using such sequences thus risk drawing inferences about the adaptation of the pathogen to its natural host from data containing laboratory artifacts. Additional problems may result from analysis of data sets that do not represent random samples of natural pathogen populations, or for which the sampling design is unknown. Here we determine the extent to which HM mutations and a known sampling bias affect studies of influenza A evolution.
Recent phylogenetic reconstruction of the evolution of human influenza A hemagglutinin (HA) of the H3 subtype revealed a 40% excess of amino acid replacements assigned to the terminal branches of the tree (6). The 40% excess of coding changes on terminal branches was calculated by using expectations based on the relative number of internal and terminal branches of the tree in Fig. 1. This observation was made in the course of identifying codons at which mutation appeared to have been adaptive in evading the human immune system. Because we used phylogenetic trees to model HA evolution (7), it was critical for our analyses that the excess mutations not be caused by evolutionary processes other than the ongoing evolution of the virus during replication in the human host. We proposed a number of hypotheses to explain the excess, but did not explore them in detail. Instead we simply deleted all mutations assigned to terminal branches from our analyses. In this paper we have tested two hypotheses that help to explain our observation.
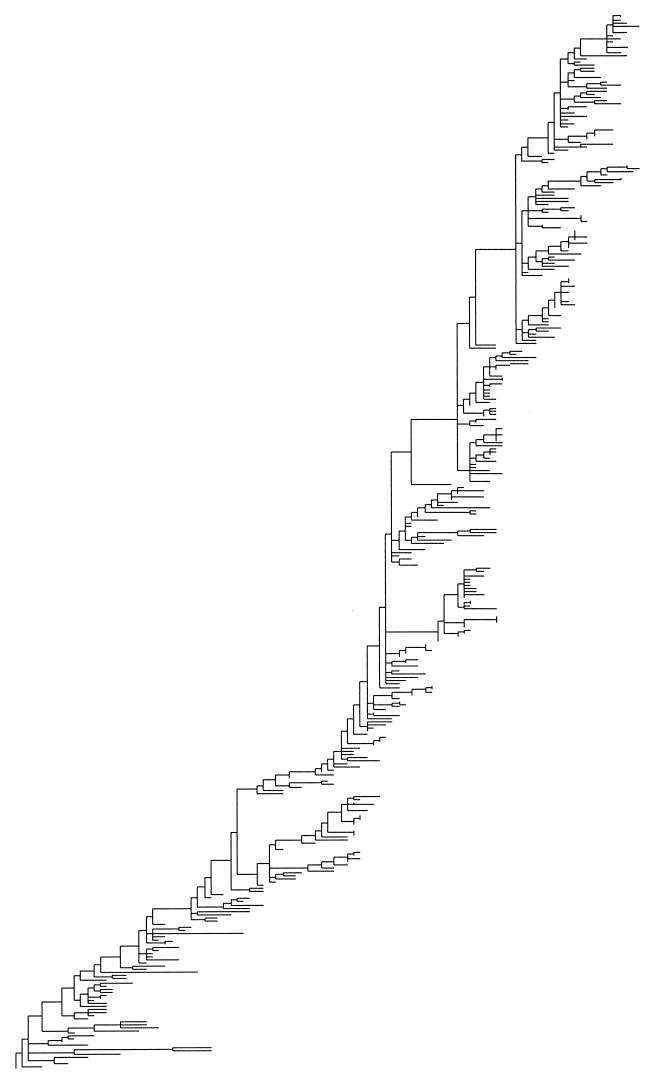
Figure 1.
Maximum parsimony tree constructed from 357 HA1 genes of the human influenza virus type A subtype H3.
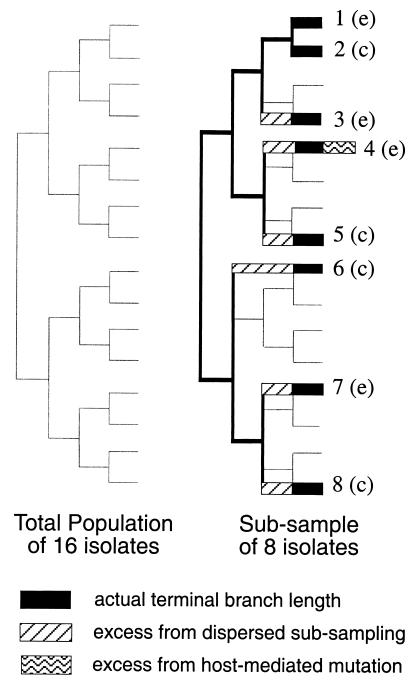
The first hypothesis is that the excess consists of mutations that were either not present or were at low frequency in the viral sample when isolated from its human host. Although such mutations may increase in frequency in the laboratory because of genetic drift, for at least 22 HA1 codons an increase in frequency is thought to reflect a response to selective pressure for growth in embryonated chicken eggs (6). Such HM mutations most likely will appear on a phylogenetic tree as an additional mutation on a terminal branch, which is the branch attaching the sequence from a viral isolate to the tree. Phylogenetic reconstruction is based on similarity at all 329 codons. A HM mutation will alter only one of the 329 codons. Thus, a sequence of an isolate containing a HM mutation would in most cases still be most similar to the sequence from that isolate's closest relative. The effect of the HM mutation on the phylogenetic tree would be an increase in the length of the terminal branch joining the sequence from the egg-cultured isolate to the tree rather than a change in the point at which the branch is attached to the tree (Fig. 2).
Figure 2.
Partitioning mutations assigned to terminal branches of a phylogenetic tree. The tree on the left represents the evolution of a population of 16 viruses that each differ from their ancestor by one unique mutation. The tree on the right is a reconstruction after (i) sampling only eight of the viruses with a bias against sequencing closely related isolates and (ii) propagating the isolates in embryonated chicken eggs (e) or cell culture (c) in the laboratory. The tree constructed of sampled sequences is shown in black, with the terminal branches as thicker lines. The branch attaching sequence 4 (an egg-cultured isolate) to the tree is one mutation longer than it should be. The additional mutation is HM, that is, a mutation not present or at low frequency in the isolate before laboratory propagation. The branches attaching sequences from isolates 3–8 to the tree are longer than they would have been if our sample had included their nearest relatives. The increased length of branch 4 is in part caused by a process other than the ongoing evolution of the virus during replication in the human host. The remaining excess is simply a reflection of sampling bias, and thus does not affect evolutionary inferences made from the tree.
The 22 suspected HM codons (Table 1) make up only 6.7% of the 329 codons in the HA1 domain, yet they account for 36.0% of the amino acid replacements across the HA tree in Fig. 1. Codons other than the set of 22 HM codons also may be found to undergo HM mutation with future study. There is thus great potential for error in inference if one assumes that HM mutations reflect evolution of influenza viruses within the human host. Here we test for the presence of HM mutations in our data set by examining the distribution of mutations in the HM and non-HM codons between branches attaching sequences from egg-cultured and cell-cultured isolates to the tree.
Table 1.
Codons known to undergo HM mutations during propagation in egg culture
| Codon | Rbs | AB | PosSel |
|---|---|---|---|
| 111 | 0 | 0 | 0 |
| 126 | 0 | 1 | 0 |
| 137 | 1 | 1 | 0 |
| 138 | 1 | 1 | 1 |
| 144 | 0 | 1 | 0 |
| 145 | 0 | 1 | 1 |
| 155 | 1 | 1 | 0 |
| 156 | 0 | 1 | 1 |
| 158 | 0 | 1 | 1 |
| 159 | 0 | 1 | 0 |
| 185 | 0 | 0 | 0 |
| 186 | 0 | 1 | 1 |
| 193 | 0 | 1 | 1 |
| 194 | 1 | 1 | 1 |
| 199 | 0 | 0 | 0 |
| 219 | 0 | 0 | 0 |
| 226 | 1 | 0 | 1 |
| 229 | 0 | 0 | 0 |
| 246 | 0 | 0 | 0 |
| 248 | 0 | 0 | 0 |
| 276 | 0 | 0 | 0 |
| 290 | 0 | 0 | 0 |
Five of the 22 HM codons known to undergo HM mutations during propagation in egg culture are associated with the HA sialic acid receptor binding site (Rbs), 12 HM codons are in or near antibody combining sites A or B (AB). Eight HM codons have been identified as having been under positive selection (PosSel) to change the amino acid they encoded in the past (6).
The second hypothesis to explain why we observed excess mutations assigned to the terminal branches of the HA tree is sampling bias. Our sequencing efforts are largely a contribution toward the World Health Organization influenza surveillance program. A priority in influenza surveillance is the identification of antigenically novel isolates from which previous infection with epidemic strains or prior immunization would not protect. The first level of screening for antigenic variants is the HA inhibition (HI) test, in which viral isolates are tested against postinfection ferret antiserum containing antibodies against HA from currently circulating strains of human influenza. We preferentially sequence the HA1 of isolates that appear, on the basis of the HI test, to be antigenically different from known circulating strains.
We illustrate how a bias against sequencing closely related viruses affects phylogenetic reconstruction in Fig. 2. In this hypothetical example, the tree on the left depicts the total population and each branch represents a single unique mutation. The tree on the right was constructed from a subset of eight relatively unrelated isolates. One of the 22 mutations used to construct the right-hand tree (on branch 4) reflects an HM change. Of the remaining 21 mutations, 15 are assigned to the eight terminal branches, and the remaining six mutations are assigned to the six internal branches. If mutations were assigned to terminal and internal branches in proportion to the relative number of each branch type, we would expect to have 12 mutations assigned to the terminal branches. However, we observed 15 mutations on the terminal branches, an excess of 25% over expectations. Thus intentionally sampling with a bias toward genetically divergent isolates results in those isolates being attached to the tree by longer branches than if their close relatives were also in the sample.
Unlike the excess mutations assigned to terminal branches that are caused by HM change, the excess caused by sampling bias is not of concern with respect to the evolutionary inference one might draw from the tree. Apportioning excess terminal branch lengths to the two different hypotheses is easily illustrated in a cartoon such as Fig. 2. In reality, we know that we have observed an excess of mutations on the terminal branches; however, we don't know precisely which branches or mutations are involved. In this paper we show how to determine the proportion, but not the actual identities of HM mutations present in such a data set. After partitioning the excess caused by HM mutations out of the data set, we can determine whether the remaining excess is consistent with what we would expect given our sampling scheme. This was done through comparison with trees produced from sampling a simulated data set.
Description of Data Set and Definition of Terms
Fig. 1 shows the phylogenetic tree for which we recently reported an excess of mutations assigned to the terminal branches (6, 7). This tree was constructed by using the maximum parsimony routine of paup* 4.0b2 (8) using 357 sequences, each 987 nt in length, produced from isolates collected between 1983 and September 30, 1997 (6, 7). The terminal nodes of a tree are the sequences obtained from isolates in the laboratory. Internal nodes are the ancestors of the terminal nodes as reconstructed by the parsimony algorithm. Terminal branches attach terminal nodes, that is, the sequence from an isolate, to the tree. All other branches are internal branches.
We use the term egg isolates when referring to the 152 isolates that were propagated in embryonated chicken eggs in the laboratory. The egg isolates also may have been previously propagated in cell culture. We use the term cell isolates to refer to the 148 isolates propagated in cell culture but never in eggs. The remaining sequences were obtained from direct PCR (n = 3) or from isolates of partially unknown passage history (n = 54). The propagation histories of these isolates (GenBank accession nos. AF008656–AF008909 and AF180564–AF180666) can be found in the curated influenza database at Los Alamos National Laboratory (http://www.flu.lanl.gov/).
For this study we constructed additional trees by using two different samples of the original data set. Trees constructed using only the 152 sequences obtained from isolates propagated in eggs or using only the 148 sequences from viruses propagated in cells are referred to as the egg tree and the cell tree, respectively. Twenty two codons (Table 1) have been reported to undergo HM replacements in influenza isolates grown in eggs (6). We refer to these 22 codons as the HM codons and the other 307 codons as the non-HM codon set. Silent and nonsilent nucleotide substitutions were abbreviated by the letters S and NS, respectively.
Comparing the Phylogenetic Distribution of Nonsilent and Silent Substitutions.
Analyses reported in this paper were performed by using all substitutions, only nonsilent substitutions, or only silent substitutions. Because the results in most cases were very similar, and because nonsilent and silent substitutions are distributed similarly across the internal and terminal branches of the tree in Fig. 1 (Table 2), all analyses reported below used only the nonsilent substitutions, unless stated otherwise.
Table 2.
The distribution of nonsilent (NS) and silent (S) substitutions
| Branch type | Number of branches | Exp NS | Obs NS | χ2 | Exp S | Obs S | χ2 |
|---|---|---|---|---|---|---|---|
| Terminal | 357 | 503.1 | 510 | 0.12 | 405.9 | 399 | 0.12 |
| Internal | 355 | 242.9 | 235 | 0.24 | 196.1 | 204 | 0.25 |
| Sum | 712 | 745.0 | 745 | 0.36 | 603.0 | 603 | 0.37 |
Results of a 2 × 2 contingency test show that nonsilent and silent substitutions are similarly distributed across the terminal and internal branches of the tree in Fig. 1. Total χ2 = 0.73, df = 1, P > 0.4. Obs, observed; Exp, expected.
Reconstructing Ancestral Character States.
In the last step of the process by which parsimony algorithms assign mutations to the branches of a tree, mutations are assigned along each lineage starting at the root and moving along the lineage toward the terminal nodes. In some lineages on the tree in Fig. 1 there was flexibility as to which branches the mutations could be assigned. In our previous work, when there was a choice, we set our algorithm to delay assigning mutations as long as possible (6, 7). That is, mutations were assigned to branches that were as far from the root as possible. We did this to minimize the extent to which HM mutations were assigned to the internal branches of the tree. In the present work, however, one goal is to identify and quantify HM mutations. HM mutations are most likely to be assigned to the terminal branches, thus we did not want to assign mutations to the terminal branches unless it was necessary to do so. Resetting our algorithm to assign mutations as close to the root as possible caused a net change of 14 replacements to be shifted from the terminal to internal branches. This reduced the excess of nonsilent substitutions on the terminal branches from 40.0% to 36.5% (Table 3). Thus our assignment procedure was not responsible for the majority of the observed excess. We retain the assignment procedure that minimizes the number of mutations assigned to the terminal branches for all analyses that follow.
Table 3.
The distribution of nonsilent (NS) substitutions across internal and terminal branches
| Branch type | Number of branches | Exp NS | Obs NS | χ2 |
|---|---|---|---|---|
| Terminal | 357 | 373.6 | 510 | 49.8 |
| Internal | 355 | 371.4 | 235 | 50.1 |
| Sum | 712 | 745.0 | 745 | 99.9 |
The tree in Fig. 1 has significantly more nonsilent substitutions assigned to its terminal branches than expected based on the relative numbers of internal and terminal branches (P < 0.05, df = 1). Exp, expected; Obs, observed.
Hypothesis 1: HM Mutations
We first determined whether there was evidence that HM mutations were contributing to the excess nonsilent substitutions on the terminal branches of the HA tree. We examined two sources of HM mutations. First, we looked for evidence that mutations were occurring at the 22 known HM codons. Second, we determined whether there were any additional codons, besides the 22 in the HM set, that showed evidence for undergoing HM mutations.
HM Mutations in the Egg and Cell Branches.
If HM mutations were occurring in the 22 HM codons, then we should see excess mutations in the HM codons on the egg branches, or the terminal branches attaching sequences from egg-cultured isolates to the tree (Fig. 1). The expectations for this test are based on the distribution of mutations in the non-HM codons across the egg and cell branches. As would be expected if HM mutations were occurring, the set of 22 HM codons underwent a significantly greater number of nonsilent substitutions on the egg branches than expected based on the distribution of mutations at the non-HM codons (Table 4). The number of excess nonsilent substitutions caused by HM change can be estimated as follows. We first assume that the distribution of nonsilent substitutions in the non-HM codons to the egg and cell branches (49.6% and 50.4%, respectively) is unaffected by HM mutation. (We verify this assumption below.) We also assume that none of the 47 nonsilent substitutions in the HM codons on the cell branches were HM. Based on these assumptions the number of nonsilent substitutions we would expect on egg branches is 46.3, which is 58.7 fewer than the 105 observed (Table 4). If the 58.7 “excess” nonsilent substitutions were indeed HM, then approximately 8% of the 745 amino acid replacements in our data set did not occur within a human host.
Table 4.
The distribution of nonsilent (NS) substitutions in HM and non-HM codons across egg and cell branches
| Branch type | Obs NS non-HM | Exp NS HM | Obs NS HM | χ2 |
|---|---|---|---|---|
| Egg branches | 138 | 75.45 | 105 | 11.57 |
| Cell branches | 140 | 76.55 | 47 | 11.40 |
| Sum | 278 | 152.00 | 152 | 22.98 |
The HM codons had significantly more nonsilent substitutions on the terminal branches attaching sequences from egg-cultured isolates to the tree in Fig. 1 than on branches attaching sequences from cell-cultured isolates (P < 0.05, df = 1). Expectations are based on the distribution of nonsilent substitutions in non-HM codons. Obs, observed; Exp, expected.
HM Mutations in the Non-HM Codon Set.
The 22 HM codons may not be the only codons undergoing HM mutation in the HA1 domain. There could be other codons that undergo HM mutation during propagation in eggs but have not as yet been identified as doing so. To explore this possibility we excluded the 22 HM codons from our data set and then contrasted the structure of two trees: one constructed by using only the 152 isolates known to have been grown in egg culture, and the other constructed by using the 148 isolates that had undergone passage in cell but not egg culture (not shown). The egg and cell data sets are similarly sized and contain isolates collected over the same range of time with the same sampling bias. If only the HM codons undergo HM mutation, the egg and cell trees should show similar distributions of replacements across the terminal and internal branches. However, if additional (non-HM) codons are accruing HM mutations, the egg tree should have a larger excess of mutations assigned to the terminal branches than the cell tree. We found a 30% excess of nonsilent substitutions on the terminal branches of both the egg tree and the cell tree (Table 5). Based on this analysis we find no evidence to support the hypothesis that codons in addition to the 22 in the HM codon set are undergoing HM mutations during laboratory passage unless they are doing so at the same rate at which they undergo mutations in response to passage in cell culture.
Table 5.
The phylogenetic distribution of nonsilent (NS) substitutions on trees constructed using only sequences from egg-cultured or cell-cultured isolates and using only non-HM codons
| Number of branches | Exp NS | Obs NS | χ2 | |
|---|---|---|---|---|
| Branches on egg tree | ||||
| Terminal | 152 | 119.3 | 155 | 10.7 |
| Internal | 150 | 117.7 | 82 | 10.8 |
| Total | 302 | 237.0 | 237 | 21.5 |
| Branches on cell tree | ||||
| Terminal | 148 | 121.3 | 158 | 11.1 |
| Internal | 146 | 119.7 | 83 | 11.2 |
| Total | 294 | 241.0 | 241 | 22.3 |
Trees constructed without the HM codons using sequences from isolates propagated in egg culture or in cell culture both showed significant excesses of nonsilent substitutions on their terminal branches (P < 0.05, df = 1 for both tests). The percent excess of nonsilent substitutions on the terminal branches was 30% for both the egg and cell trees.
Hypothesis 2: Sampling Bias
We have shown that HM mutations in the 22 HM codons appear to be responsible for some of the excess mutations on the terminal branches of the HA1 tree in Fig. 1. We also have demonstrated that the non-HM codons do not appear to be undergoing HM mutations. We are now left to explain why there is still, after partitioning out the HM mutations, a 30% excess of mutations on the terminal branches of the egg and cell trees.
As illustrated in Fig. 2, a sampling scheme biased against sequencing closely related viruses will cause an excess of mutations to be assigned to the terminal branches of a tree. We preferentially sequence isolates that we do not believe, based on HI tests, to be closely related to isolates already sequenced. For instance, in the 1996–1997 influenza season we sequenced only 7% of the isolates on which we performed HI assays. Because the isolates sent to the Centers for Disease Control and Prevention from the World Health Organization collaborating laboratories may themselves already be biased against commonly occurring isolates, the bias against sequencing closely related viruses is even greater than 7%. Based on this bias we expect the terminal branches on our trees to be longer on average than they would have been had we sampled randomly. Because we do not know the genetic structure of the influenza population circulating in nature, we cannot know how we actually sampled it. Thus, we cannot calculate the exact distribution of mutations we should expect on the terminal and internal branches of the tree constructed by using our sample. We can, however, determine whether the excess we have observed is consistent with what we would expect based on our sampling protocol.
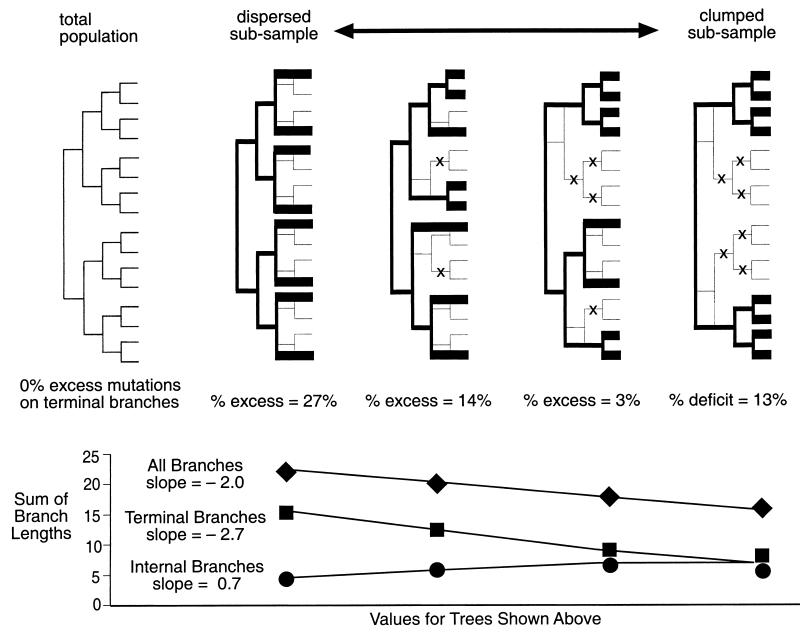
We sampled a simulated viral population using various sampling schemes to determine the extent to which our observation is consistent with this hypothesis. We constructed a hypothetical population of 16 viral isolates and sampled it as illustrated in Fig. 3. The samples consisted of eight relatively unrelated isolates (the dispersed sample), eight closely related isolates (the clumped sample), and two collections of eight isolates sampled in an intermediate manner. To ensure that samples all included the total range of variation in the population, each included the upper-most and bottom-most isolate on the 16-isolate tree. The percent excess of mutations assigned to the terminal branches of the eight-isolate trees was greatly influenced by the degree to which the sampled isolates were dispersed or clustered. The dispersed sample shows a 27.3% excess of mutations assigned to the terminal branches, the clumped sample has a 13% deficit.
Figure 3.
The effects of sampling bias on phylogenetic reconstruction. The tree on the left shows a hypothetical population of 16 isolates that each differ from their ancestor by one unique mutation. The four trees to the right show the original tree overlaid with the tree that would result from sampling only half of the total population. The tree constructed of sampled sequences is shown in black, with the terminal branches as thicker lines. Clumped sampling causes a decrease in the total genetic variation sampled. The mutations not captured in the sample would have been assigned only to internal branches, as shown by the symbol X. As a result, the proportion of mutations assigned to the internal and terminal branches changes with sampling dispersion, but not at the same rate (shown in the line plot at the bottom). Without knowledge of where a sample lies on such a continuum, there is no way to derive the expected proportion of mutations that should be assigned to the terminal and internal branches of a phylogenetic tree.
The magnitude of the excess or deficit depends not only on the degree of dispersion, but also on the proportion of the total population sampled. In the example in Fig. 3, 50% of the total population was sampled. If we were to increase the size of the total population from 16 to 64 and again sample only eight isolates, we would be sampling 12.5% of the total population. This is close to the percent of isolates (7%) that we sequenced based on results from HI tests. Sampling eight dispersed isolates of 64 results in a 47.5% excess of mutations on the terminal branches, a much greater excess than the 27.3% shown in Fig. 3. Thus, even though we do not know the actual distribution of genetic variation present in nature during the time span included in our study, and therefore do not know exactly how we sampled that variation, the magnitude of excess mutations assigned to the terminal branches of the tree in Fig. 1 is consistent with our sampling bias: we have sampled only a fraction of circulating viral strains and have done so in a consciously dispersed manner.
Discussion
We found evidence suggesting that approximately 59 nonsilent substitutions assigned to the terminal branches of the HA tree in Fig. 1 were caused by HM mutations occurring in the set of 22 codons known to undergo HM mutation in chicken eggs in the laboratory. We have no way of identifying which 59 particular substitutions were HM except that they are among the 105 nonsilent substitutions assigned to branches attaching sequences of egg-cultured isolates to the tree. We found no evidence to suggest that HM mutations are occurring at the other 307 codons in the HA1. The majority of the excess mutations that were assigned to terminal branches of the HA tree are most likely simply the result of sampling bias. Detailed antigenic and genetic analysis of viruses collected during influenza surveillance is purposefully biased toward sequencing antigenically dissimilar strains in an effort to identify new antigenic and genetic variants that may signal the need to update the vaccine. Thus, viral isolates that are antigenically very similar to the predominant antigenic variant that circulates during a particular influenza season are sequenced less often than are antigenically variant strains.
The 59 apparently HM mutations represent 7.9% of the 745 nonsilent substitutions that occurred over the time period sampled. Thus, there is good reason for concern about HM mutations if one wants to draw inferences about evolution from this or any similarly affected data set. Culture in live cells is necessary for the propagation not only of viruses, but for many bacteria, such as the obligately intracellular rickettisal and chlamydial bacteria, as well. Laboratories involved in influenza surveillance have long been attuned to the presence of HM mutations. However, people obtaining influenza sequences from public databases might not suspect that the sequences could contain laboratory artifacts. In our previous analyses of these data (6, 7) we dealt with this problem by removing all mutations assigned to terminal branches (70% of the total) from our analyses. Our results indicate that HM mutations are confined to the 22 HM codons, thus, we could take a less drastic approach in the future. For instance, we could assign missing data codes to the HM codons when sequences are obtained from egg-cultured isolates.
We have shown that the excess mutations that remain on the terminal branches after accounting for HM mutations is of a magnitude consistent with expectations given our sampling protocol. Despite our bias toward dispersed sampling, examination of Fig. 1 shows that our data set does contain a number of closely related isolates. To get an idea of how sensitive the calculation of percent excess mutations on the terminal branches is to the degree to which we sampled in a dispersed manner as opposed to clumped manner, we removed 10 of 357, or 2.8%, of the most genetically divergent isolates from our original data set, and constructed a new tree (not shown). The excess of replacements on the terminal branches was reduced from 40% to 32%. Removing 38, or 10.6%, of the most genetically divergent isolates reduced the excess to 28%. Thus the presence of even small numbers of genetically divergent isolates accounts for much of the excess of mutations assigned to the terminal branches of the HA tree.
Unlike the excess mutations on terminal branches caused by sampling bias, the excess caused by HM change could cause problems in studies of how influenza viruses evolve as they replicate in their human hosts. For instance, in our previous work identifying codons under positive selection, we examined the ratio of nonsilent to silent substitutions (6). If an isolate were sequenced shortly after a new amino acid replacement became fixed in a laboratory culture, sequencing viruses from that culture might fail to show silent substitutions that also had occurred during passage but that had been lost in the selective sweep. After fixation of the HM nonsilent substitution, silent substitutions once again would begin to accumulate. Because we do not know the exact circumstances under which HM mutations occurred in our data set relative to the time at which particular isolates were sequenced, we cannot make any predictions about the relative frequencies of nonsilent or silent substitutions in the HM codons as compared with the non-HM codons. However, we can examine the frequencies of nonsilent and silent substitutions in the two codon sets to learn more about how HM codons differ from the non-HM codon set. The HM codons showed significantly greater numbers of nonsilent substitutions than expected (Table 6). As shown in Table 1, eight of the 22 HM codons are among those we previously identified as being under positive selection to change the amino acid they encode. One interpretation of this result is that some of the HM codons are under selection to change the amino acid they encode to adapt to growth in egg culture in addition to being under selection to evade the human immune response.
Table 6.
The distribution of silent and nonsilent substitutions in the HM vs. non-HM codons
| All branches | Obs non-HM | Exp HM | Obs HM | χ2 |
|---|---|---|---|---|
| Nonsilent sub | 478 | 142.76 | 268 | 109.88 |
| Silent sub | 560 | 167.24 | 42 | 93.79 |
| Sum | 1038 | 310.00 | 310 | 203.67 |
The HM codons showed a significant excess of nonsilent substitutions as opposed to silent substitutions compared to expectations based on the non-HM codon set (P < 0.05, df = 1).
The observation of excess mutations assigned to the terminal branches of the HA tree is consistent with expectations based on two very different hypotheses. HM mutations appear to account for part of the excess. The majority of the excess is of a magnitude consistent with expectations based on our sampling protocol, which is biased against sequencing closely related viruses. Unlike the excess caused by sampling bias, excess mutations attributable to HM change reflect processes other than the ongoing evolution of the virus during replication in the human host, and thus should be identified and extracted before making evolutionary inference based on phylogenetic reconstruction of influenza evolution.
Acknowledgments
We gratefully acknowledge the technical expertise of Huang Jing and critical reviews by C. Bergstrom, B. Levin, A. Moya, and K. Subbarao. This work was supported by National Institutes of Health Grant 1R01AI44474–01 and by funds provided by the University of California for the conduct of discretionary research by Los Alamos National Laboratory, conducted under the auspices of the U.S. Department of Energy.
Abbreviations
- HA
hemagglutinin
- HI
HA inhibition
- HM
host-mediated
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Robertson J S. Rev Med Virol. 1993;3:97–106. [Google Scholar]
- 2.Sawyer L S W, Wrin M T, Crawford-Miksza L, Potts B, Wu Y, Weber P A, Alfonso R D, Hanson C V. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J X, Ni H, Wills M R, Campbell G A, Sil B K, Ryman K D, Kitchen I, Barrett A D. J Gen Virol. 1995;76:2757–2764. doi: 10.1099/0022-1317-76-11-2757. [DOI] [PubMed] [Google Scholar]
- 4.Graff J, Normann A, Feinstone S M, Flehmig B. J Virol. 1994;68:548–554. doi: 10.1128/jvi.68.1.548-554.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh M, Isegawa Y, Hotta H, Homma M. J Gen Virol. 1997;78:3207–3215. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 6.Bush R M, Fitch W M, Bender C A, Cox N J. Mol Biol Evol. 1999;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- 7.Bush R M, Bender C A, Subbaro K, Cox N J, Fitch W M. Science. 1999;286:1921–1925. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 8.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1999. , Version 4. [Google Scholar]