Abstract
HLA-G is a nonclassical, class I major histocompatibility complex (MHC) gene that exhibits immunomodulatory properties and likely plays a role in the maintenance of successful pregnancy. In this study, we investigated the role of HLA-G polymorphisms on risk for recurrent pregnancy loss (RPL) and on circulating levels of soluble (s)HLA-G in Iraqi women. DNA and plasma were obtained from blood samples collected at 9 to 12 weeks gestation from 50 women with RPL and 50 healthy pregnant women in Basrah province, Iraq. As measured by ELISA, median sHLA-G levels were significantly lower in the RPL cases compared to healthy controls (21.4 vs. 38.8 U/ml, respectively; P = 0.025), and decreased with increasing maternal age (P = 0.0051). However, HLA-G allele and haplotype frequencies did not differ significantly between cases and controls (P values ≥ 0.12 for all tests). In contrast, homozygosity for the C allele (CC) at a tri- allelic promoter polymorphism, −725C/G/T, was associated with lower concentrations of sHLA- G compared to the CG or CT genotypes (median levels 21.1 vs. 40.1 vs. 42.6 U/ml, respectively; P = 0.0089). These results demonstrate that HLA-G genotype influences circulating sHLA-G levels during pregnancy but is not significantly associated with risk of RPL.
Keywords: HLA-G, recurrent pregnancy loss, soluble HLA-G, HLA-G genotype
1. Introduction
Maternal and fetal immune cells are in close contact during pregnancy, with apparent tolerance of maternal cells toward the fetus and vice versa. It has been suggested that the immunomodulatory molecule, human leukocyte antigen G (HLA-G), contributes to this tolerance, although the precise mechanisms by which this occurs are not completely known [1–3]. HLA-G is considered a “non-classical” class I HLA due to its limited coding region polymorphism and restricted tissue distribution compared to classical class I HLA (HLA-A, HLA-B, HLA-C). Moreover, HLA-G transcripts have the unique property among HLA genes in that they undergo alternative splicing to generate at least seven transcripts and four protein isoforms [4–6].
Despite its restricted tissue distribution, HLA-G protein is expressed under normal physiological conditions, as circulating sHLA-G has been detected in the peripheral blood of healthy men and non-pregnant women [7, 8]. During pregnancy, relatively high levels of circulating sHLA-G have been observed in maternal plasma and/or serum [9–11]; these elevated levels likely reflect HLA-G protein expression from both fetal trophoblast cells and maternal cells, such as monocytes or other immune cells [12, 13]. Decreased levels of sHLA-G have been associated with poor implantation rates in in vitro fertilization [14], increased risk for pregnancy loss [15], and preeclampsia [10, 11, 16, 17] in most, but not all [18], studies. In addition, specific polymorphisms in the HLA-G gene have been associated with sHLA-G levels [8, 19] and with pregnancy outcomes (sporadic miscarriage, RPL, preeclampsia) [20–27] in some, but not all [28– 31], studies.
To date, however, comparisons of multiple HLA-G polymorphisms, sHLA-G concentration, and clinical outcomes of pregnancy have rarely been conduced in the same study population. In addition, the majority of studies of HLA-G and clinical outcomes have focused on women of European or European American descent. That focus results in an incomplete view of HLA-G function because the minor allele frequencies of several HLA-G polymorphisms are too rare in European populations to assess their effects on sHLA-G levels or pregnancy outcome [32, 33]. Therefore, combined studies of HLA-G genotypes, sHLA-G levels, and clinical outcomes in ethnically diverse populations are required to assess the full spectrum of functional or clinical effects of HLA-G variation.
To address these gaps and to better understand the effects of genetic variation in HLA-G on circulating levels of sHLA-G in the first trimester of pregnancy in RPL cases and controls, we initiated studies in women from the Basrah province of Iraq, a population that has not previously been studied for RPL, HLA-G polymorphisms, or sHLA-G concentrations. Specifically, we sought to answer three main questions: i) Do HLA-G allele or haplotype distributions differ between Iraqi women with RPL and control women?, ii) Does genetic variation at HLA-G influence circulating sHLA-G levels in the first trimester of pregnancy?, and iii) How do the observed HLA-G allele and haplotype distributions in the Iraqi women compare to other global populations?
2. Subjects and methods
2.1. Sample Composition
All participants were of Iraqi ancestry and lived in the Basrah province in Iraq. Women (N=50) classified as having recurrent pregnancy loss (RPL), defined by two or more consecutive miscarriages of less than 20 weeks gestation, were recruited from the Obstetrics Unit at the Basrah Maternity and Childrens Hospital. These women presented to the Emergency Room between 9 and 12 weeks gestation with vaginal bleeding and no fetal heart tones; all miscarried while still at the hospital. Prior to the index pregnancy, the RPL women had normal levels of serum progesterone levels in the luteal phase (>10 ng/ml) and normal thyroid function (T3 between 0.9–2.5 nmol/L; T4 between 60–120 nmol/L). In addition, antiphospholipid antibodies, anticardiolipin antibodies, antinuclear antibodies, and TORCH (toxoplasmosis, rubella, cytomegalovirus, herpes simplex virus) studies were negative. These women were treated with either low dose aspirin (75 mg; administered in single dose) or progesterone (5 mg; administered orally twice per day) during the index pregnancy. Control women (N=50) were healthy pregnant women who were recruited between 9 and 12 weeks gestation from the same hospital as the women with RPL. The control women, who were age-matched to the RPL women, were normotensive and delivered a single infant of normal birth weight for gestational age at term. Control women had at least one child and no previous miscarriages, preterm deliveries, or stillbirths. The period of recruitment was from January 2008 to March 2010.
Three milliliters of EDTA-anticoagulated venous blood was collected from each woman with RPL prior to the infusion of intravenous fluids and from each control woman. Plasma was removed following centrifugation (5 minutes at 3000 rpm) of freshly-drawn blood and immediately frozen in 100 L aliquots at −18 C. DNA was extracted from the remaining cells, using the phenol-chloroform extraction protocol of Sambrook et al. [34], and stored frozen until use. Frozen plasma and DNA were shipped on dry ice from Basrah, Iraq to Chicago for genotyping and sHLA-G studies.
All women provided written informed consent. This study was approved by the Training and Development Division in the Basrah Health Office and the University of Basrah, and the Institutional Review Board at the University of Chicago.
2.2. HLA-G Genotyping
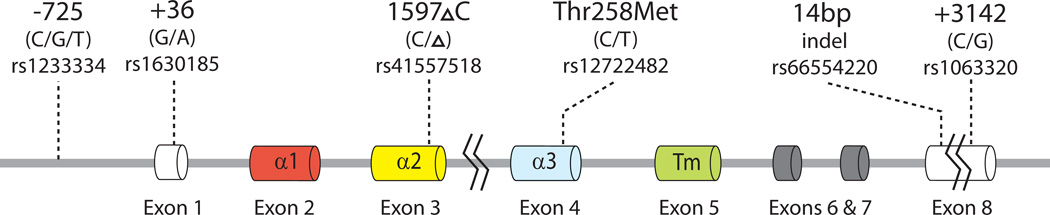
DNA samples were genotyped for six polymorphisms (Fig. 1) by SNaPshot (Applied Biosystems, Carlsbad, CA), using a modification of the protocol of Tan et al. [23] that included the +3142G/C variant [35]. The six polymorphisms genotyped were (i) the −725C/G/T promoter variant (rs1233334) that has been associated with sporadic miscarriage [22] and expression differences in reporter assays [36], (ii) the +36G/A variant (rs1630185) in the untranslated exon 1 that tags two major HLA-G promoter haplotypes [37] (iii) the 3’UTR 14bp insertion/deletion (indel) polymorphism (rs66554220) that has been associated with HLA-G transcript levels [38], circulated sHLA-G levels [8], and preeclampsia [24, 39], (iv) the 1597 C indel (1 bp deletion; rs41557518) in exon 3 that results in a null allele with respect to protein expression [40] and has been associated with RPL [20, 21], (v) the Thr258Met variant (rs12722482) that has been associated with preeclampsia [23, 39], and (vi) the +3142G/C variant (rs1063320) in the 3’UTR that disrupts a micro(mi)RNA target site and influences HLA-G expression levels in the presence of the miRNAs [35].
1.
HLA-G gene structure and location of the six polymorphisms included in this study. Exon 1 encodes a signal peptide, exons 2 and 3 encode the 1 and 2 domains that form the peptide-binding cleft, exon 4 encodes the 3 domain that is involved in receptor binding, exon 5 encodes the transmembrane domain (Tm), and exons 6–8 encode the cytoplasmic domains.
These six variants define the following seven HLA-G alleles or allelic groups (referred to here as haplotypes) (Table 1): G*0101 (G*010101, G*010104, and G*010108 haplotypes cannot be differentiated by the six genotyped variants so will be referred to here as G*0101), G*010101b,c (distinguished from G*0101 by the presence of the −725G, as described in [22, 37]), G*010102/G*010103 (these highly similar haplotypes cannot be differentiated by the genotyped variants), G*0103, G*0104, G*0105N, and G*0106. HLA-G haplotypes were assigned to each woman manually based on the known allelic composition of the six variants on each haplotype [24, 37, 41]. In three women (one case, two controls), the genotypes at the six polymorphic sites were not consistent with known haplotypes, representing either genotyping error or identification of previously unreported haplotypes. Because of limited DNA availability we could not differentiate between these two possibilities, and these samples were excluded from analyses of alleles and haplotypes.
Table 1.
Associations between six genotyped variants and seven imputed HLA-G haplotypes.
| Haplotype | HLA-G Polymorphisms | |||||
|---|---|---|---|---|---|---|
| −725C/G/T | +36G/A | 1597 C | Thr258Met | 14bp indel |
+3142G/C | |
| G*0101^ | C | G | C | Thr | Del | C |
| G*010101b,c | G | G | C | Thr | Del | C |
|
G*010102/ G*010103 |
C | A | C | Thr | Ins | G |
| G*0103 | T | G | C | Thr | Ins | G |
| G*0104 | C | A | C | Thr | Del | G |
| G*0105N | C | A | Thr | Ins | G | |
| G*0106 | C | A | C | Met | Ins | G |
Includes the 010101, 010104, and 010108 haplotypes that cannot be differentiated by these six polymorphisms.
2.3. Measurements of sHLA-G Concentrations
sHLA-G ELISA kits were purchased from EXBIO (Vestec, Czech Republic)/BioVendor (Brno, Czech Republic) for measurement of soluble G5 and shed transmembrane G1 in plasma samples. ELISAs were run according to the manufacturer’s instructions. All samples were run in duplicate; and mean absorbance, measured at a wavelength of 450 nm, were determined for each subject. Calibration curves based on the absorbance of calibrators of known concentration were used to determine the concentration of sHLA-G in each sample.
2.4. Statistical Analyses
Frequencies of alleles at each of the six polymorphisms and for each of the seven haplotypes were compared between RPL cases and healthy controls by the Pearson 2 test, or a Fisher exact test if cell counts were <5. Associations between individual polymorphisms or haplotypes and sHLA-G levels were evaluated by nonparametric methods, using either the Wilcoxon rank sum test (to compare 2 groups) or the Kruskal-Wallis test (to compare 3 or more groups). The effective number of independent tests performed in the analysis of genetic associations with sHLA-G was estimated given the correlation structure among polymorphisms using Li and Ji’s method [42], as implemented in the matrix spectral decomposition (matSpD) program [43]. Results from Li and Ji’s method indicated that the six polymorphisms in HLA-G represented 5.19 independent variables. Significant P-values from the tests of genetic associations were therefore corrected for 5.19 tests using the Bonferroni correction and are presented as Pcorrected. Linear regression was used to test for an association between log sHLA-G concentration and maternal age, and the direction and strength of this association was estimated using the Pearson product-moment correlation coefficient (r). Multivariate analysis of the combined effect of maternal age, −725 genotype, and RPL status on sHLA-G levels was performed on log-transformed sHLA-G measurements using a standard least squares regression. Analyses were performed using JMP software (SAS Institute Inc., Cary, NC), version 8.0.2.2. P-values < 0.05 were considered significant.
3. Results
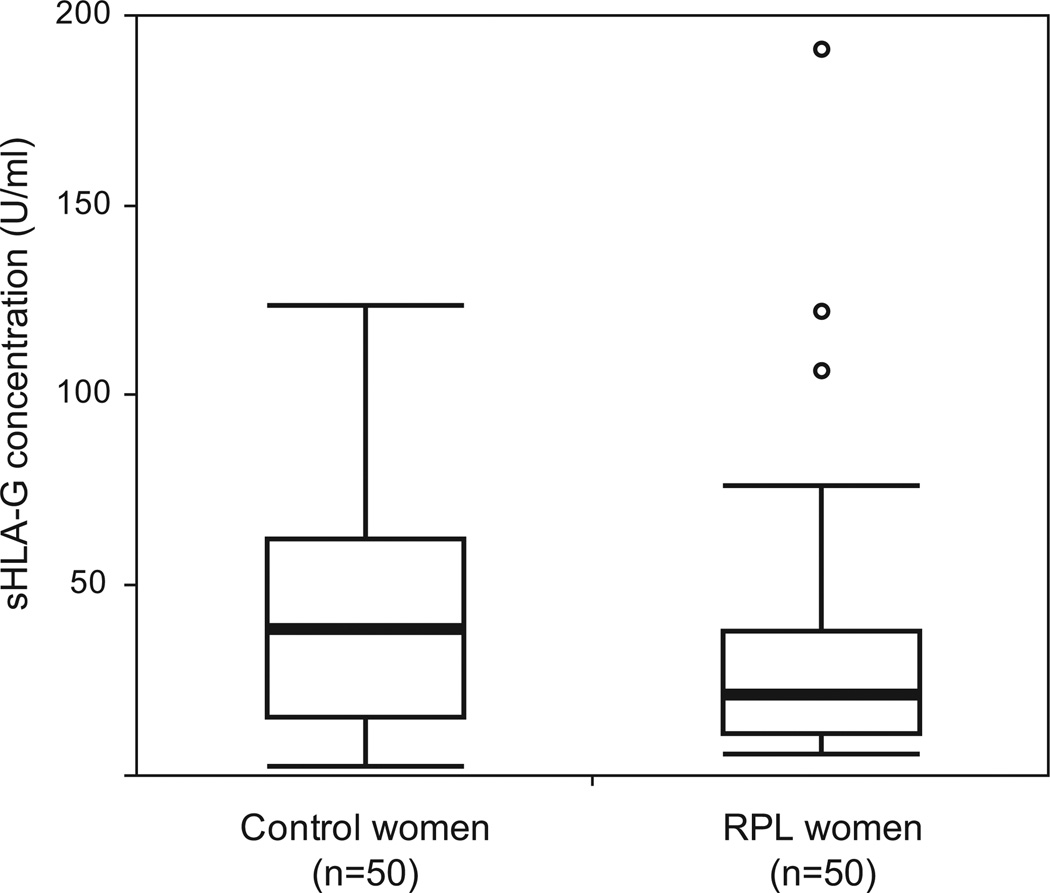
The RPL cases and healthy controls included in this study are described in Table 2. Cases had significantly more pregnancies on average compared to the control women (mean 4.3 vs. 3.4, respectively), whereas control women had significantly more live born children on average compared to the RPL cases (mean 2.4 vs. 0.42, respectively). Circulating levels of sHLA-G in the first trimester were significantly lower in RPL cases compared to controls (median concentration = 21.3 U/ml and 38.8 U/ml, respectively; Wilcoxon rank sum test, P = 0.025) (Fig. 2). Among the RPL cases, sHLA-G levels in women who experienced exactly two previous miscarriages were not significantly different from the levels in women who experienced more than 2 previous miscarriages (Wilcoxon rank sum test, P = 0.33). Log sHLA-G concentration was also significantly negatively correlated with maternal age (F ratio = 8.2, r = −0.28, P = 0.0051) (Supplementary data, Fig. S1), but not with number of prior pregnancies or live births (P> 0.50; data not shown).
Table 2.
Characteristics of the study sample. Means were compared by t-test; P-values are shown.
| RPL Cases (N=50) |
Controls (N=50) |
P-value | |
|---|---|---|---|
| Mean Age ± SD (Range) |
28.4 ± 6.2 yrs (20–40) |
26.7 ± 6.0 yrs (17–39) |
0.16 |
| Mean Number of Pregnancies ± SD (Range) |
4.3 ± 1.9 (2–14) |
3.4 ± 1.4 (2–9) |
0.0097 |
| Mean Number of Prior Live Births ± SD (Range) |
0.42 ± 0.64 (0–2) |
2.4 ± 1.4 (1–8) |
7.8×10−14 |
2.
Soluble HLA-G plasma concentrations in RPL cases and controls. sHLA-G concentration differed significantly between RPL cases and controls (Wilcoxon rank sum test, P = 0.025). Boxes show interquartile range, horizontal lines show the median values and whiskers extend an additional 1.5 interquartile ranges from the boxes. Dots represent outliers.
We were able to successfully genotype 49 RPL cases and 48 controls. All polymorphisms were in Hardy-Weinberg equilibrium in cases only, in controls only, and in the combined sample (data not shown). The frequencies of alleles at each polymorphic site in the cases and controls are shown in Table 3. While none of the allele frequencies were significantly different between the two groups (P 0.12 for all comparisons), we did observe a higher frequency of the insertion allele at the 14 bp indel in the RPL cases compared to controls (0.62 versus 0.51, respectively).
Table 3.
Frequencies of alleles for six HLA-G polymorphisms in 49 RPL cases and 48 healthy controls. Minor allele frequency differences between RPL cases and controls were assessed using the Pearson chi-square test in 2×2 contingency tables.
| Polymorphism | Allele | RPL Cases | Controls | P-value |
|---|---|---|---|---|
| −725C/G/T | C | 0.84 | 0.84 | |
| G | 0.082 | 0.10 | 0.59 | |
| T | 0.082 | 0.052 | 0.41 | |
| +36G/A | A | 0.57 | 0.61 | |
| G | 0.43 | 0.39 | 0.54 | |
| 1597 C | C | 0.93 | 0.91 | |
| 0.071 | 0.094 | 0.57 | ||
| Thr258Met | Thr | 0.87 | 0.86 | |
| Met | 0.13 | 0.14 | 1.00 | |
| 14bp indel | Ins | 0.62 | 0.51 | |
| Del | 0.38 | 0.49 | 0.12 | |
| +3142G/C | G | 0.69 | 0.68 | |
| C | 0.31 | 0.32 | 0.81 | |
The frequencies of each of the seven haplotypes in cases, controls and the combined sample of Iraqi women are shown in Table 4. Overall, the G*0101 and G*010102/G*010103 haplotypes were the most common haplotypes, similar to other populations. However, all other haplotypes were relatively more frequent in the Iraqi women compared to many other populations [3, 22, 37]. For example, the frequency of the G*0105N haplotype (carrying the null 1597 C allele) was 0.082, among the highest frequency reported in populations of non-African ancestry [3, 44, 45], and the frequency of the G*0103 haplotype (carrying the −725T and the Thr31Ser alleles) was 0.072, also among the highest ever reported [3]. Nonetheless, haplotype frequencies did not significantly differ between cases and controls; although the G*0104 haplotype was relatively more common in controls compared to cases (P = 0.12).
Table 4.
Frequencies of seven HLA-G haplotypes in RPL cases (N=49), healthy controls (N=48), and the pooled sample (N=97). Frequency differences between cases and controls were assessed using the Pearson chi-square test in 2×2 contingency tables.
| Haplotype | RPL Cases | Controls | Pooled Sample | P-value |
|---|---|---|---|---|
| G*0101 | 0.22 | 0.20 | 0.21 | 0.65 |
| G*010101b,c | 0.092 | 0.13 | 0.11 | 0.46 |
|
G*010102/ G*010103 |
0.30 | 0.25 | 0.24 | 0.92 |
| G*0103 | 0.092 | 0.052 | 0.072 | 0.29 |
| G*0104 | 0.092 | 0.17 | 0.13 | 0.12 |
| G*0105N | 0.071 | 0.094 | 0.082 | 0.57 |
| G*0106 | 0.12 | 0.11 | 0.12 | 0.86 |
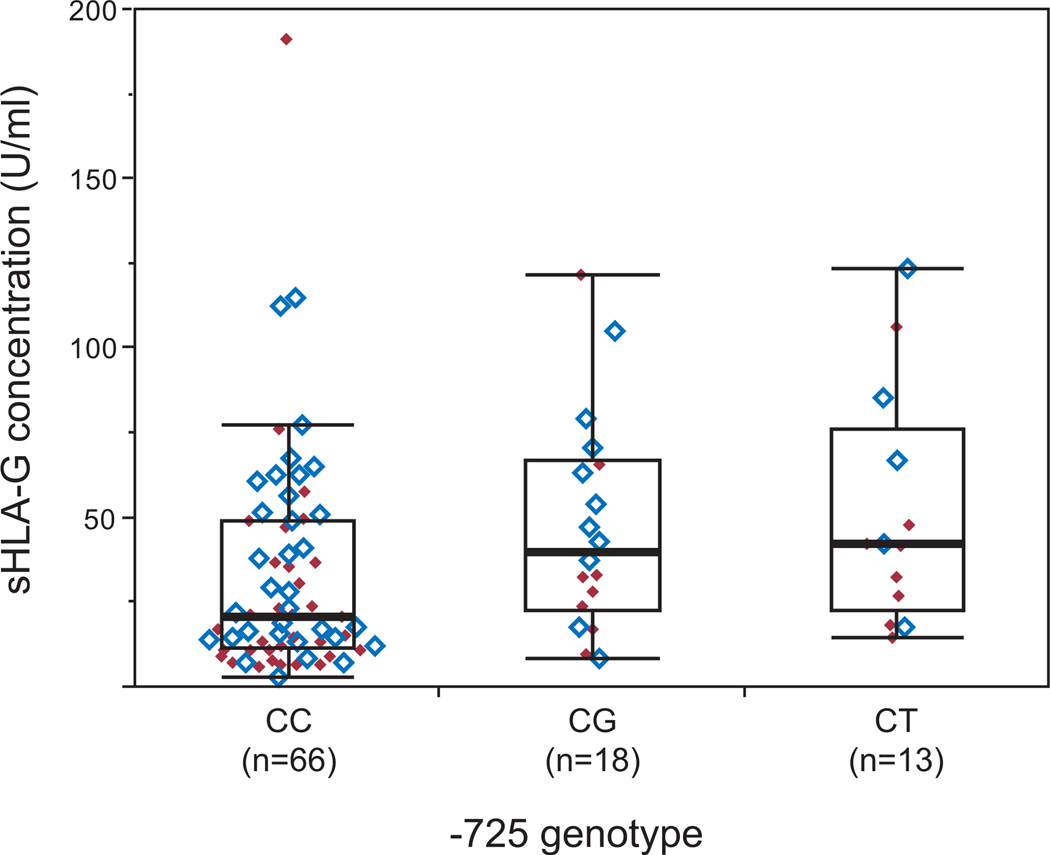
We next examined associations between HLA-G genotype and sHLA-G concentrations (Table 5). Genotype at one polymorphism, −725C/G/T, was significantly associated with sHLA- G levels in the combined sample (Kruskal-Wallis test; P = 0.0089); this association remained significant after correction for multiple tests (Pcorrected = 0.046) (Fig. 3). The concentration of sHLA-G was lowest among women with the common CC genotype (median = 21.1 U/ml) compared to women with the CG (median = 40.1 U/ml) and CT (median 42.6 U/ml) genotypes (no women were homozygous for the G or T alleles). This pattern of association was similar in both the RPL cases and healthy controls (Table 5).
Table 5.
HLA-G genotype association with sHLA-G concentration in the combined, controls-only, and cases-only cohorts. P-values were obtained for each polymorphism using either the Wilcoxon rank sum test or the Kruskal-Wallis test.
| Combined sample (n=97) | Controls-only (n=48) | Cases-only (n=49) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | Genotype | |||||||||
| n | Median sHLA-G, U/ml (interquartile range) |
P | n | Median sHLA-G, U/ml (interquartile range) |
P | n | Median sHLA-G, U/ml (interquartile range) |
P | ||
| −725C/G/T | CC | 66 | 21.1 (11.9–49.1) | 0.0089 | 33 | 28.1 (14.7–58.8) | 0.076 | 33 | 15.6 (10.2–36.2) | 0.053 |
| CG | 18 | 40.1 (22.6–67.0) | 10 | 50.8 (32.3–72.8) | 8 | 30.7 (19.1–57.6) | ||||
| CT | 13 | 42.6 (22.7–76.3) | 5 | 66.8 (30.4–104.8) | 8 | 37.2 (20.6–46.6) | ||||
| +36G/A | AA | 37 | 23.6 (13.5–50.4) | 0.87 | 18 | 38.8 (14.5–54.6) | 0.89 | 19 | 17.1 (11.1–49.0) | 0.81 |
| AG | 41 | 32.7 (13.6–61.6) | 23 | 43.0 (15.7–65.1) | 18 | 23.9 (11.1–36.1) | ||||
| GG | 19 | 28.1 (17.3–47.3) | 7 | 28.1 (18.0–67.5) | 12 | 26.4 (15.3–40.6) | ||||
| 1597 C | CC | 81 | 28.1 (14.6–55.5) | 0.73 | 39 | 41.3 (16.5–65.1) | 0.51 | 42 | 22.5 (11.1–38.1) | 0.99 |
| C | 16 | 28.0 (11.8–47.5) | 9 | 39.2 (13.9–46.1) | 7 | 17.1 (11.2–49.0) | ||||
| Thr258Met | Thr/Thr | 74 | 26.2 (14.3–48.3) | 0.48 | 36 | 37.8 (15.9–62.1) | 0.17 | 38 | 22.6 (11.2–38.1) | 0.67 |
| Thr/Met | 20 | 34.4 (15.1–69.7) | 11 | 51.9 (29.6–70.7) | 9 | 20.9 (10.1–51.7) | ||||
| Met/Met | 3 | 21.3 (14.0–49.7) | 1 | 14.0 | 2 | 35.5 (21.2–49.7) | ||||
| 14bp indel | Ins/Ins | 36 | 25.2 (13.8–49.6) | 0.92 | 14 | 45.9 (17.9–63.6) | 0.83 | 22 | 19.7 (11.2–38.6) | 0.57 |
| Ins/Del | 38 | 28.0 (14.3–62.7) | 21 | 23.2 (15.3–64.2) | 17 | 32.4 (11.9–45.3) | ||||
| Del/Del | 23 | 28.6 (16.9–28.58) | 13 | 38.4 (17.5–62.1) | 10 | 22.8 (9.2–32.2) | ||||
| +3142G/C | GG | 49 | 26.8 (13.7–51.5) | 0.95 | 23 | 41.3 (14.8–66.8) | 0.72 | 26 | 19.9 (11.1–47.7) | 0.74 |
| CG | 35 | 32.4 (14.6–56.7) | 19 | 37.2 (15.7–62.5) | 16 | 31.6 (13.1–40.7) | ||||
| CC | 13 | 24.2 (17.1–42.0) | 6 | 37.7 (17.8–76.9) | 7 | 21.4 (10.0–28.6) | ||||
3.
Soluble HLA-G plasma concentrations by −725C/G/T genotype. sHLA-G concentration was significantly associated with −725 genotype (Kruskal-Wallis test, P = 0.0089). Boxes show interquartile range, horizontal lines show the median values and whiskers extend an additional 1.5 interquartile ranges from the boxes. RPL cases are indicated by solid red diamonds and controls by open blue diamonds.
Lastly, we assessed the combined effects of −725C/G/T genotype, maternal age, and case/control status on sHLA-G concentrations by multivariate linear regression. The model that included all three of these predictor variables was highly significant (F ratio = 7.57, P-value = 1.37x10−4), explaining 19.6% of the variance in log sHLA-G levels. Notably, both −725 genotype and maternal age were significant predictors in the multivariate model (P = 0.012 and P = 0.0017, respectively), whereas the effect of RPL status on log sHLA-G levels failed to achieve statistical significance (P = 0.066). In the multivariate model, maternal age and −725 genotype account for 5.7% and 9.0%, respectively, of the total variance in log sHLA-G concentration. Overall, these results indicate that HLA-G genotype and maternal age independently influence circulating concentrations of sHLA-G during the first trimester of pregnancy in the Iraqi women.
4. Discussion
We present here the first study of HLA-G in Iraqi women and examine the effects of genotypes on RPL and circulating levels of sHLA-G. We report here for the first time a significant association between the HLA-G promoter polymorphism, −725C/G/T, and sHLA-G concentrations in the first trimester of pregnancy in women with RPL and in healthy controls, and show that sHLA-G concentrations significantly decrease with increasing maternal age. Perhaps not surprisingly, sHLA-G concentrations were lower in women experiencing a miscarriage. However, because −725C/G/T genotype frequencies did not differ between RPL cases and healthy controls and the effect of pregnancy status (RPL case versus control) was reduced when genotype and maternal age were included in a multivariate model, we suggest that reduced concentrations of circulating sHLA-G in the RPL cases resulted from the miscarriage but were not necessarily the cause of the pregnancy loss.
There have been few studies of the HLA-G −725C/G/T polymorphism, but as part of a comprehensive study of variation in the promoter region of HLA-G, we previously reported an association between the −725G allele and sporadic miscarriage in fertile couples (the −725T allele was not surveyed at that time) [22]. We subsequently demonstrated that the G allele was associated with increased HLA-G expression in a reporter assay [36], consistent with the findings in the current study (Fig. 3). In our earlier study, we reported that constructs carrying the T allele were not higher expressers in untreated JEG3 (placental) cells or in cells treated with IFN- or with a CpG methylase (M. SssI). However, luciferase expression was 2–3 times higher for constructs carrying either the −725G or −725T alleles compared to those with the −725C allele in JEG3 cells treated with both IFN- and M. SssI (see Figure 2D in Ober et al. 2006). That is, similar to the results in the current study, all promoters carrying the common −725C allele were associated with lower expression compared to promoters carrying either the −725G or −725T alleles, under specific conditions. Although we do not know the mechanism accounting for the genotype-specific effects on circulating sHLA-G concentrations in the current study, these earlier experiments indicate that −725 alleles influence gene expression differently in different cellular environments. The current study further shows that women who are homozygous for the C allele are the lowest expressers of sHLA-G in the first trimester of pregnancy.
Most previous studies of HLA-G genotypes and sHLA-G or HLA-G genotypes and pregnancy outcomes were conducted in populations of European descent. Yet, most of the known HLA-G alleles and haplotypes are relatively rare in European populations. As a result, assessing the effects of the less common haplotypes or alleles on clinical phenotypes requires studying non-European populations. This fact has been appreciated in studies of the null 1597 C allele, which defines the G*0105N haplotype, because this allele occurs at relatively high frequencies (0.05–0.12) in populations of African descent, but is quite rare in European and east Asian populations (0–0.05) [20, 40, 46–49]. Surprisingly, the frequency of this allele is also quite high in Iraqi women (0.083). This is consistent with recent reports of strikingly high frequencies (0.18 and 0.14) of the 1597 C allele in Iranian and east Indian populations, respectively [50, 51]. It is possible, therefore, that this variant occurs at highest frequency in the Middle East and South Asia, and not in Africa, as previously thought [48, 52].
Given the high frequency of the 1597 C allele in Iraqi women, it was unexpected that this allele was not associated with sHLA-G concentrations because it is a proven null allele for the G1 and G5 isoforms [40], which are the two isoforms measured by the HLA-G ELISA used in this study. It is possible that maternal genotype for the null allele is not fully predictive of circulating levels during pregnancy, for example due to compensation by the second, non- G*10105N allele in heterozygous individuals or due to the contribution of the fetal-origin sHLA- G, both of which would reduce power to detect associations between the null allele and plasma sHLA-G levels. Because of the low frequency of the 1597 C null allele in most populations, there have been no previous studies with adequate power to detect associations between the null allele and circulating sHLA-G concentration. However, the null allele has been associated with RPL in European and European American women [20, 21], but not in Han Chinese women [53], reflecting potential heterogeneity in genetic associations with RPL. Additional studies of both maternal and fetal genotypes, and in larger sample sizes, will be necessary to evaluate these hypotheses.
One haplotype and one allele occur at high frequencies in Iraqi women compared to European populations. The G*0106 haplotype, carrying the Met258Thr variant, occurs at frequencies <0.07 in most European populations [22, 39, 54, 55], whereas the frequency of this haplotype in Iraqi women was observed to be 0.12. The −725T allele occurs at frequencies of 0.022 in European Americans, 0.102 in African Americans [37], and 0.12 in Iraqi women, whereas, the −725G allele occurs at frequencies of 0.12 in European Americans, 0.068 in African Americans [37], and 0.11 in Iraqi women. Therefore, non-C alleles occur at 23% frequency in Iraqi women and, as a result, the −725 CC genotype is less common in Iraqi women compared to either European American or African American subjects.
A limitation of our study is that our sample size does not provide adequate power to identify genetic variants with small to moderate effect sizes. In the present study, the 14bp indel polymorphism showed the largest allele frequency difference between RPL cases and controls. Given the observed 14bp indel allele frequencies and sample size, our study had approximately 80% power to detect an effect size corresponding to an odds ratio of 3.6 or larger. Although we observed a non-significant increase in the 14bp insertion allele frequency in Iraqi RPL cases and an odds ratio (OR=1.69; 95%CI 0.87–2.2) similar to that reported in previous studies [26, 27], we were underpowered to detect an association at a P value less than 0.05. Thus, a larger sample size would be needed to detect the small to moderate effect sizes typical of genetic variants associated with complex diseases such as RPL.
This study provides novel insights into the regulation of circulating levels of sHLA-G in early pregnancy and clinical outcome. First, the lack of an association between the −725 CC genotype and RPL, despite the observation of significantly lower sHLA-G concentrations in CC women, suggests that constitutively low levels of sHLA-G due to −725 genotype is not a cause of recurrent pregnancy loss in these women. This would be consistent with the observation that CC is the most common genotype at this site, and indicates that the genotype-specific differences observed in this study are likely well within the range required for successful implantation and maintenance of pregnancy. Interestingly, even at 9–12 weeks of gestation in spontaneously terminating pregnancies, the genotype effects on sHLA-G concentrations are still apparent. Whether these genotype effects on sHLA-G concentrations remain throughout pregnancy, or whether they predict outcomes later in pregnancy remains to be determined. Second, we report an unexpected association between sHLA-G concentrations and maternal age that is independent of the HLA-G genotype effect. This observation needs to be replicated in additional studies, and examined in later trimesters of pregnancy. If confirmed, reduced concentrations of sHLA-G in the first trimester in older mothers may be a contributory mechanism, and a possible marker, for the increased risk for adverse pregnancy outcomes in later pregnancy, such as preeclampsia and preterm birth, among these women [56–58], a hypothesis that can be examined in future prospective studies.
In summary, the relationship between HLA-G genotype, circulating sHLA-G concentrations in the first trimester, and adverse pregnancy outcomes remains complex. This study identifies two independent determinants of sHLA-G concentrations in the first trimester of pregnancy, including genotype at a promoter polymorphism, −725C/G/T, that was previously demonstrated to have functional effects on transcription [36], and maternal age, which is itself a well established risk factor for adverse pregnancy outcomes throughout pregnancy. Further studies are required to elucidate the clinical effects of these observations throughout pregnancy and in ethnically diverse women.
Supplementary Material
Acknowledgements
We thank Joan Hunt, Margaret Petroff, and Mary Stephenson for extensive discussions and helpful comments on this manuscript; and K.S. Dhamia, Adnan Al-Badran, and Ayat Al- Laaeiby for isolating cell samples at Basrah University. This work was supported by P01 HD049480. R.M.J.’s research was supported in part by the Ministry of Higher Education in Iraq; D.A.L. was supported in part by F32 HL095268 and T32 HL007605.
Appendix. Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 2.Carosella ED, Favier B, Rouas-Freiss N, Moreau P, LeMaoult J. Beyond the increasing complexity of the immunomodulatory HLA-G molecule. Blood. 2008;111:4862–4870. doi: 10.1182/blood-2007-12-127662. [DOI] [PubMed] [Google Scholar]
- 3.Hviid TVF. HLA-G in human reproduction: aspects of genetics, function and pregnancy complications. Hum Reprod Update. 2006;12:209–232. doi: 10.1093/humupd/dmi048. [DOI] [PubMed] [Google Scholar]
- 4.Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class-I and class-II antigens. Proc Natl Acad Sci U S A. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morales PJ, Pace JL, Platt JS, Phillips TA, Morgan K, Fazleabas AT, et al. Placental cell expression of HLA-G2 Isoforms is limited to the invasive trophoblast phenotype. J Immunol. 2003;171:6215–6224. doi: 10.4049/jimmunol.171.11.6215. [DOI] [PubMed] [Google Scholar]
- 6.Fujii T, Ishitani A, Geraghty DE. A soluble form of the HLA-G antigen is encoded by a messenger ribonucleic acid containing intron 4. J Immunol. 1994;153:5516–5524. [PubMed] [Google Scholar]
- 7.Rebmann V, van der Ven K, Passler M, Pfeiffer K, Krebs D, Grosse-Wilde H. Association of soluble HLA-G plasma levels with HLA-G alleles. Tissue Antigens. 2001;57:15–21. doi: 10.1034/j.1399-0039.2001.057001015.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen XY, Yan WH, Lin A, Xu HH, Zhang JG, Wang XX. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens. 2008;72:335–341. doi: 10.1111/j.1399-0039.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- 9.Hunt JS, Jadhav L, Chu W, Geraghty DE, Ober C. Soluble HLA-G circulates in maternal blood during pregnancy. Am J Obstet Gynecol. 2000;183:682–688. doi: 10.1067/mob.2000.106762. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo R, Andersen AS, Lassen MR, Sorensen HC, Bergholt T, Larsen MH, et al. Soluble human leukocyte antigen-G isoforms in maternal plasma in early and late pregnancy. Am J Reprod Immunol. 2009;62:320–338. doi: 10.1111/j.1600-0897.2009.00742.x. [DOI] [PubMed] [Google Scholar]
- 11.Steinborn A, Varkonyi T, Scharf A, Bahlmann F, Klee A, Sohn C. Early detection of decreased soluble HLA-G levels in the maternal circulation predicts the occurrence of preeclampsia and intrauterine growth retardation during further course of pregnancy. Am J Reprod Immunol. 2007;57:277–286. doi: 10.1111/j.1600-0897.2007.00475.x. [DOI] [PubMed] [Google Scholar]
- 12.Feger U, Tolosa E, Huang YH, Waschbisch A, Biedermann T, Melms A, et al. HLA-G expression defines a novel regulatory T-cell subset present in human peripheral blood and sites of inflammation. Blood. 2007;110:568–577. doi: 10.1182/blood-2006-11-057125. [DOI] [PubMed] [Google Scholar]
- 13.Alegre E, Diaz-Lagares A, LeMaoult J, Lopez-Moratalla N, Carosella ED, Gonzalez A. Maternal antigen presenting cells are a source of plasmatic HLA-G during pregnancy: Longitudinal study during pregnancy. Hum Immunol. 2007;68:661–667. doi: 10.1016/j.humimm.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Rizzo R, Melchiorri L, Stignani M, Baricordi OR. HLA-G expression is a fundamental prerequisite to pregnancy. Hum Immunol. 2007;68:244–250. doi: 10.1016/j.humimm.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Pfeiffer KA, Rebmann V, Passler M, van der Ven K, van der Ven H, Krebs D, et al. Soluble HLA levels in early pregnancy after in vitro fertilization. Hum Immunol. 2000;61:559–564. doi: 10.1016/s0198-8859(00)00123-3. [DOI] [PubMed] [Google Scholar]
- 16.Yie SM, Taylor RN, Librach C. Low plasma HLA-G protein concentrations in early gestation indicate the development of preeclampsia later in pregnancy. Am J Obstet Gynecol. 2005;193:204–208. doi: 10.1016/j.ajog.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 17.Hackmon R, Koifman A, Hyobo H, Glickman H, Sheiner E, Geraghty DE. Reduced third-trimester levels of soluble human leukocyte antigen G protein in severe preeclampsia. Am J Obstet Gynecol. 2007;197 doi: 10.1016/j.ajog.2007.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Steinborn A, Rebmann V, Scharf A, Sohn C, Grosse-Wilde H. Placental abruption is associated with decreased maternal plasma levels of soluble HLA-G. J Clin Immunol. 2003;23:307–314. doi: 10.1023/a:1024592901663. [DOI] [PubMed] [Google Scholar]
- 19.Hviid TVF, Rizzo R, Christiansen OB, Melchiorri L, Lindhard A, Baricordi OR. HLA-G and IL-10 in serum in relation to HLA-G genotype and polymorphisms. Immunogenetics. 2004;56:135–141. doi: 10.1007/s00251-004-0673-2. [DOI] [PubMed] [Google Scholar]
- 20.Aldrich CL, Stephenson MD, Karrison T, Odem RR, Branch DW, Scott JR, et al. HLA-G genotypes and pregnancy outcome in couples with unexplained recurrent miscarriage. Mol Hum Reprod. 2001;7:1167–1172. doi: 10.1093/molehr/7.12.1167. [DOI] [PubMed] [Google Scholar]
- 21.Pfeiffer KA, Fimmers R, Engels G, van der Ven H, van der Ven K. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol Hum Reprod. 2001;7:373–378. doi: 10.1093/molehr/7.4.373. [DOI] [PubMed] [Google Scholar]
- 22.Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, et al. Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet. 2003;72:1425–1435. doi: 10.1086/375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan CY, Ho JFV, Chong YS, Loganath A, Chan YH, Ravichandran J, et al. Paternal contribution of HLA-G*0106 significantly increases risk for pre-eclampsia in multigravid pregnancies. Mol Hum Reprod. 2008;14:317–324. doi: 10.1093/molehr/gan013. [DOI] [PubMed] [Google Scholar]
- 24.Larsen MH, Hylenius S, Andersen AMN, Hviid TVF. The 3'-untranslated region of the HLA-G gene in relation to pre-eclampsia: revisited. Tissue Antigens. 2010;75:253–261. doi: 10.1111/j.1399-0039.2009.01435.x. [DOI] [PubMed] [Google Scholar]
- 25.Yie SM, Li LH, Xiao R, Librach CL. A single base-pair mutation in the 3'-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol Hum Reprod. 2008;14:649–653. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
- 26.Hviid TVF, Hylenius S, Lindhard A, Christiansen OB. Association between human leukocyte antigen-G genotype and success of in vitro fertilization and pregnancy outcome. Tissue Antigens. 2004;64:66–69. doi: 10.1111/j.1399-0039.2004.00239.x. [DOI] [PubMed] [Google Scholar]
- 27.Yan WH, Lin A, Chen XJ, Dai MZ, Gan LH, Zhou MY, et al. Association of the maternal 14-bp insertion polymorphism in the HLA-G gene in women with recurrent spontaneous abortions. Tissue Antigens. 2006;68:521–523. doi: 10.1111/j.1399-0039.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 28.Aldrich C, Verp MS, Walker MA, Ober C. A null mutation in HLA-G is not associated with preeclampsia or intrauterine growth retardation. J Reprod Immunol. 2000;47:41–48. doi: 10.1016/s0165-0378(00)00052-8. [DOI] [PubMed] [Google Scholar]
- 29.Iversen AC, Nguyen OTD, Tommerdal LF, Eide IP, Landsem VM, Acar N, et al. The HLA-G 14bp gene polymorphism and decidual HLA-G 14bp gene expression in pre-eclamptic and normal pregnancies. J Reprod Immunol. 2008;78:158–165. doi: 10.1016/j.jri.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Vianna P, Dalmaz CA, Veit TD, Tedoldi C, Roisenberg I, Chies JAB. Immunogenetics of pregnancy: Role of a 14-bp deletion in the maternal HLA-G gene in primiparous pre-eclamptic Brazilian women. Hum Immunol. 2007;68:668–674. doi: 10.1016/j.humimm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Lin A, Yan WH, Dai MZ, Chen XJ, Li BL, Chen BG, et al. Maternal human leukocyte antigen-G polymorphism is not associated with pre-eclampsia in a Chinese Han population. Tissue Antigens. 2006;68:311–316. doi: 10.1111/j.1399-0039.2006.00667.x. [DOI] [PubMed] [Google Scholar]
- 32.Aruna M, Sudheer PS, Andal S, Tarakeswari S, Reddy AG, Thangaraj K, et al. HLA-G polymorphism patterns show lack of detectable association with recurrent spontaneous abortion. Tissue Antigens. 2010;76:216–222. doi: 10.1111/j.1399-0039.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 33.Hviid TV, Hylenius S, Hoegh AM, Kruse C, Christiansen OB. HLA-G polymorphisms in couples with recurrent spontaneous abortions. Tissue Antigens. 2002;60:122–132. doi: 10.1034/j.1399-0039.2002.600202.x. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ober C, Billstrand C, Kuldanek S, Tan Z. The miscarriage-associated HLA-G-725G allele influences transcription rates in JEG-3 cells. Hum Reprod. 2006;21:1743–1748. doi: 10.1093/humrep/del036. [DOI] [PubMed] [Google Scholar]
- 37.Tan Z, Shon AM, Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet. 2005;14:3619–3628. doi: 10.1093/hmg/ddi389. [DOI] [PubMed] [Google Scholar]
- 38.Hviid TV, Hylenius S, Rorbye C, Nielson LG. HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics. 2003;55:63–79. doi: 10.1007/s00251-003-0547-z. [DOI] [PubMed] [Google Scholar]
- 39.Moreau P, Contu L, Alba F, Lai S, Simoes R, Orru S, et al. HLA-G gene polymorphism in human placentas: Possible association of G*0106 allele with preeclampsia and miscarriage. Biol Reprod. 2008;79:459–467. doi: 10.1095/biolreprod.108.068874. [DOI] [PubMed] [Google Scholar]
- 40.Ober C, Aldrich C, Rosinsky B, Robertson A, Walker MA, Willadsen S, et al. HLA-G1 protein expression is not essential for fetal survival. Placenta. 1998;19:127–132. doi: 10.1016/s0143-4004(98)90000-5. [DOI] [PubMed] [Google Scholar]
- 41.Ober C, Rosinsky B, Grimsley C, van der Ven K, Robertson A, Runge A. Population genetic studies of HLA-G: allele frequencies and linkage disequilibrium with HLA-A. J Reprod Immunol. 1996;32:111–123. doi: 10.1016/s0165-0378(96)01000-5. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 43.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin A, Li M, Xu DP, Zhang WG, Yan WH. Ethnic variation of the HLA-G*0105N allele in two Chinese populations. Tissue Antigens. 2009;73:270–274. doi: 10.1111/j.1399-0039.2008.01196.x. [DOI] [PubMed] [Google Scholar]
- 45.Ober C, Aldrich CL. HLA-G polymorphisms: neutral evolution or novel function? J Reprod Immunol. 1997;36:1–21. doi: 10.1016/s0165-0378(97)00062-4. [DOI] [PubMed] [Google Scholar]
- 46.van der Ven K, Skrablin S, Engels G, Krebs D. HLA-G polymorphisms and allele frequencies in Caucasians. Hum Immunol. 1998;59:302–312. doi: 10.1016/s0198-8859(98)00021-4. [DOI] [PubMed] [Google Scholar]
- 47.Tian W, Cai JH, Wang F, Li LX, Cao Y. HLA-G*0105N and HLA-G 14 bp dimorphisms in exon 8 in four distinct populations in mainland China. Tissue Antigens. 2010;75:227–234. doi: 10.1111/j.1399-0039.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 48.Ishitani A, Kishida M, Sageshima N, Yashiki S, Sonoda S, Hayami M, et al. Re-examination of HLA-G polymorphism in African Americans. Immunogenetics. 1999;49:808–811. doi: 10.1007/s002510050555. [DOI] [PubMed] [Google Scholar]
- 49.Matte C, Lacaille J, Zijenah L, Ward B, Roger M, et al. ZVITAMBO Study Group. HLA-G and HLA-E polymorphisms in an indigenous African population. Hum Immunol. 2000;61:1150–1156. doi: 10.1016/s0198-8859(00)00200-7. [DOI] [PubMed] [Google Scholar]
- 50.Rahimi R, Hosseini AZ, Yari F. The polymorphism of human leucocyte antigen-G gene in a healthy population of Iran. International journal of immunogenetics. 2010;37:269–272. doi: 10.1111/j.1744-313X.2010.00919.x. [DOI] [PubMed] [Google Scholar]
- 51.Abbas A, Tripathi P, Naik S, Agrawal S. Analysis of human leukocyte antigen (HLA)-G polymorphism in normal women and in women with recurrent spontaneous abortions. Eur J Immunogenet. 2004;31:275–278. doi: 10.1111/j.1365-2370.2004.00487.x. [DOI] [PubMed] [Google Scholar]
- 52.Aldrich CL, Wambebe C, Odama L, Di Rienzo A, Ober C. Linkage disequilibrium and age estimates of a deletion polymorphism (1597deltaC) in HLA-G suggest non-neutral evolution. Hum Immunol. 2002;63:405–412. doi: 10.1016/s0198-8859(02)00377-4. [DOI] [PubMed] [Google Scholar]
- 53.Yan WH, Fan LA, Yang JQ, Xu LD, Ge Y, Yao FJ. HLA-G polymorphism in a Chinese Han population with recurrent spontaneous abortion. Int J Immunogenet. 2006;33:55–58. doi: 10.1111/j.1744-313X.2006.00567.x. [DOI] [PubMed] [Google Scholar]
- 54.Sipak-Szmigiel O, Cybulski C, Wokolorczyk D, Lubinski J, Kurzawa R, Baczkowski T, et al. HLA-G polymorphism and in vitro fertilization failure in a Polish population. Tissue Antigens. 2009;73:348–352. doi: 10.1111/j.1399-0039.2008.01205.x. [DOI] [PubMed] [Google Scholar]
- 55.Hviid TV, Christiansen OB, Johansen JK, Hviid UR, Lundegaard C, Moller C, et al. Characterization of a new HLA-G allele encoding a nonconservative amino acid substitution in the alpha 3 domain (exon 4) and its relevance to certain complications in pregnancy. Immunogenetics. 2001;53:48–53. doi: 10.1007/s002510100296. [DOI] [PubMed] [Google Scholar]
- 56.Cleary-Goldman J, Malone FD, Vidaver M, Ball RH, Nyberg DA, Comstock CH, et al. Impact of maternal age on obstetric outcome. Obstet Gynecol. 2005;105:983–990. doi: 10.1097/01.AOG.0000158118.75532.51. [DOI] [PubMed] [Google Scholar]
- 57.Fretts RC, Schmittdiel J, Mclean FH, Usher RH, Goldman MB. Increased maternal age and the risk of fetal death. N Engl J Med. 1995;333:953–957. doi: 10.1056/NEJM199510123331501. [DOI] [PubMed] [Google Scholar]
- 58.Cnattingius S, Forman MR, Berendes HW, Isotalo L. Delayed childbearing and risk of adverse perinatal outcome - a population-based study. JAMA. 1992;268:886–890. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.