Abstract
Recent studies have identified genes involved in high-altitude adaptation in Tibetans. Genetic variants/haplotypes within regions containing three of these genes (EPAS1, EGLN1, and PPARA) are associated with relatively decreased hemoglobin levels observed in Tibetans at high altitude, providing corroborative evidence for genetic adaptation to this extreme environment. The mechanisms that afford adaptation to high-altitude hypoxia, however, remain unclear. Considering the strong metabolic demands imposed by hypoxia, we hypothesized that a shift in fuel preference to glucose oxidation and glycolysis at the expense of fatty acid oxidation would improve adaptation to decreased oxygen availability. Correlations between serum free fatty acids and lactate concentrations in Tibetan groups living at high altitude and putatively selected haplotypes provide insight into this hypothesis. An EPAS1 haplotype that exhibits a signal of positive selection is significantly associated with increased lactate concentration, the product of anaerobic glycolysis. Furthermore, the putatively advantageous PPARA haplotype is correlated with serum free fatty acid levels, suggesting a possible decrease in the activity of fatty acid oxidation. Although further studies are required to assess the molecular mechanisms underlying these patterns, these associations suggest that genetic adaptation to high altitude involves alteration in energy utilization pathways.
Introduction
Native high-altitude populations have persisted for hundreds of generations in oxygen-deprived (hypoxic) environments. Recent genome-wide scans of positive selection in Tibetans have identified hypoxia-sensing and -regulated genes as candidates for high-altitude adaptation [1–10]. A few of the candidate regions thought to underlie Tibetans’ adaptation were previously shown to be associated with their hemoglobin (Hb) levels [1, 7, 10], which are lower than those of native highland groups from the Andes or visitors to high altitude [1, 7, 10, 11]. Positively selected regions containing the EPAS1 gene, which encodes the HIF-2α subunit of the hypoxia inducible factor (HIF) complex, were associated with Hb concentration in Tibetan populations examined in two separate studies [1, 12]. Two additional genomic regions were associated with Hb concentration in a Qinghai Tibetan population. One contains EGLN1, which encodes the proline hydroxylase PHD2 that negatively regulates HIFs’ α subunits in an oxygen-dependent manner [12]. The second is the genomic region containing PPARA, which encodes the nuclear peroxisome proliferator activated receptor alpha (PPARα) that regulates fatty acid metabolism and is in turn regulated by HIF [7].
It is unclear whether these previously identified associations reflect the action of selection directly on Hb level or are consequences of natural selection acting on other advantageous traits [13, 14]. Members of the hypoxia inducible factor (HIF) pathway orchestrate molecular responses during hypoxic stress through a complex series of cellular metabolites [15]. In the absence of adequate oxygen, energy production from oxidative metabolism may be diminished. At the same time, if oxidative metabolism proceeds under hypoxia, reactive oxidative intermediates will accumulate in mitochondria. Either of these conditions can result in cell death. Recent work on the whole-organism level has revealed that HIF plays a major role in regulating metabolism, highlighting a strong relationship between HIF and metabolic demands in humans [16].
The hypoxia signaling system triggers a pleiotropic response that increases tissue oxygenation by increasing vascularization and oxygen carrying capacity of the blood [1,2]. Simultaneously, HIF signaling triggers alterations in metabolism to decrease tissue oxygen demand [17, 18]. Oxidation of fatty acids yields less ATP per molecule of oxygen consumed than oxidation of carbohydrates, suggesting that decreased fatty acid oxidation could also be a favorable adaptation to hypoxia [19]. Several studies have demonstrated decreased reliance on fat metabolism at high altitude, both in people living habitually at high altitude [19] and in those not dwelling at high altitude but acclimatized [20, 21]. Another metabolic change induced by hypoxia is a conversion from oxidative glucose metabolism to glycolysis in order to maintain energy production. This occurs through up-regulation of glucose uptake and glycolysis and down-regulation of mitochondrial glucose oxidation [22, 23].
These observations suggest that natural selection on HIF-related genetic variants in a hypoxic environment could be associated with significant metabolic changes in high-altitude Tibetans. If natural selection on genes in the HIF pathway has reduced fatty acid oxidation and increased glycolysis, then we predict that individuals who have the favorable HIF-related haplotypes should exhibit metabolic profiles consistent with these adaptations. Specifically, they should manifest increased levels of lactate and serum free fatty acids and/or triglycerides.
Here we examine relationships that provide insight into this hypothesis by comparing genetic variation in a Tibetan population with concentrations of several serum metabolites. We show that the putatively advantageous EPAS1 haplotype previously associated with [Hb] [1, 10] is also highly associated with changes in serum lactate concentration. In addition, the putatively adaptive PPARA haplotype exhibit an association with serum free fatty acid concentration in this population.
Results and Discussion
We employed the analytic strategies (iHS [24] and XP-EHH [25] statistics) used in our previous study of Tibetan high-altitude adaptation [7] to identify targets of natural selection in a different Tibetan population from the Tuo Tuo River region in the Qinghai-Tibetan Plateau (Simonson et al. unpublished data). Both EPAS1 and EGLN1 gene regions exhibit significant signals of selection for the iHS test [24] (p < 0.004 and p < 0.01, respectively), although the genomic region containing PPARA did not reach significance (p < 0.14) in this population for tests of selection performed (see Simonson et al. 2011 for discussion of differences among selection signals in Tibetan populations). Interestingly, a recent study of adaptation in Ethiopian highlanders shows that variants within the PPARA genomic region exhibits a signal of selection in Ethiopian highlanders; furthermore, this putatively selected haplotype is associated with decreased [Hb] in this population [26]. Considering variants/haplotypes in these three regions have been associated with [Hb] in one or more populations from the Qinghai-Tibetan Plateau [3, 5, 6], we examined whether these regions were associated with variation in metabolites.
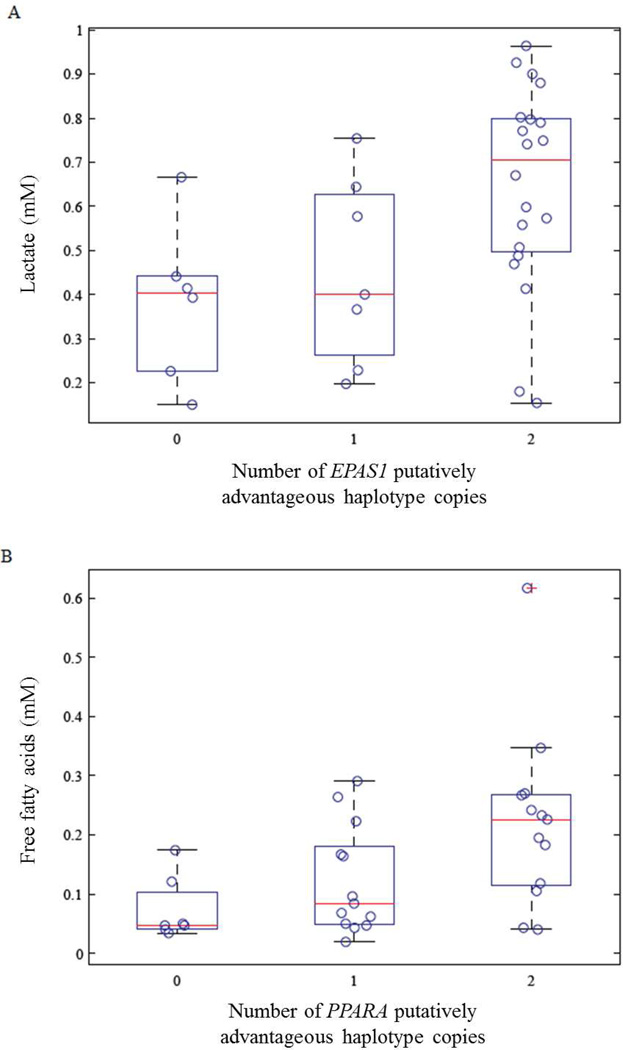
We measured serum concentration of triglycerides, free fatty acids, β-hydroxybutyrate, and lactate in non-fasted serum samples from 36 individuals from the Tuo Tuo River region (Simonson et al. unpublished data). We performed Spearman rank-order correlation analysis between the serum concentration and the selected haplotypes (0, 1, or 2 copies; see Table 1). Lactate concentration was positively associated with the putatively adaptive EPAS1 haplotype (p<0.003; Figure 1, Table 1). Additionally, the previously identified PPARA haplotype of interest [7] exhibited a significant positive relationship with serum free fatty acids (p < 0.01; Figure 1, Table 1). The putatively advantageous EGLN1 gene region was not associated with any of the serum metabolite levels measured.
Table 1.
Haplotype-phenotype significance values for Spearman rank-order correlation analysis of metabolites measured in Tibetans living at 4500 meters.
| EGLN1 | EPAS1 | PPARA | ||||
|---|---|---|---|---|---|---|
| P value | Rho | P value | Rho | P value | Rho | |
| Triglycerides | 0.150 | 0.245 | 0.307 | 0.175 | 0.973 | −0.006 |
| Free fatty acids | 0.505 | −0.115 | 0.860 | −0.031 | 0.014 | 0.406 |
| Three hydroxybutyrate | 0.230 | −0.205 | 0.590 | 0.093 | 0.182 | 0.227 |
| Lactate | 0.070 | 0.305 | 0.003 | 0.482 | 0.424 | 0.137 |
Fig. 1.
Association of previously identified adaptive haplotypes and metabolites. A) Lactate concentration is plotted against the group of putatively advantageous haplotypes (0, 1, or 2) at the EPAS1 locus. B) Serum free fatty acid concentration (FFA) is plotted against the number of putatively advantageous PPARA haplotype copies (0–2). A box-and-whisker plot overlying the data points shows the median, upper and lower quartiles, and extreme measurements as a red line, boxed ends, and dashed lines, respectively.
Both EPAS1 and PPARA are involved in hypoxia signaling [27]. We observed that the putatively adaptive EPAS1 haplotypes were associated with increased lactate concentration, which is consistent with decreased glucose oxidation. Humans acutely exposed to hypoxia consequently exhibit increased anaerobic glucose metabolism [28], and HIF-2α’s metabolic activity has been shown to be required for the shift to anaerobic metabolism that facilitates adaptation to hypoxia in skeletal muscle of mice [15]. Additionally, individuals with Chuvash polycythemia, an autosomal recessive disorder in which HIF degradation is impaired, exhibit higher lactate levels during exercise than do normal individuals, illustrating a metabolic regulatory role of HIF under exercise conditions [16]. Mice lacking EPAS1, however, exhibit lactic acidosis [29], implying that the adaptive Tibetan polymorphism could be associated with either increased or decreased HIF-2α activity.
Because fatty acid oxidation consumes more oxygen than glycolysis for energy production, it is conceivable that a decrease in fatty acid oxidation activity is preferred and adaptive to the hypoxia environment. Indeed, a previous report demonstrates a high prevalence of hypertriglyceridemia in Tibetan highlanders [30]. We measured fasting triglyceride concentration in a separate cohort of Tibetans and Han Chinese living in an urban environment near sea level, and confirmed this observation of significantly higher triglyceride concentration in Tibetans (298±23 compared to 139±8 mg/dl in the Han, p=0.006; data not shown). We therefore examined the relationship of the putatively adapted haplotypes with serum free fatty acid and triglyceride concentrations in the high-altitude Tibetan samples. Free fatty acid concentration was positively associated with the PPARA putatively adapted haplotype, although serum triglyceride concentration was not. It should be pointed out, however, that the sera were not collected in the fasting state, and serum triglyceride concentration varies acutely with fasting, feeding, and composition of the diet.
PPARA encodes the nuclear receptor protein PPARα, a major regulator of fatty acid oxidation [31, 32]. Down regulation of several genes involved in fatty acid oxidation, including PPARA, has been observed in rats exposed to hypoxia [33]. Previous studies have shown that PPARα activation is associated with lower serum free fatty acid and triglyceride concentration [34]; hence the putatively adaptive haplotype is consistent with decreased expression or activity of PPARα. We did not, however, observe decreased β-hydroxybutyrate, which is indicative of fatty acid breakdown, with the PPARA haplotype (Table 1), so there is no direct evidence of decreased fatty acid oxidation. Another explanation for increased lipid concentration would be increased lipid synthesis. Fat anabolic pathways are up regulated in hypoxia, mediated at least in part by up regulation of PPARγ [32, 35] and SREBP-1 [36], but we have no data from our studies to determine the degree to which increased synthesis, increased lipolysis, or decreased metabolism of fatty acids contribute to the observed phenotype.
Recent studies suggest that some of the PPARα-dependent effects of hypoxia on fat metabolism may be mediated through HIF-2α [27]. Paradoxically, mice with either deletion of Epas1 or liver-specific over expression of Epas1 exhibit hepatic steatosis and both models show evidence of decreased fatty acid oxidation [29, 37]. Thus, the interrelationships among the status of the primary hypoxia signaling pathways, their downstream metabolic effectors, and the final metabolic phenotype of the organism are highly complex. In addition, environmental factors are crucial in determining metabolic status. Thus, determining the specific roles of changes in EPAS1, and PPARA genomic regions on the observed changes in metabolites will require further study.
Hypoxia-induced regulation of metabolism and its alteration in adapted populations may carry implications for the risks of diabetes and obesity. The putatively adaptive haplotypes may result in a relative inability to shift between fat and glucose oxidation, so-called metabolic inflexibility [38]. Such inflexibility and fatty acid oxidation capacities [39, 40] are both implicated in the pathogenesis of type 2 diabetes mellitus. Tibetan highlanders have a relatively low prevalence of diabetes [41], but the diet is also relatively low-calorie [42] and high altitudes are associated with lower body weights among Tibetans [43]. As populations move to lower altitudes and encounter a more industrialized lifestyle and higher calorie diets, however, the metabolic adaptations to altitude could have health implications. For example, increasing total fat and calorie consumption with a metabolic profile that will not support fat oxidation could result in accumulation of lipid intermediates thought to play a role in diabetes pathogenesis [39, 40]. Further study of the metabolic implications of high altitude adaptation may allow interventions to ameliorate this risk and also identify potential new targets to treat obesity and diabetes.
Our results demonstrate increased lactate and free fatty acid concentrations in Tibetans with putatively advantageous EPAS1 and PPARA haplotypes. This pattern provides insight consistent with the hypothesis that anaerobic glucose metabolism is increased and fatty acid oxidation may be decreased in the Tibetans with the putatively adaptive haplotypes compared to Tibetans without the adapted haplotypes living at the same altitude. Controlled studies including more dynamic metabolic analyses and studies at different altitudes will be required to better understand the physiological significance of these patterns.
Materials and Methods
DNA sample collection
DNA was extracted from whole blood samples for individuals (non-smokers, no chronic diseases). This population, who speak the Kham dialect, is from the Tuo Tuo River area in the Qinghai-Tibetan Plateau (~4,500 m), People’s Republic of China. Informed consent was obtained for all participants according to guidelines approved by the Institutional Review Boards at the High Altitude Medical Research Institute (Xining, Qinghai, People’s Republic of China).
SNP genotyping
DNA samples were genotyped using Affymetrix 6.0 SNP Array technology (>900,000 SNPs) at Capital Bio Corporation (Beijing, China). We used default parameters for the Birdseed algorithm (version 2) to determine genotypes for all samples (Affymetrix, Santa Clara, CA, USA). Genotypic data were analyzed using the Affymetrix Genotyping Console 3.1 (Affymetrix).
Estimates of relatedness
We used unrelated samples as described in Simonson et al. [7] and the program ERSA [44] to exclude related individuals in the second set of Tibetan samples, excluding one member of the pair if their relationship exceeded that expected for first cousins. Based on these criteria, a total of 36 unrelated individuals were included in the genotype-phenotype analyses.
Phenotype collection
We measured metabolites in a cohort of Tibetans, excluding related individuals (N=36) using kit protocols for lactate (Point Scientific, Inc., Lincoln Park, MI), β-hydroxybutyrate (WAKO Diagnostics, Richmond, VA), free fatty acids (Half Micro Test, Roche, Penzberg Germany) and triglycerides (Sigma Chemical, St Louis, MI).
Genotype-phenotype association
We determined whether the selection candidate regions that were previously associated with [Hb] (Table 1) are also associated with metabolites. We performed selection scans using the iHS (Voight et al. 2006) and XP-EHH (Sabeti et al. 2007) test statistics, and defined three-SNP haplotypes as previously described [7]: SNPs within 200 kb genomic region with the most extreme iHS scores comprise each of the putatively advantageous haplotypes (Hg 18 build haplotype-defining SNPs: chromosome 1 positions 229601735, 229604075, and 229793717 for EGLN1; chromosome 2 positions 46490868, 46590298, and 46592661 for EPAS1; chromosome 22 positions 44807657, 44827140, and 44866192 for PPARA).
We tested for an association between these haplotypes and free fatty acid, lactate, triglyceride, and β-hydroxybutyrate concentrations in 36 Tibetan individuals from the Tuo Tuo River area on the Qinghai-Tibetan Plateau (Simonson et al. unpublished data) using a nonparametric Spearman rank test. The number of putatively advantageous haplotype copies (0, 1, or 2) was determined using SNP haplotypes described above for EPAS1, EGLN1, and PPARA regions and compared to metabolite concentrations.
Highlights.
We examined relationships between genes involved in high-altitude adaptation and metabolite levels in a native Tibetan population residing at 4500m.
We identified correlations between genomic regions containing EPAS1 and PPARA and lactate and serum free fatty acid levels, respectively.
These changes suggest that genetic adaptation to high altitude involves alteration in energy utilization pathways.
Acknowledgments
This work was supported by the NIDDK (1R01 DK081842, DAM), NCRR (UL1 RR025764, DAM), the Research Service of the Veterans Administration (DAM), the Robinson Family Foundation (DAM), the National Basic Research Program of China (No. 2012CB518200, L-RG), Program of International S&T Cooperation of China (No: 0S2012GR0195, L-RG) and National Natural Science Foundation of China (No. 30393133, L-RG), and the National Institutes of Health (NIH GM-59290). T.S.S. was supported by the National Institute of Health (NIH T32 GM-007464). JX is supported by the National Human Genome Research Institute, National Institute of Health (K99HG005846). The authors would like to thank the high-altitude inhabitants for participating in this study. We also thank Drs. Xue-Feng, Yong Mei, Zhaxi, Zhuruma, He Long, Dan Ba at the hospitals of Maduo and Tuo Tuo River for their cooperation and hospitality during data collection, and J. Lindsley for insightful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beall CM, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigham A, et al. Identifying Signatures of Natural Selection in Tibetan and Andean Populations Using Dense Genome Scan Data. PLoS Genet. 2010;6(9):e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacInnis MJ, Rupert JL. ’ome on the Range: Altitude Adaptation, Positive Selection, and Himalayan Genomics. High Altitude Medicine & Biology. 2011;12(2):133–139. doi: 10.1089/ham.2010.1090. [DOI] [PubMed] [Google Scholar]
- 4.Peng Y, et al. Genetic Variations in Tibetan Populations and High-Altitude Adaptation at the Himalayas. Molecular Biology and Evolution. 2011;28(2):1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 5.Scheinfeldt LBT, SA Living the high life: high-altitude adaptation. GenomeBiology.com. 2010;11(9) doi: 10.1186/gb-2010-11-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonson T, et al. Genetic determinants of Tibetan high-altitude adaptation. Human Genetics. :1–7. doi: 10.1007/s00439-011-1109-3. [DOI] [PubMed] [Google Scholar]
- 7.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329(5987):72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, et al. On the Origin of Tibetans and Their Genetic Basis in Adapting High-Altitude Environments. PLoS ONE. 2011;6(2):e17002. doi: 10.1371/journal.pone.0017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu S, et al. A Genome-Wide Search for Signals of High-Altitude Adaptation in Tibetans. Molecular Biology and Evolution. 2011;28(2):1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 10.Yi X, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi Z, et al. Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic minority group. Leg Med (Tokyo) 2010;12(2):108–111. doi: 10.1016/j.legalmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Storz JF, Scott GR, Cheviron ZA. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 2010;213(Pt 24):4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tissot van Patot MC, Gassmann M. Hypoxia: Adapting to High Altitude by Mutating EPAS-1, the Gene Encoding HIF-2alpha. High Alt Med Biol. 2011;12(2):157–167. doi: 10.1089/ham.2010.1099. [DOI] [PubMed] [Google Scholar]
- 15.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formenti F, et al. Regulation of human metabolism by hypoxia-inducible factor. Proceedings of the National Academy of Sciences. 2010;107(28):12722–12727. doi: 10.1073/pnas.1002339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8(9):705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. Perspectives on oxygen sensing. Cell. 1999;98(3):281–284. doi: 10.1016/s0092-8674(00)81957-1. [DOI] [PubMed] [Google Scholar]
- 19.Holden JE, et al. Enhanced cardiac metabolism of plasma glucose in high-altitude natives: adaptation against chronic hypoxia. J Appl Physiol. 1995;79(1):222–228. doi: 10.1152/jappl.1995.79.1.222. [DOI] [PubMed] [Google Scholar]
- 20.Roberts AC, et al. Acclimatization to 4,300-m altitude decreases reliance on fat as a substrate. J Appl Physiol. 1996;81(4):1762–1771. doi: 10.1152/jappl.1996.81.4.1762. [DOI] [PubMed] [Google Scholar]
- 21.Brooks GA, et al. Increased dependence on blood glucose after acclimatization to 4,300 m. J Appl Physiol. 1991;70(2):919–927. doi: 10.1152/jappl.1991.70.2.919. [DOI] [PubMed] [Google Scholar]
- 22.Papandreou I, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Kim JW, et al. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Voight BF, et al. A Map of Recent Positive Selection in the Human Genome. PLoS Biology. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheinfeldt L, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biology. 2012;13(1):R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aragones J, et al. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40(2):170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- 28.Kelly KR, et al. Acute altitude-induced hypoxia suppresses plasma glucose and leptin in healthy humans. Metabolism. 2010;59(2):200–205. doi: 10.1016/j.metabol.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scortegagna M, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat Genet. 2003;35(4):331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 30.Sherpa LY, et al. Lipid Profile and Its Association with Risk Factors for Coronary Heart Disease in the Highlanders of Lhasa, Tibet. High Altitude Medicine & Biology. 2011;12(1):57–63. doi: 10.1089/ham.2010.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narravula S, Colgan SP. Hypoxia-inducible factor 1-mediated inhibition of peroxisome proliferator-activated receptor alpha expression during hypoxia. J Immunol. 2001;166(12):7543–7548. doi: 10.4049/jimmunol.166.12.7543. [DOI] [PubMed] [Google Scholar]
- 32.Piguet AC, et al. Hypoxia aggravates non-alcoholic steatohepatitis in mice lacking hepatocellular PTEN. Clin Sci (Lond) 2010;118(6):401–410. doi: 10.1042/CS20090313. [DOI] [PubMed] [Google Scholar]
- 33.Kennedy SL, et al. Alterations in enzymes involved in fat metabolism after acute and chronic altitude exposure. J Appl Physiol. 2001;90(1):17–22. doi: 10.1152/jappl.2001.90.1.17. [DOI] [PubMed] [Google Scholar]
- 34.Barbier O, et al. Pleiotropic actions of peroxisome proliferator-activated receptors in lipid metabolism and atherosclerosis. Arterioscler Thromb Vasc Biol. 2002;22(5):717–726. doi: 10.1161/01.atv.0000015598.86369.04. [DOI] [PubMed] [Google Scholar]
- 35.Krishnan J, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell Metab. 2009;9(6):512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. Altered metabolic responses to intermittent hypoxia in mice with partial deficiency of hypoxia-inducible factor-1alpha. Physiol Genomics. 2006;25(3):450–457. doi: 10.1152/physiolgenomics.00293.2005. [DOI] [PubMed] [Google Scholar]
- 37.Rankin EB, et al. Hypoxia-Inducible Factor 2 Regulates Hepatic Lipid Metabolism. Mol. Cell. Biol. 2009;29(16):4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storlien L, Oakes ND, Kelley DE. Metabolic flexibility. Proc Nutr Soc. 2004;63(2):363–368. doi: 10.1079/PNS2004349. [DOI] [PubMed] [Google Scholar]
- 39.Holland WL, et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5(3):167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Koves TR, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 41.Matsubayashi K, et al. Comprehensive geriatric assessment of elderly highlanders in Qinghai, China I: activities of daily living, quality of life and metabolic syndrome. Geriatr Gerontol Int. 2009;9(4):333–341. doi: 10.1111/j.1447-0594.2009.00548.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Dang S, Yan H. Nutrient intakes of rural Tibetan mothers: a cross-sectional survey. BMC Public Health. 2010;10(1):801. doi: 10.1186/1471-2458-10-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherpa LY, et al. Obesity in Tibetans aged 30–70 living at different altitudes under the north and south faces of Mt. Everest. Int J Environ Res Public Health. 2010;7(4):1670–1680. doi: 10.3390/ijerph7041670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huff CD, et al. Maximum-likelihood estimation of recent shared ancestry (ERSA) Genome Res. 2011;21(5):768–774. doi: 10.1101/gr.115972.110. [DOI] [PMC free article] [PubMed] [Google Scholar]