Abstract
Escape behaviors are crucial to survive predator encounters. Touch to the head of C. elegans induces an escape response where the animal rapidly backs away from the stimulus and suppresses foraging head movements. The coordination of head and body movements facilitates escape from predacious fungi that cohabitate with nematodes in organic debris. An appreciation of the natural habitat of laboratory organisms, like C. elegans, enables a comprehensive neuroethological analysis of behavior. In this review we discuss the neuronal mechanisms and the ecological significance of the C. elegans touch response.
Introduction
“Eat but don’t get eaten” is a prevailing motto that guides animal behavior. However, this principle presents a dilemma since foraging often increases the risk of predation. Animals can offset part of these risks by trying to survive an confrontation with a predator. Run, dart, jump, fly, burrow and hide can all improve the prey’s odds in these life or death encounters. Time is of the essence so the animal needs to quickly translate sensory information into action. As a consequence, these escape responses are typically robust, use dedicated neuronal structures and have a clear evolutionary purpose, making them favorite subjects for laboratory study [1]. The tail-flip escape in the crayfish [2], the C-start escape in goldfish [3] and the mollusk withdrawal response [4] have provided crucial insights into fundamental neuronal processes as diverse as synaptic transmission, sensory transduction, decision making, and learning and memory. The study of these relatively simple circuits has provided some of the rare examples where we know the complete path from sensory input to a motor output. However, genetic analyses in these organisms are difficult, leaving the molecular coding of these behaviors relatively unexplored. Studies in genetically tractable organisms, like the fruit fly Drosophila melanogatser and the roundworm Caenorhabditis elegans, have provided some insight into the molecular basis of escape behaviors. In the fly, the giant fiber (GF) neurons coordinate leg extension and wing depression, which are critical for fast flight initiation when a fly is startled by a strong visual stimulus [5] (See review G. Card, this issue). A number of genes have been identified from genetic screens which play a role in the development of the giant fiber circuit, identifying molecular mechanisms that control outgrowth of GF axons, and the formation and maturation of synapses [6]. The neuronal pathway from the GF to motor neurons is well defined, but relatively little is known about its sensory inputs.
The C. elegans touch response
The complete wiring diagram of the C. elegans nervous system is known [7]. This framework is a tremendous asset for understanding sensory processing, including the escape response. C. elegans moves on its side by propagating a sinusoidal wave of dorsal ventral flexures along the length of its body [8]. Locomotion is accompanied by exploratory head movements, where the head of the animal sways rapidly from side to side (Figure 1). Head and body movements are controlled independently by distinct classes of motor neurons and muscles. While body bends are restricted to the dorsal-ventral plane, the animal can flex its head in three dimensions. Head movements most likely allow the animal to explore its immediate environment and aid in the search for food, as the tip of worms nose contains the sensory endings that smell, taste and sense touch. Gentle touch to the body of the animal induces an escape response where the animal moves away from the stimulus. Touch to the tail of the animal causes the nematode to speed up, while touch to the anterior half of the animal induces a quick reversal during which foraging head movements are suppressed [9], [10]. Much like the coordination of leg extension and wing depression during a fly escape, the worm coordinates backward locomotion with the suppression of foraging head movements in response to anterior touch.
Figure 1. Escape responses.
Silhouettes of animal escape responses. Arrows indicate the direction of the threatening stimulus. Crayfish tail-flip (top): Tail touch in the crayfish induces powerful abdominal flexures that are spatially and temporally controlled to propel the animal through the water away from the stimulus. Time from first to last frame is approximately 15 ms [2] [48]. Tritonia swim reflex: Upon touch to the body, Tritonia initiates a series of coordinated dorsal and ventral body flexures to swim away from predators. Time from first to last frame is approximately 5 s [49]. Goldfish C-start: Lateral stimulation causes the animal to coordinate both the strength and the timing of agonist and antagonist muscle contractions on either side of the body to quickly change direction to move away from the stimulus. Time from first to last frame is approximately 50 ms [50]. Drosophila startle response: A strong visual stimulus induces fast flight initiation, where the fly couples leg extension and wing depression to quickly fly away. Time from first to last frame is approximately 25 ms [5]. C. elegans anterior touch response: Gentle touch to the anterior of the body of the worm induces a reversal coupled with the suppression of foraging head movements followed by a deep ventral bend (omega turn) and a 180° change in the direction of locomotion. Time from first to last frame is approximately 10s [10].
The neural circuit of escape
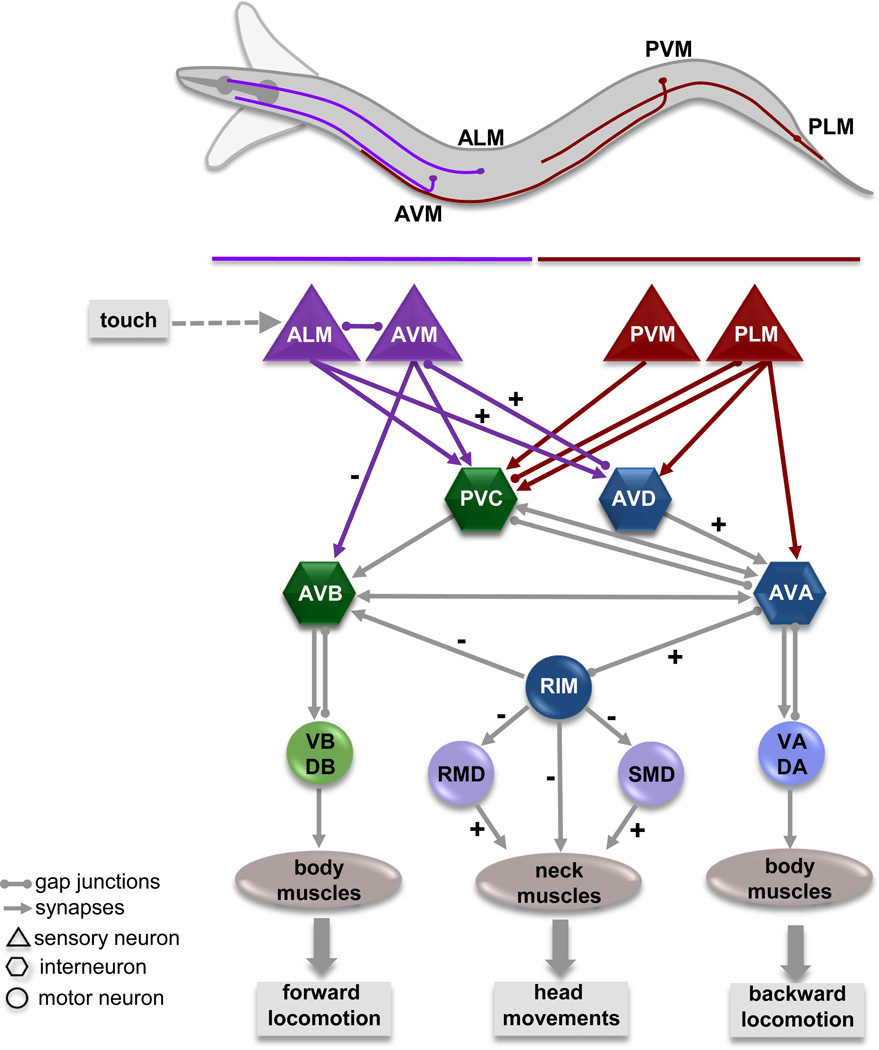
In the worm, gentle touch to the body is sensed by six mechanosensory neurons; the ALM and AVM neurons sense touch to the anterior half, while the PLM and PVM neurons sense touch to the posterior half (Figure 2) [11]. Optogenetic activation of the ALM/AVM or PLM/PVM neurons in transgenic animals that express the light activated channelrhodopsin in the mechanosensory neurons induces a reversal or a forward acceleration, respectively [12], [13]. All six neurons send anteriorly directed processes that run close to the cuticle and can sense touch over their entire length. Touch reception is mediated by a DEG/ENaC channel complex that can sense the application and removal of forces as small as 100 nN with a latency of less than 5 ms, allowing the animal to quickly respond to even the lightest of touches [•14], [15]. The touch sensory neurons make chemical synapses and electrical gap junctions with a set of command like interneurons that control locomotion. The PVC and AVB locomotion command neurons provide inputs into the VB and DB motorneurons which synapse onto body wall muscles and drive forward movement; the AVD and AVA neurons provide inputs into the VA and DA motor neurons that drive backward locomotion [7]. Laser ablation studies and genetic perturbations indicate that the activity of the interneurons establishes the direction of locomotion [9], [16]. Recent calcium imaging studies support this notion, indicating that the reciprocal activation of AVB and AVA command neurons correlates with forward and backward locomotion respectively [17], [18], [19].
Figure 2. Neural circuit of the C. elegans escape response.
C. elegans moves its head rapidly from side to side during forward locomotion as indicated by the outline. The relative cell body position and neuronal process of neurons responsible for sensing touch (purple: anterior touch; red: posterior touch) are depicted. The circuit diagram illustrates the connections between the sensory neurons and those that control locomotion and head movements. Synaptic connections (arrows) and gap junctions (spheres) are as described by White, et al. (1986). Sensory neurons are shown as triangles, command neurons required for locomotion are shown as hexagons, and motor neurons as circles. Excitatory (+) and inhibitory (−) connections involved in the anterior touch escape response are noted. Anterior touch activates the AVA backward locomotion command neuron, which in turn activates the tyraminergic RIM motor neuron. Tyramine release from the RIM neurons activates the tyramine gated chloride channel LGC-55, which is expressed in the AVB forward locomotion command neuron and cells of the head movement circuit: RMD, SMD, and neck muscles. Activation of LGC-55 causes hyperpolarization of neck muscles and the AVB neuron inducing the suppression of head movements and sustained backward locomotion in response to anterior touch [9], [10], [•22].
Activation of the anterior touch sensory neurons inhibits command neurons that drive forward locomotion (PVC, AVB) and activates those, which promote backward locomotion (AVD, AVA), causing the animal to back away from the stimulus (Figure 2). During this reversal, the animal also suppresses foraging head movements. Animals that lack tyrosine decarboxylase (tdc-1), the biosynthetic enzyme that makes tyramine, or animals in which the single pair tyraminergic RIM neurons are ablated fail to suppress these head movements and back up less far in response to touch [10]. The RIM neuron is connected to the backward locomotion circuitry via a gap junction with the AVA command neuron. Optogenetic activation of the RIM neurons causes an increase in intracellular calcium in the AVA [20], suggesting that the AVA and RIM are coactivated during a backing response. The RIM has synaptic outputs onto the forward locomotion command neuron, AVB, as well as the motor neurons and neck muscles that facilitate foraging head movements. The identification of the tyramine-gated chloride channel, LGC-55 [21], [•22] demonstrated that tyramine can act as a classical neurotransmitter. LGC-55 is expressed post-synaptically to the RIM, consistent with a model where activation of LGC-55 by tyramine release in response to anterior touch induces hyperpolarization of neck muscles and the AVB leading to the suppression of head movements and sustained backward locomotion [•22]. The role of tyramine in the C. elegans escape response illustrates how biogenic amines can coordinate the output of independent motor programs in the generation of complex behaviors.
C. elegans ecology
What does C. elegans need to escape from? Natural populations of C. elegans are found world wide in decomposing organic material such as compost, decaying leaves and rotting fruit [23], [••24]. C. elegans, can survive less favorable conditions as a dauer larvae, an alternative developmental stage that is long lived. Dauer larvae can climb onto protrusions, stand on their tail and wave in the air, a process known as nicatation. This behavior may promote dispersal to new more favorable habitats. Dauer larvae have been found in association with insects and snails [25], and have been shown to hitch a ride on fruit flies in the labortory [26]. Once a favorable habitat is found, a viable evolutionary option is to produce many offspring quickly. C. elegans reaches adulthood in 3 to 4 days and under laboratory conditions produces approximately 300 offspring. The prolific nature of C. elegans was one of the genetic selection criteria that eased its transition from English compost in the 1950s to luscious laboratory life [27]. The laboratory strain N2 spends its life on agar plates with abundant amounts of E. coli as a food source. The domestication of C. elegans has brought a different set of selective pressures. Laboratory strains of C. elegans have different preferences for oxygen [• 28], [• 29], foraging behavior [30], and response to dauer pheromone [•• 31] than animals recovered from the wild, indicating an adaptation to laboratory life. These behavioral adaptations to changing environmental conditions emphasize the importance of the natural history of the animal. Its natural habitat, with fluctuating temperature, humidity and osmotic pressure and other microbes, presents a plethora of behavioral challenges to C. elegans. It needs to forage for bacteria and slime molds that compose a majority of its diet [32], [33]. It also needs to be selective in its dietary choices, since some bacteria and fungi can infect and kill the animal [34]. Knowing what has been on the menu helps C. elegans make healthy choices, since it can learn to avoid eating bacteria that are hard to digest or are pathogenic [35], [36].
While nourishment is essential for the animal’s survival and reproduction, foraging behavior incurs costs that extend beyond the energy used for finding food. Foraging can expose C. elegans to encounters with natural predators. Nematophagous mites [37], springtails [38], water bears [39] and other predacious nematodes like Pristionchus pacificus [40] can actively kill and eat nematodes. Nematodes face another lurking foe in their habitat: predacious fungi. These carnivorous fungi, from the family of Orbiliales (Ascomycota), possess hyphal structures to trap nematodes in the soil and decaying organic debris [41], [42]. One method of capturing nematodes employs adhesive branches, knobs or hyphal nets that stick to, and entangle nematodes. By far the most sophisticated trapping mechanism is the constricting ring. When a worm crawls through the ring gentle friction induces the ring to rapidly inflate and lasso its prey (Figure 3). Predacious fungi form few, if any traps in the absence if nematodes. Trap formation is induced by the presence of nematodes, including C. elegans [43], and stimulates the growth of a variety of hyphal traps. Once a nematode is caught by a hyphal trap, death does not come quickly. A prolonged struggle usually only ends when fungal hyphae perforate the cuticle, and absorb the contents of the nematode.
Figure 3. C. elegans caught by a constricting ring of a nematophagous fungus.
Scanning electron micrograph of C. elegans L2 larvae caught in the constricting rings of the nematode trapping fungi, Drechslerella doedycoides. Image Sean M. Maguire.
Out of the jaws of death
Since the C. elegans mechanosensory neurons are very sensitive, the touch of a mite, a sticky hyphae or a tightening fungal noose could provide enough force to trigger an escape response. An acceleration or quick reversal may allow the worm to survive such an encounter with a predator. The wide array of available mutants in C. elegans provides unique opportunities to directly test if behavioral traits contribute to the odds of surviving predator encounters. Studies of predator-prey relationships between C. elegans and the constricting ring fungus Drechslerella doedycoides showed that the fungus is most efficient in entrapping the early larval stages with a diameter similar to that of the ring [••44]. Young larvae are small enough to move far into the ring and wide enough to wedge themselves and trigger trap inflation. There is a delay between the animal moving into a trap and trap closure allowing some nematodes to enter and withdraw from the ring before getting caught. While wild-type animals survive most encounters with the hyphal noose, mutants that fail to sense touch or that cannot reverse, are trapped more frequently. Moreover, mutants that fail to suppress the foraging head movements in response to touch are caught more often than the wild type. The ability to tag different genotypes with transgenic fluorescent markers allows one to test whether coordination of a touch-induced reversal with the suppression of swaying head movements provides a selective advantage. Animals that fail to suppress foraging head movements are caught more efficiently by the fungus than the wild type animals in direct competition experiments in with mixed populations of wild-type and mutant animals. Thus, the coordination of motor programs in the C. elegans escape response is vitally important to evade fungal predation.
Other soil nematodes from the Eurhabditis clade, close relatives of C. elegans, display a similar escape response. Interestingly, several species from the Pleiorhabditis clade, which have been found in association with insects, fail to coordinate head and body movements in their escape response (J.K.P and M.J.A unpublished observation). This may indicate that the touch-induced suppression of head movements has evolved from selective pressures imposed by predacious fungi. Nematodes and predacious fungi that use rings as trapping devices have been found in 100 million year old amber indicating an ancient predator-prey relationship [45]. The phylogeny of predacious fungi and morphology of their trapping devices suggest that constricting rings have evolved from non-constricting rings [46]. This raises the tantalizing possibility that constricting rings and the suppression of head movements are the result of an evolutionary arms race. An extensive analysis of the ecology of predatory fungi and cohabitating nematodes is required to fully comprehend the dynamics of this predator-prey relationship. Since we know the molecular and neuronal basis of the escape response in C. elegans, comparative studies with other nematodes should allow us to find selection pressures that shape behavioral adaptation.
Conclusions
While escape responses have been a favorite subject for neuroethological studies of behavior, it has been hard to unify the analysis of proximate and ultimate causes of behavior. A renewed appreciation for the natural history of laboratory animals may allow us to bridge a gap between the molecular “how” and behavioral “why”. Furthermore, even though genomic approaches have been instrumental in studies of genotypic selection, our understanding is largely correlative. The experimental manipulation of behavioral traits provides unique inroads to define causative forces of natural selection. The ability to connect the path from molecule to neuron to neural circuit to behavior, ecology, and evolution fulfils the true goals of neuroethology set out by Niko Tinbergen half a century ago [47].
Highlights.
► C. elegans coordinates head and body movements during a touch induced escape response. ► The complete sensory-motor circuit that controls the C. elegans escape response is known. ► Predacious fungi catch nematodes using hyphal trapping devices. ► The C. elegans escape response increases its odds of surviving encounters with predacious fungi. ► Study of the natural habitat of genetic model organisms allows for a comprehensive neuroetholigical analysis of behavior.
Acknowledgements
We would like to thank Claire Benard and Scott Waddell for comments on the manuscript. This work is supported by a grant from the National Institutes of Health grant GM084491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eaton RC. Neural Mechanisms of Startle Behavior. New York: Plenum Press; 1984. [Google Scholar]
- 2.Edwards DH, Heitler WJ, Krasne FB. Fifty years of a command neuron: the neurobiology of escape behavior in the crayfish. Trends Neurosci. 1999;22:153–161. doi: 10.1016/s0166-2236(98)01340-x. [DOI] [PubMed] [Google Scholar]
- 3.Korn H, Faber DS. The Mauthner cell half a century later: a neurobiological model for decision-making? Neuron. 2005;47:13–28. doi: 10.1016/j.neuron.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Katz PS. Neuromodulation intrinsic to the central pattern generator for escape swimming in Tritonia. Ann N Y Acad Sci. 1998;860:181–188. doi: 10.1111/j.1749-6632.1998.tb09048.x. [DOI] [PubMed] [Google Scholar]
- 5.Card G, Dickinson M. Performance trade-offs in the flight initiation of Drosophila. J Exp Biol. 2008;211:341–353. doi: 10.1242/jeb.012682. [DOI] [PubMed] [Google Scholar]
- 6.Allen MJ, Godenschwege TA, Tanouye MA, Phelan P. Making an escape: development and function of the Drosophila giant fibre system. Semin Cell Dev Biol. 2006;17:31–41. doi: 10.1016/j.semcdb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 7.White JG, Southgate E, Thomson JN, Brenner S. The structure of the of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 8.Croll NA. Behavioural analysis of nematode movement. Adv Parasitol. 1975;13:71–122. doi: 10.1016/s0065-308x(08)60319-x. [DOI] [PubMed] [Google Scholar]
- 9.Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 12.Stirman JN, Crane MM, Husson SJ, Wabnig S, Schultheis C, Gottschalk A, Lu H. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362.. • This study describes recordings from the touch receptor neurons in live animals demonstrate that the DEG/ENaC channel proteins MEC-4 and MEC-10 transduce mechanical signals. The MEC-4/MEC-10 channel is opened by the application and removal of a mechanical stimulus indicating that movement of the channel out of the plane of the membrane leads to channel opening.
- 15.Geffeney SL, Cueva JG, Glauser DA, Doll JC, Lee TH, Montoya M, Karania S, Garakani AM, Pruitt BL, Goodman MB. DEG/ENaC but Not TRP Channels Are the Major Mechanoelectrical Transduction Channels in a C. elegans Nociceptor. Neuron. 2011;71:845–857. doi: 10.1016/j.neuron.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 17.Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 18.Ben Arous J, Tanizawa Y, Rabinowitch I, Chatenay D, Schafer WR. Automated imaging of neuronal activity in freely behaving Caenorhabditis elegans. J Neurosci Methods. 2010;187:229–234. doi: 10.1016/j.jneumeth.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Kawano T, Po MD, Gao S, Leung G, Ryu WS, Zhen M. Gap junctions regulate directional movement in Caenorhabditis elegans. Neuron. 2011 doi: 10.1016/j.neuron.2011.09.005. In Press. [DOI] [PubMed] [Google Scholar]
- 20.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringstad N, Abe N, Horvitz HR. Ligand-gated chloride channels are receptors for biogenic amines in C. elegans. Science. 2009;325:96–100. doi: 10.1126/science.1169243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pirri JK, McPherson AD, Donnelly JL, Francis MM, Alkema MJ. A tyramine-gated chloride channel coordinates distinct motor programs of a Caenorhabditis elegans escape response. Neuron. 2009;62:526–538. doi: 10.1016/j.neuron.2009.04.013.. • This study shows that tyramine can act as genuine neurotransmitter. Activation of a tyramine-gated chloride channel coordinates backward locomotion with the suppression of head movements during an escape response.
- 23.Haber M, Schungel M, Putz A, Muller S, Hasertet B. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- 24. Kiontke KC, Felix MA, Ailion M, Rockman MV, Braendle C, Penigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339.. •• Natural isolates of Caenorhabditis species were relatively rare. In this study the authors describe many new Caenorhabditis species that were discovered in rotten fruits and plant material. This indicates that Caenorhabditis are not soil nematodes but rather thrive in rotting organic matter. The ecology and phylogeny of these Caenorhabditis species provide new insight into their natural habitat and provide a framework for further evolutionary analysis.
- 25.Barriere A, Felix MA. WormBook. 2005. Natural variation and population genetics of Caenorhabditis elegans; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee H, Choi MK, Lee D, Kim HS, Hwang H, Kim H, Park S, Paik YK, Lee J. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci. 2011 doi: 10.1038/nn.2975.. • The authors define sensory neurons that are required for nictation in C.elegans. Nictating worms can hitch a ride with fruit flies in the laboratory, indicating that this behavior allows dispersal to new environments.
- 27.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012.. • In this study, the authors characterize the behavioral differences of two C. elegans isolates in response to changing oxygen levels. Polymorphisms in the globin gene, glb-5 and neuropeptide receptor gene, npr-1 in the Bristol strain, N2 and the Hawaiian strain, CB4856 result in opposite responses to changes in environmental oxygen. The authors trace the history of the laboratory strain N2 and their results indicate that the Bristol strain has adapted to high oxygen concentrations under laboratory conditions.
- 29. Persson A, Gross E, Laurent P, Busch KE, Bretes H, de Bono M. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009 doi: 10.1038/nature07820.. • See above.
- 30.Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature. 2011;472:313–318. doi: 10.1038/nature09821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378.. •• The authors show the adaptation of two Caenorhabditis species to growth at high density in liquid media occurs through independently acquired resistance to pheromone induced dauer formation. These results demonstrate that C. elegans can adapt to specific environments through a reproducible change in its chemoreceptor repertoire.
- 32.Grewal PS. Influence of Bacteria and Temperature On the Reproduction of Caenorhabditis Elegans (Nematoda: Rhabditidae) Infesting Mushrooms (Agaricus Bispor Us) Nematologica, 37. 1991;37(1):72–82. [Google Scholar]
- 33.Kessin RH, Gundersen GG, Zaydfudim V, Grimson M. How cellular slime molds evade nematodes. Proc Natl Acad Sci U S A. 1996;93:4857–4861. doi: 10.1073/pnas.93.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 37.Karagoz M, Gulcu B, Cakmak I, Kaya HK, Hazir S. Predation of entomopathogenic nematodes by Sancassania sp. (Acari: Acaridae) Exp Appl Acarol. 2007;43:85–95. doi: 10.1007/s10493-007-9105-y. [DOI] [PubMed] [Google Scholar]
- 38.Read DS, Sheppard SK, Bruford MW, Glen DM, Symondson WO. Molecular detection of predation by soil micro-arthropods on nematodes. Mol Ecol. 2006;15:1963–1972. doi: 10.1111/j.1365-294X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 39.Hohnberg K, Traunspurger W. Foraging theory and partial consumption in a tardigrade–nematode system. Behav. Ecol. 2009;20:884–890. [Google Scholar]
- 40.Bento G, Ogawa A, Sommer RJ. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature. 2010;466:494–497. doi: 10.1038/nature09164. [DOI] [PubMed] [Google Scholar]
- 41.Duddington CL. The ecology of predacious fungi:: I. Preliminary survey. Transactions of the British Mycological Society. 1951;34:322–331. [Google Scholar]
- 42.Thorn RG, Barron GL. Carnivorous Mushrooms. Science. 1984;224:76–78. doi: 10.1126/science.224.4644.76. [DOI] [PubMed] [Google Scholar]
- 43.Xie H, Aminuzzaman FM, Xu L, Lai Y, Li F, Liu X. Trap induction and trapping in eight nematode-trapping fungi (Orbiliaceae) as affected by juvenile stage of Caenorhabditis elegans. Mycopathologia. 2010;169:467–473. doi: 10.1007/s11046-010-9279-4. [DOI] [PubMed] [Google Scholar]
- 44. Maguire SM, Clark CM, Nunnari J, Pirri JK, Alkema MJ. The C. elegans touch response facilitates escape from predacious fungi. Curr Biol. 2011;21:1326–1330. doi: 10.1016/j.cub.2011.06.063.. •• The authors study the ecological relevance of the nematode touch response in predator-prey relationships between C. elegans and nematophagous fungi that entrap nematodes using hyphal constricting rings. Using competition experiments with behavioral mutants the authors show that the suppression of foraging head movements allows C. elegans to smoothly retract from fungal constricting rings without triggering trap inflation.
- 45.Schmidt AR, Dorfelt H, Perrichot V. Carnivorous fungi from Cretaceous amber. Science. 2007;318:1743. doi: 10.1126/science.1149947. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Hyde KD, Jeewon R, Cai L, Vijaykrishna D, Zhang K. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia. 2005;97:1034–1046. doi: 10.3852/mycologia.97.5.1034. [DOI] [PubMed] [Google Scholar]
- 47.Tinbergen N. On aims and methods of ethology. Zeitschrift für Tierpsychologie. 1963;20:410–433. [Google Scholar]
- 48.Herberholz J, Sen MM, Edwards DH. Escape behavior and escape circuit activation in juvenile crayfish during prey-predator interactions. J Exp Biol. 2004;207:1855–1863. doi: 10.1242/jeb.00992. [DOI] [PubMed] [Google Scholar]
- 49.Willows AO, Dorsett DA, Hoyle G. The neuronal basis of behavior in Tritonia. 3. Neuronal mechanism of a fixed action pattern. J Neurobiol. 1973;4:255–285. doi: 10.1002/neu.480040308. [DOI] [PubMed] [Google Scholar]
- 50.Foreman MB, Eaton RC. The direction change concept for reticulospinal control of goldfish escape. J Neurosci. 1993;13:4101–4113. doi: 10.1523/JNEUROSCI.13-10-04101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]