Abstract
Purpose of review
Treatment-related myelodysplastic syndrome (t-MDS) is a serious complication of cancer treatment. Here we review recent advances in knowledge of the risk factors, pathogenesis and treatment of t-MDS.
Recent findings
Recent studies have provided important new information regarding genetic risk factors that may predispose individual patients to develop t-MDS after exposure to cytotoxic therapeutic agents and that may be used to predict individuals at enhanced risk for this complication. The role of specific candidate genes associated with commonly involved genetic lesions in the pathogenesis of t-MDS has also been investigated. Finally, factors determining outcomes of transplantation treatment for this disorder have been elucidated. HCT provides potentially curative therapy for t-MDS but additional improvements are necessary to improve outcomes.
Summary
Improved understanding of genetic risk factors is expected to facilitate early identification of patients at risk for t-MDS, guiding therapeutic decision making, and allowing early application of preventive or therapeutic strategies.
Keywords: therapy related MDS, genetic polymorphisms, genetic instability, hematopoietic cell transplantation
Introduction
Therapy-related myelodysplasia (t-MDS) is a lethal complication of cytotoxic therapy given for cancer or for non-malignant disorders. Two distinct cytogenetic and clinical types of t-MDS are recognized in the World Health Organization classification, depending on the causative therapeutic exposure.[1]In patients exposed to alkylating agents, t-MDS is associated with abnormalities involving chromosomes 5 (−5/del [5q]) and 7 (−7/del 7q]), a latency of 4 to 7 years after exposure, trilineage dysplasia and an insidious onset. In contrast, the disorder developing secondary to topoisomerase II inhibitors presents as overt acute myeloid leukemia (AML), associated with balanced translocations involving chromosome bands 11q23 or 21q22. The incidence of t-MDS is rising, as a result of the increasing number of patients who survive their primary cancer following cytotoxic therapy with more intensive regimens. Since the nature, dose and timing of exposure to mutagens is known, t-MDS represents an important model for studying the pathogenesis of leukemia in humans. Knowledge of mechanisms of leukemogenesis gained from study of t-MDS may be applicable to de novo MDS and AML in the elderly, which share similar genetic abnormalities. Here we will review recent advances in our understanding of the risk factors, pathogenesis and treatment of t-MDS, specifically genetic risk factors that may predispose individual patients to develop t-MDS after exposure to cytotoxic therapeutic agents, the role of specific candidate genes associated with commonly involved genetic lesions in the pathogenesis of t-MDS, and factors determining outcomes of transplantation treatment for this disorder.
Genetic lesions associated with therapy-related MDS
Loss of chromosome 5 or del (5q) and loss of chromosome 7 and del (7q) are recurring abnormalities in t-MDS.[2] The same abnormalities are also seen in AML evolving from MDS and de novo AML in elderly patients. This observation has led to a search for candidate tumor suppressor genes in these chromosomal regions. The majority of patients with 5q deletions exhibit losses at the 5q31 locus, with deletions in 5q33 seen in some patients. Chromosomal segment 7q22 is a common site of chromosome 7 deletions. Although several candidate genes have been identified in these regions, including genes that regulate hematopoietic cell growth and differentiation, identification of a commonly deleted tumor suppressor gene has been elusive. It is possible that haploinsufficiency and reduced gene dosage for critical genes involved in hematopoiesis on 5q may sufficiently alter the balance between growth and differentiation to induce dysplastic hematopoiesis.[3] Alternatively, epigenetic inactivation of the remaining allele or alterations in gene expression through loss of miRNA loci could play a role.
Graubert et al. systematically evaluated all 28 chromosome 5q31.2 genes for heterozygous nucleotide mutations or microdeletions using total exonic gene resequencing and array comparative genomic hybridization (CGH) of paired bone marrow and germline DNA samples.[4] Forty six patients with de novo MDS, including 37 without a cytogenetically recognizable 5q31.2 deletion were studied. No somatic nucleotide changes were found in the 46 MDS samples, and no cytogenetically silent 5q31.2 deletions were found in 20 samples analyzed by array CGH. The mRNA levels of seven genes in the commonly deleted interval were reduced by 50% in CD34+ cells from del (5q) MDS samples, but complete loss of expression was not seen. Therefore, haploinsufficiency of multiple genes appears to be the major consequence of this deletion, and additional deletions or point mutations in individual 5q31.2 genes are uncommon.
The role of the APC gene located in the commonly deleted 5q region has been studied. Mice with the Apcmin allele that results in a premature stop codon and loss of APC function showed no abnormality in steady-state hematopoiesis.[5] Bone marrow from Apcmin mice showed enhanced repopulation potential, but did not repopulate secondary recipients. Apcmin mice developed an MDS/myeloproliferative phenotype over time, indicating that haploinsufficiency of Apc causes insidious loss of hematopoietic stem cell (HSC) function and could contribute to the MDS phenotype in patients with del (5q).
Molecular analysis of a 2.5-Mb commonly deleted segment of chromosome band 7q22 has failed to uncover a candidate tumor suppressor gene. Shannon and colleagues evaluated whether haploinsufficiency for the 7q22 deleted segment contributes to myeloid leukemogenesis by flanking a region of orthologous synteny on mouse chromosome band 5A3 with loxP sites.[6] Mice with in vivo deletion of the targeted segment showed normal hematologic parameters and did not spontaneously develop myeloid malignancies. The targeted deletion did not cooperate with oncogenic Kras, Nf1 inactivation, or retroviral mutagenesis to accelerate leukemia development. These results fail to support the hypothesis that the 7q22 deletion contains a tumor suppressor gene.
Genetic susceptibility to t-MDS
Underlying genetic characteristics may modify the effects of therapeutic exposures and increase the risk of therapy-related leukemia. Genetic polymorphisms of enzymes capable of metabolic activation or detoxification of anticancer drugs, such as NAD (P) H: quinone oxidoreductase (NQO1), glutathione-S-transferase-M1, -T1, and -P1, and CYP3A4, have been associated with risk of development of therapy-related leukemia or MDS.[7] Other genotyping studies support the role of the p53 pathway in protection against t-MDS/AML. An interactive effect was detected between two common functional p53-pathway variants, the MDM2 SNP309 and the TP53 codon 72 polymorphisms, and increased risk of t-AML/MDS. This effect was observed in patients treated with chemotherapy but not radiotherapy and in patients with loss of chromosome 5 or 7.[8] A genome-wide association study to identify novel loci associated with t-MDS/AML susceptibility in 80 cases and 150 controls using Affymetrix Mapping 10K arrays, followed by testing of identified single nucleotide polymorphisms (SNPs) in an independent cohort of 70 patients, found evidence of association of 3 SNPs with t-MDS/AML with chromosome 5 or 7 lesions. The SNPs identified have not been previously studied in t-MDS/AML and their biological significance remains unknown.[9].
An analysis of mouse strains resistant or susceptible to t-AML induced by the alkylator ethyl-N-nitrosourea(ENU) was performed to identify genes that regulate t-AML susceptibility. Mice used for these studies were in a PML-RARA background. Thirteen quantitative trait loci (QTL) on 8 chromosomes were significantly associated with leukemia-free survival, white blood cell count (WBC) and spleen weight. The identified QTL regions contained several genes with potential roles in leukemogenesis, including p53, DNA repair and apoptosis-regulating genes. These results suggest that susceptibility to alkylator-induced leukemia in mice may be a complex trait related to genes at multiple loci.[10]
Genetic instability in t-MDS
DNA repair mechanisms have a major role in maintaining genomic integrity. The major repair pathways include mismatch repair, base excision repair, nucleotide excision repair, and DNA double-strand break repair. Defects in repair proteins have been associated with therapy-related leukemia.[11] The p53 gene has a critical role in DNA damage response signaling, affecting cell cycle, cell death, and DNA repair pathways. Abnormal p53 activity could lead to reduced ability to repair DNA damage, resulting in genomic instability and increased susceptibility to leukemogenesis. Although p53 mutations are seen in less than 10% of patients with de novo MDS and AML, they are more common in patients with t-MDS, seen in 28% to 38% of patients in different studies, often associated with deletions in chromosome 5 or 7 (or both) or complex karyotypes. [12]
Telomeres are noncoding regions of DNA that provide a cap at the ends of chromosomes and prevent dicentric fusion and other chromosomal aberrations. Each somatic cell division is associated with a loss of telomere length. Cumulative telomere shortening can impose a limit on cell divisions and lead to cell senescence. Telomere shortening is also associated with genetic instability. Short and dysfunctional telomeres may predispose to leukemia by selection of HSC that are prone to genome instability.[13] Analysis of changes in telomere length in serial blood samples from patients who developed t-MDS/AML after autologous HCT revealed accelerated telomere shortening in t-MDS/AML patients when compared with matched controls who did not develop t-MDS/AML.[14] These telomere alterations preceded the onset of t-MDS and were independent of other known risk factors for t-MDS/AML on multivariate analysis. Patients who developed t-MDS/AML also showed reduced generation of committed hematopoietic progenitors suggesting reduced regenerative capacity of HSC. Constitutional hypomorphic telomerase mutations have been observed at increased frequency in patients with AML, suggesting that inherited mutations in TERT that reduce telomerase activity are risk factors for AML. In a cohort of patients with severe aplastic anemia receiving immunosuppressive therapy, telomere length was associated with risk of relapse, clonal evolution, and lower overall survival. The probability of clonal evolution, and evolution to monosomy 7 or complex cytogenetics was more common in patients with shorter telomeres (first quartile) compared with longer telomeres (second to fourth quartiles) [15]
Hematopoietic abnormalities in t-MDS
Extensive proliferation of stem cells bearing genotoxic damage may play a role in the establishment and amplification of an abnormal clone. Gruschkus et al. evaluated the association between colony-stimulating factor (CSF) use and the risk of developing t-MDS in a cohort of elderly patients with non-Hodgkin lymphoma (NHL) who were treated with chemotherapy. CSF use was independently associated with a 53% increased risk of t-MDS.[16] Recent studies have indicated the importance of the hematopoietic microenvironment in the pathogenesis of MDS. Repeated sub-lethal irradiation of mice resulted in bone marrow (BM) dysfunction with clinical features of MDS including low WBC counts, reduced megakaryocyte and platelet levels, and macrocytic anemia. Mice with deletion of tumor necrosis factor (TNF)-α were protected from the irradiation effects:[17] Irradiated wild type (WT) mice with long term BM dysfunction had increased BM angiogenesis, production of matrix metalloproteinases (MMPs) and vascular endothelial growth factor (VEGF) and activation of the transcription factor NFkB/ p65, suggestive of disease progression. These data suggest that TNF-α induction following irradiation may contribute to BM dysfunction and secondary MDS onset and progression. This observation is of particular interest in view of the fact that patients with de novo MDS express high levels of TNF-α in the marrow, and TNF-α induces profound alterations in gene expression in marrow stroma cells.[18] Raaijmakers et al. showed that deletion of Dicer1 specifically in mouse osteoblast progenitors, but not in mature osteoblasts, resulted in hematopoietic abnormalities and the development of MDS and AML.[19] There was reduced expression of the Sbds gene, which is mutated in Schwachman-Bodian-Diamond syndrome, a human marrow failure syndrome associated with predisposition to MDS and leukemia. Deletion of Sbds in mouse osteoblast progenitors induced bone marrow dysfunction with myelodysplasia. These results indicate that perturbation of microenvironmental cells can induce defects in hematopoietic cells and result in MDS.
Gene expression profiling in t-MDS
Gene expression profiling of CD34+ hematopoietic progenitor cells from t-MDS patients identified different subtypes of t-MDS with specific gene expression patterns. Common to each subgroup were gene expression patterns characteristic of arrested differentiation in early progenitor cells. Li et al. performed microarray analysis of gene expression in patients who developed t-MDS after autologous hematopoietic cell transplantation (HCT) for Hodgkin lymphoma or NHL and controls who did not develop t-MDS after autologous HCT.[20] Peripheral blood stem cell (PBSC) samples obtained from t-MDS cases and controls before autologous HCT were studied. This approach allowed identification of gene expression changes pre-HCT in patients who subsequently developed t-MDS. Patients who developed t-MDS showed gene expression profiles significantly different from controls. Genetic alterations in pre-HCT samples were related to mitochondrial function, protein synthesis, DNA repair, and regulation of hematopoiesis. Progression to overt t-MDS was associated with additional alterations in cell-cycle regulatory genes. PBSC CD34+ cells from cases that developed t-MDS demonstrated altered mitochondrial function, increased generation of reactive oxygen species (ROS), reduced ROS detoxification, and enhanced DNA damage after therapeutic exposure. These results also support previous studies indicating a potential role for abnormalities in stem cell growth, protein synthesis, DNA repair and checkpoint response in t-MDS pathogenesis. Importantly these results indicate that genetic programs associated with t-MDS/AML are perturbed long before disease onset. These changes may represent factors predisposing to t-MDS or effects related to pre-HCT therapy.
Therapy of t-MDS
The prognosis of t-MDS is poor, with a life expectancy of typically less than a year. As many as 90% of patients with t-MDS have high risk clonal karyotypes. While various strategies with classic chemotherapy or hypomethylating agents have been pursued, the only currently available therapy that has been shown to have curative potential is allogeneic HCT. A recent study summarized results in 257 patients, 3–72 (median 41) years of age, with “secondary” MDS who had undergone allogeneic HCT; among those, 192 had received various types of cytotoxic therapy.[21] The remaining patients had an antecedent hematologic disorder. Preparation for HCT consisted of either chemotherapy only (n=148; generally busulfan plus Cyclophosphamide (CY)) or chemotherapy plus total body irradiation (TBI; n=109; mostly in combination with CY or fludarabine). Donors were either related (HLA identical siblings in 108, HLA non-identical family members in 25, identical twins, in 2) or unrelated (HLA identical in 98 and non-identical in 24). Five-year relapse-free survival was highest (43%) and non-relapse mortality lowest (28%) in patients conditioned with a regimen that consisted of busulfan (with dose adjustments to a predetermined plasma level) plus CY. Comparison with results in 339 patients with de novo MDS (including transformation to AML) failed to show significant differences in outcome between the two cohorts when adjusted for disease stage and karyotypes, both of which affected significantly relapse probability and relapse-free survival (p=<0.001 for both parameters). Thus, importantly, the driving force behind outcome was not the disease etiology but rather the “severity” of the disease, suggesting that with a given karyotype, the overall prognosis post-transplant is similar regardless of whether the disease occurs de novo or in a patient who has received cytotoxic treatment.
A subsequent analysis of data from the CIBMTR (Center for International Bone Marrow Transplantation Research) database included 323 patients with t-MDS and 545 patients with t-AML who had been transplanted from 1990 through 2004.[22] Treatment given for the original disease consisted of chemotherapy regimens in 385, of radiation alone in 39, and of a combination of radiation plus chemotherapy in 444 patients. The conditioning regimen used in preparation for HCT consisted of various high-dose regimens in 670 patients, and reduced-intensity regimens in 198 patients. Donors were related in 329 patients (HLA-identical siblings in 282 and other family members in 47) and unrelated in the remaining patients (HLA-matched in 204, partially matched/mismatched in 335 patients). Treatment-related non-relapse mortality was 41%, and relapse-related mortality 27% at one year; the corresponding figures at 5 years were 48% and 31%, respectively. The one-year overall survival and relapse-free survival were 37% and 32%, respectively; the corresponding figures at 5 years were 22% and 21%, respectively. The four major risk factors that adversely impacted relapse-free and overall survival were age older than 35 years, poor risk cytogenetics, non-remission/advanced disease stage at HCT, and donors other than HLA-identical siblings or partially or well matched unrelated donors. The probabilities of 5-year survival for patients with none, one, two, three, or four of these risk factors were 50%, 26%, 21%, 10%, and 4%, respectively (p<0.001).
Similar data were reported by the European Group for Blood and Marrow Transplantation in an analysis including 461 patients, 3 – 69 years of age, with t-MDS.[23] Donors were related in 325 patients (HLA identical siblings in 290, other in 35) and unrelated in 136 patients (HLA matched in 95, and mismatched in 41). The 3-year probabilities of relapse and non-relapse mortality were 31% and 37%, respectively. The major risk factors for relapse were not being in remission at the time of HCT (p= 0.002), karyotype (p=0.005), older age (p= 0.03) and t-MDS (p=0.04), while older age correlated with non-relapse mortality. The 3-year overall and relapse-free survival rates were 35% and 33%, respectively. Results improved in recent transplant years.
These data show, therefore, that HCT provides potentially curative therapy for t-MDS as it does for de novo MDS. The risk factors for inferior outcome appear to be the same for both categories of patients, the major ones being disease stage and cytogenetics, with the addition of patient age and donor HLA match or mismatch. It is clear, however, from these analyses, that additional developments are necessary to improve the overall outcome both in regards to non-relapse mortality and relapse.
Conclusion
Recent studies have resulted in improved understanding of genetic risk factors for t-MDS and have elucidated the potential role of specific genetic lesions in the pathogenesis of t-MDS. Continued advances in understanding the genetic basis for t-MDS susceptibility and progression are expected to facilitate early identification of patients at risk for t-MDS, guide therapeutic decision making, and allow early application of preventive or therapeutic strategies, in particular HCT.
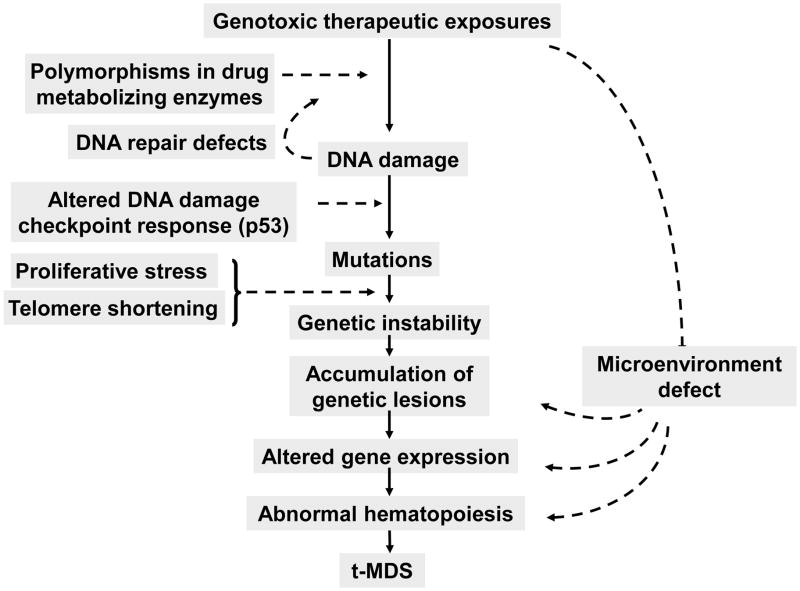
Figure 1. Pathogenesis of t-MDS.
Cytotoxic exposures to hematopoietic stem cells lead to DNA damage. Genetic risk factors including polymorphisms leading to altered drug metabolism, detoxification, DNA repair, DNA damage checkpoint response predispose individual patients to increased risk of developing mutations following cytotoxic exposure. Genetic instability is enhanced by increased proliferative stress related to hematopoietic regeneration and dysregulation of cell cycle, and by excessive telomere shortening. Accumulation of specific lesions in hematopoietic stem cell regulatory genes and damage to the hematopoietic microenvironment by cytotoxic exposures lead to hematopoietic dysfunction and eventually to development of MDS or AML.
Key points.
The role of haploinsufficiency of multiple genes related to the major deletions associated with t-MDS continues to be elucidated
Susceptibility to t-MDS appears to be a complex trait related to genes at multiple loci
Gene expression programs associated with t-MDS are perturbed long before disease onset
HCT provides potentially curative therapy for treatment-related MDS as it does for de novo MDS, but additional developments to improve outcomes are necessary
Acknowledgments
Supported in part by grants R01HL083050 (RB), P01HL036444 (HJD), and P50CA107399 (RB)
References
- *1.Vardiman JW. The World Health Organization (WHO) classification of tumors of the hematopoietic and lymphoid tissues: an overview with emphasis on the myeloid neoplasms. Chem Biol Interact. 2010;184:16–20. doi: 10.1016/j.cbi.2009.10.009. Summarizes the current WNO classification of myeloid malignancies and where t-MDS falls within this. [DOI] [PubMed] [Google Scholar]
- 2.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, Vardiman JW, Rowley JD, Larson RA. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102:43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- *3.Qian Z, Joslin JM, Tennant TR, Reshmi SC, Young DJ, Stoddart A, Larson RA, Le Beau MM. Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia. Chem Biol Interact. 2010;184:50–57. doi: 10.1016/j.cbi.2009.11.025. Reviews cytogenetic and molecular lesions associated with t-MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **4.Graubert TA, Payton MA, Shao J, Walgren RA, Monahan RS, Frater JL, Walshauser MA, Martin MG, Kasai Y, Walter MJ. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS One. 2009;4:e4583. doi: 10.1371/journal.pone.0004583. Systematic evaluation of all genes associated with chromosome 5q31.2 deletions. Results indicate that additional small deletions, point mutations or both in individual 5q31.2 genes are uncommon and that haploinsufficiency of multiple genes is major genetic consequence of this deletion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *5.Lane SW, Sykes SM, Al-Shahrour F, Shterental S, Paktinat M, Lo Celso C, Jesneck JL, Ebert BL, Williams DA, Gilliland DG. The Apc(min) mouse has altered hematopoietic stem cell function and provides a model for MPD/MDS. Blood. 2010;115:3489–3497. doi: 10.1182/blood-2009-11-251728. Shows that haploinsufficiency of Apc gene within the commonly deleted 5q segment causes insidious loss of HSC function and may contribute to the MDS phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **6.Wong JC, Zhang Y, Lieuw KH, Tran MT, Forgo E, Weinfurtner K, Alzamora P, Kogan SC, Akagi K, Wolff L, et al. Use of chromosome engineering to model a segmental deletion of chromosome band 7q22 found in myeloid malignancies. Blood. 2010;115:4524–4532. doi: 10.1182/blood-2009-07-232504. A mouse model of the 7q22 deletion was generated by flanking a region of orthologous synteny on mouse chromosome band 5A3 with loxP sites. Mice with in vivo deletion of the targeted segment showed normal hematologic parameters and did not spontaneously develop myeloid malignancies. These results do not support the presence of a tumor suppressor gene in the commonly deleted segment and the mechanism of its contribution to MDS remains unclear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolufer P, Collado M, Barragan E, Calasanz MJ, Colomer D, Tormo M, Gonzalez M, Brunet S, Batlle M, Cervera J, et al. Profile of polymorphisms of drug-metabolising enzymes and the risk of therapy-related leukaemia. Br J Haematol. 2007;136:590–596. doi: 10.1111/j.1365-2141.2006.06469.x. [DOI] [PubMed] [Google Scholar]
- 8.Ellis NA, Huo D, Yildiz O, Worrillow LJ, Banerjee M, Le Beau MM, Larson RA, Allan JM, Onel K. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–749. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Knight JA, Skol AD, Shinde A, Hastings D, Walgren RA, Shao J, Tennant TR, Banerjee M, Allan JM, Le Beau MM, et al. Genome-wide association study to identify novel loci associated with therapy-related myeloid leukemia susceptibility. Blood. 2009;113:5575–5582. doi: 10.1182/blood-2008-10-183244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Funk RK, Maxwell TJ, Izumi M, Edwin D, Kreisel F, Ley TJ, Cheverud JM, Graubert TA. Quantitative trait loci associated with susceptibility to therapy-related acute murine promyelocytic leukemia in hCG-PML/RARA transgenic mice. Blood. 2008;112:1434–1442. doi: 10.1182/blood-2008-01-132084. The above two papers report analyses of genes that regulate t-AML susceptibility within different mouse strains and suggest that susceptibility to leukemia development is a complex, multigenic trait. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillem V, Tormo M. Influence of DNA damage and repair upon the risk of treatment related leukemia. Leuk Lymphoma. 2008;49:204–217. doi: 10.1080/10428190701769657. [DOI] [PubMed] [Google Scholar]
- 12.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19:1405–1413. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- 13.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14.Chakraborty S, Sun CL, Francisco L, Sabado M, Li L, Chang KL, Forman S, Bhatia S, Bhatia R. Accelerated telomere shortening precedes development of therapy-related myelodysplasia or acute myelogenous leukemia after autologous transplantation for lymphoma. J Clin Oncol. 2009;27:791–798. doi: 10.1200/JCO.2008.17.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Scheinberg P, Cooper JN, Sloand EM, Wu CO, Calado RT, Young NS. Association of telomere length of peripheral blood leukocytes with hematopoietic relapse, malignant transformation, and survival in severe aplastic anemia. JAMA. 2010;304:1358–1364. doi: 10.1001/jama.2010.1376. These two papers shows that telomere shortening precedes and predicts for development of t-MDS in lymphoma patients undergoing autologous HCT or in patients with aplastic anemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruschkus SK, Lairson D, Dunn JK, Risser J, Du XL. Use of white blood cell growth factors and risk of acute myeloid leukemia or myelodysplastic syndrome among elderly patients with non-Hodgkin lymphoma. Cancer. 2010;116:5279–5289. doi: 10.1002/cncr.25525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Cachaco AS, Carvalho T, Santos AC, Igreja C, Fragoso R, Osorio C, Ferreira M, Serpa J, Correia S, Pinto-do OP, et al. TNF-alpha regulates the effects of irradiation in the mouse bone marrow microenvironment. PLoS One. 2010;5:e8980. doi: 10.1371/journal.pone.0008980. Indicates a role for inflammatory mediators in the microenvironment in the pathogenesis of t-MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stirewalt DL, Mhyre AJ, Marcondes M, Pogosova-Agadjanyan E, Abbasi N, Radich JP, Deeg HJ. Tumour necrosis factor-induced gene expression in human marrow stroma: clues to the pathophysiology of MDS? Br J Haematol. 2008;140:444–453. doi: 10.1111/j.1365-2141.2007.06923.x. [DOI] [PubMed] [Google Scholar]
- *19.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 464:852–857. doi: 10.1038/nature08851. Shows that deletion of Dicer1 specifically in mouse osteoblasts resulted in development of MDS and AML related to reduced expression of the Schwachman-Bodian-Diamond syndrome gene. These results further indicate that perturbations of the microenvironment can result in MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Li L, Li M, Sun C-L, Sabado MD, Francisco L, McDonald T, Chang KL, Wang S, Radich J, Zhao LP, et al. Gene Expression Changes in CD34+ Cells Precede Development of Therapy-Related Leukemia (t-MDS) After Autologous Hematopoietic Cell Transplantation (aHCT) for Hodgkin (HL) or Non-Hodgkin Lymphoma (NHL) Blood. 2009;114(22):677. This study shows that gene expression changes associated with t-MDS after HCT are detectable several months tro years prior to development of clinically overt disease. [Google Scholar]
- 21.Chang C, Storer BE, Scott BL, Bryant EM, Shulman HM, Flowers ME, Sandmaier BM, Witherspoon RP, Nash RA, Sanders JE, et al. Hematopoietic cell transplantation in patients with myelodysplastic syndrome or acute myeloid leukemia arising from myelodysplastic syndrome: similar outcomes in patients with de novo disease and disease following prior therapy or antecedent hematologic disorders. Blood. 2007;110:1379–1387. doi: 10.1182/blood-2007-02-076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Litzow MR, Tarima S, Perez WS, Bolwell BJ, Cairo MS, Camitta BM, Cutler CS, de Lima M, Dipersio JF, Gale RP, et al. Allogeneic transplantation for therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 2010;115:1850–1857. doi: 10.1182/blood-2009-10-249128. This paper describes the largest series of transplantation for patients with therapy-related MDS/AML. The report confirms the relevance of risk factors previously described in smaller series, in particular cytogenetics and disease stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroger N, Zabelina T, van Biezen A, Brand R, Niederwieser D, Martino R, Lim ZY, Onida F, Schmid C, Garderet L, et al. Allogeneic stem cell transplantation for myelodysplastic syndromes with bone marrow fibrosis. Haematologica. 2010 doi: 10.3324/haematol.2010.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]