Abstract
Purpose
Mesalazine (5-aminosalicylic acid, 5-ASA) has chemopreventive properties in colitis-associated cancer. In vitro, it improves replication fidelity at (CA)13 microsatellites independent of mismatch repair proficiency. Therefore, 5-ASA might be advantageous in patients with hereditary nonpolyposis colorectal cancer. At this point, however, it is uncertain whether this improvement of replication fidelity is specific for (CA)13 repetitive sequences. Here, we tested the effect of 5-ASA on replication fidelity in mononucleotide, dinucleotide, and tetranucleotide repeats.
Experimental Design
HCT116 and HCT116+chr3 cells were transfected with pIREShyg2-EGFP reporter plasmids harboring the following microsatellites: A10, G10, (CA)13, (CA)26, (AAAG)17, poly-A tracts, and their flanking sequences of transforming growth factor β receptor II (TGFBR2; A10) and activin type II receptor (ACVR2; A8). Stably transfected single-cell clones were selected, characterized by Southern blotting, sorted into six-well plates, and cultured with or without 5-ASA. Frameshift mutations that shift the enhanced green fluorescence protein into its proper reading frame were quantified by flow cytometry.
Results
In HCT116, 5-ASA reduced the mutant fraction at (CA)13 by 48.3%, at A10 by 35.6–43.6%, at G10 by 74.9–83.6%, and at (AAAG)17 by 37.6–44.4%. Similar results were observed in hMLH1-proficient HCT116+chr3 cells. Moreover, the presence of 5-ASA significantly reduced mutations in TGFBR2 (A10) and ACVR2 (A8) by 39.9% and 46.2%, respectively.
Conclusions
5-ASA increases replication fidelity in mononucleotide, dinucleotide, and tetranucleotide repeats and reduces mutations in tumor suppressor genes TGFBR2 and ACVR2, a finding that may provoke in vivo studies for the prevention of colorectal cancer in hereditary nonpolyposis colorectal cancer.
Colorectal cancer is a possible outcome of chronic bowel inflammation, particularly in patients with ulcerative colitis (1). Preventive measures such as surveillance colonoscopy and drug treatment are considered to reduce cancer incidence and mortality (2, 3). Mesalazine (5-aminosalicylic acid, 5-ASA), commonly used in the treatment of ulcerative colitis, seems to have cancer-preventive properties (3). Epidemiologic data supporting this concept are not without uncertainty (3–8), although the antiinflammatory activity of 5-ASA seems to reduce the driving force behind colitis-associated cancer. Additionally, studies have identified important molecular properties of 5-ASA that may counteract colon carcinogenesis independent of its antiinflammatory properties. Improvement of DNA replication fidelity was one of such properties (9). The fidelity of DNA replication is a product of polymerase accuracy, its proofreading activity, and the proficiency of the postreplicational mismatch repair (MMR) system (5). The inefficiency of one of these processes can be a key that lead to the development of human cancer, best illustrated by the familial cancer syndrome hereditary nonpolyposis colorectal cancer (HNPCC, also called Lynch syndrome). In HNPCC, loss-of-function mutations in DNA MMR proteins reduce the activity of postreplicational DNA MMR and strongly elevate the mutation rate, consistent with the mutator phenotype hypothesis as origin of cancer (6). This loss of MMR activity leads to frameshift mutations at repetitive DNA sequences that is termed microsatellite instability (MSI).
We recently used a flow cytometry–based assay to study replication fidelity under 5-ASA. Frameshift mutations were quantified at a (CA)13 microsatellite that shifted an enhanced green fluorescence protein (EGFP) into a +2 bp position, thereby leading to the expression of a truncated nonfluorescent peptide (8, 9). 5-ASA lowered the amount of frameshift mutations at this (CA)13 microsatellite in a dose-dependent fashion (9). Biologically (CA)n repeats are common in noncoding DNA sequences and participate in DNA recombination. For carcinogenesis, frameshift mutations in exonic microsatellites within certain genes are more relevant. Such genes include TGFBR2, BAX, or IGF-2 and harbormostly poly-A and poly-G tracts (10). To better judge the 5-ASA's properties on improvement of replication fidelity and its possible chemopreventive application in HNPCC families, we tested its ability to stabilize other microsatellites including poly-A, poly-G, and portions of exon 3 of transforming growth factor β receptor II (TGFBR2) and exon 10 of activin type II receptor (ACVR2) sequences in vitro.
Materials and Methods
Cell culture
HCT116 (hMLH1 mutant) and LoVo (hMSH2 mutant) colorectal cancer cells were obtained from the American Type Culture Collection. HCT116+chr3 (hMLH1 wild-type; ref. 11) and Lovo+chr2 (hMSH2 wildtype) were kind gifts from Dr. Boland and Dr. Koi (Baylor University Medical Center, Dallas, Texas). Cells were grown in Iscove'smodifiedDulbecco'smedium(Life Technologies - Invitrogen) containing 10% fetal bovine serum (Biochrom). The medium for HCT116+chr3 additionally contained 400 µg/mL and the medium for Lovo+chr2 contained 700 µg/mL Geneticin (G418, Life Technologies -Invitrogen). HCT116 reporter cell lines harboring poly-A tracts surrounded by flanking regions of exon 3 and exon 10 of TGFBR2 and ACVR2 were kindly provided by Dr. Carethers and Dr. Chung (University of California San Diego, La Jolla, California; ref. 10) and were grown in Iscove's modified Dulbecco's medium containing 200 µg/mL hygromycin B (Life Technologies). All cells were maintained without antibiotics at 37°C, full humidity, and 5% CO2.
Plasmid construction and sequence analysis
Using the restriction enzymes PmeI and AscI (New England Biolabs), the previously established pIREShyg2-EGFP vector (12) was linearized. Sense and antisense oligonucleotides A10, (AAAG)17, (CA)26, and G10 with a 5′-CGCG overhang to create a compatible site for the linearized pIREShyg2-EGFP were annealed and directionally cloned into the PmeI-AscI site of pIREShyg2-EGFP to generate the plasmids pIREShyg2-EGFP-(oligonucleotide). The ligation products pIREShyg2-EGFP(A)10, pIREShyg2-EGFP(G)10, pIREShyg2-EGFP(CA)26, and pIREShyg2-EGFP(AAAG)17 as well as the previously established pIREShyg2-EGFP (CA)13 (12) were transformed into Stbl2 (Life Technologies) and the cells were grown at 30°C on selection agar plates. Ampicillin-resistant colonies were selected and the correct sequence of the microsatellite insert was confirmed by PCR amplification with 5′-EGFP (5′-CCCACTGCTTACTGGCTTATCfG-3′) and 3′-EGFP (3′-CCTGAAGTTCATCTGCACCACC-5′). Subsequent sequencing with the same primer set on an ABI Prism 310 (Applied Biosystems, Inc.) using the DyEx 2.0 Spin kit (Qiagen) was done according to the manufacturer's manual.
Transfection and stable clone selection
Cells were transfected with the above-described reporter plasmids using Effectene (Qiagen) according to the manufacturer's manual. Twenty-four hours after transfection, cells were selected with 200 µg/mL hygromycin B (HCT116 and LoVo) or, 150 µg/mL hygromycin B and 400 or 700 µg/mL geneticin (HCT116+chr3 and LoVo+chr2 cells), respectively. Stable cell clones were analyzed by PCR and subsequent cycle sequencing.
Southern blot analysis
The number of integrated plasmid into the genome of the clones was identified by Southern blotting. Total cellular DNA from stably transfected cells that had been grown from single-cell colonies was isolated by phenol/chloroform/isoamylalcohol extraction. Twenty micrograms of total DNA were digested with BamHI, EcoRV, or both (New England Biolabs), resolved on a 0.8% agarose gel, and transferred onto a nylon membrane (Hybond N, Amersham Pharmacia) by vacuum blotting (Pharmacia-LKB Vacugene 2016). EGFP cDNA labeling, hybridization, and washing conditions were done as previously described (13).
Analysis of mutant fractions by flow cytometry
Nonfluorescent HCT116, HCT116+chr3, Lovo, and Lovo+chr2 frameshift-reporter cell lines were sorted into six-well plates on a FACSAria cell sorter by using the CloneCyt Plus software and sorting technology (Becton Dickinson Immunocytometry Systems). After 8 d of culture with or without 5 mmol/L 5-ASA, cells were rinsed with cold Ca2+/Mg2+-free PBS (Life Technologies-Invitrogen) and detached with 320 µL Accutase (PAA Laboratories). Eighty microliters of the cell suspension were analyzed on a FACScan using Cyflogic flow cytometry data analysis software (ver. 1.2.1, CyFlo Ltd.) and cell counts were multiplied by four to quantify the total number of cells per well. The EGFP-positive mutant fraction was expressed as percentage of the total cell number.
Statistical analysis
Experiments were carried out in triplicates and repeated twice. Data are represented as mean with the SD and compared by using the Student's t test or one-way ANOVA. P values of <0.05 were considered to be statistically significant.
Results
Establishment and characterization of clones
Various mononucleotide, dinucleotide, and tetranucleotide DNA repeat sequences were inserted after the translation initiation codon of the EGFP gene in the plasmid pIREShyg2-EGFP, resulting in a frameshift that leads to a truncated, nonfluorescent protein. Frameshift mutations within the repetitive sequence will restore proper EGFP protein expression. Compatible DNA oligo-nucleotides [A10, G10, (CA)26, and (AAAG)17] were cloned into pIREShyg2-EGFP as described in ref. (12). HCT116 and HCT116+chr3 cells were transfected with these pIREShyg2-EGFP reporter plasmids and stable single-cell clones were selected. Additionally, hMHS2-proficient and hMSH2-deficient cells (LoVo+chr2, LoVo) were transfected with pIREShyg2-EGFP-(CA)13. Clones were characterized by sequencing, Southern blotting, and flow cytometry (data not shown). HCT116 cells harboring A10 and A8 DNA repeats within flanking regions of exon 3 and exon 10 of TGFBR2 and ACVR2, respectively, were previously established (10).
Effect of 5-ASA on cell growth
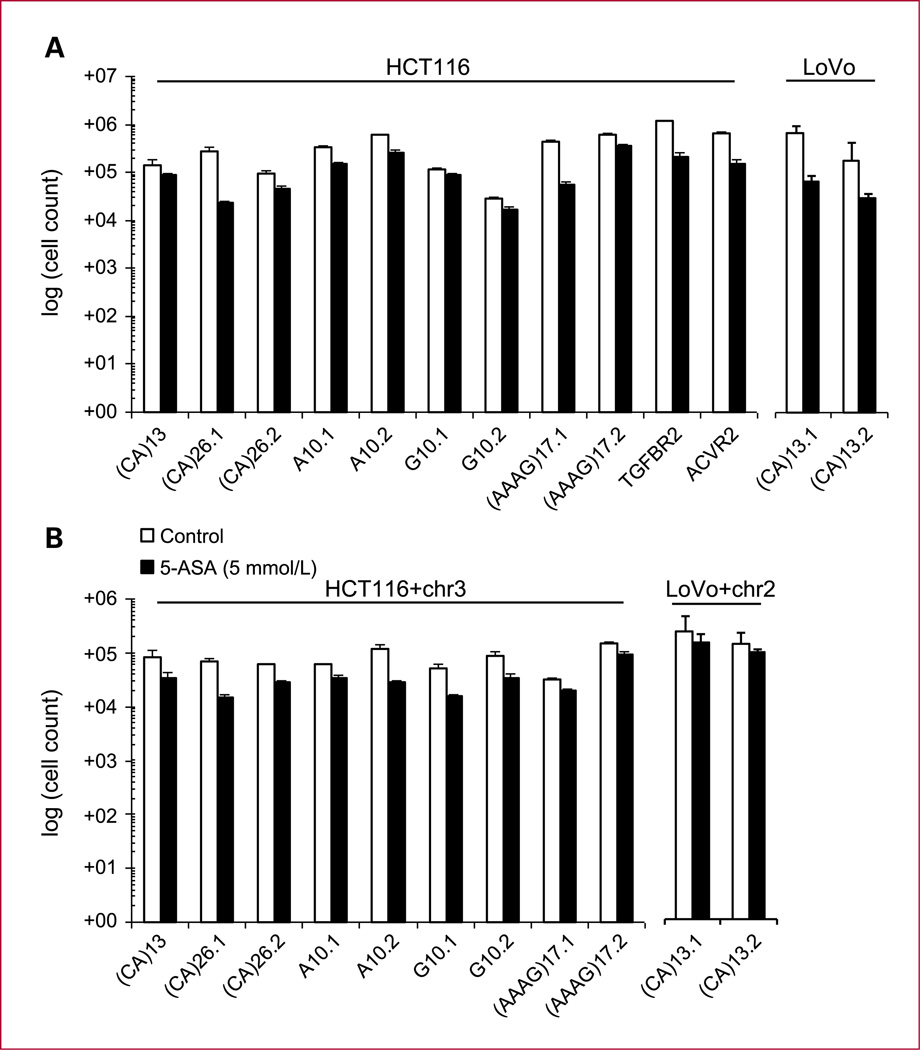
We have previously reported on 5-ASA activating a replication checkpoint and thereby reducing the proliferation speed (14). To control for such antiproliferative properties of 5-ASA, we analyzed the total number of MMR-deficient HCT116 and LoVo and MMR-proficient HCT116+chr3 and LoVo+chr2 cells harboring the above-described reporter plasmids. We observed a significant reduction of cell growth upon treatment with 5-ASA (P < 0.001 by ANOVA; Fig. 1A and B). This suggests that 5-ASA inhibits cell proliferation in colon epithelial cells.
Fig. 1.
Effect of 5-ASA on the number of cultured cells. To test the effect of 5-ASA (5 mmol/L) on cell growth, stably transfected cell lines were cultured for 8 d and the total cell count was analyzed. Except for LoVo+chr2 cells a reduction in the total cell count was seen for all clones (P < 0.001). One-way ANOVA method was used for statistical analysis; columns, mean of triplicate cultures for each clone; bars, SEM.
Effect of 5-ASA on poly-CA tracts
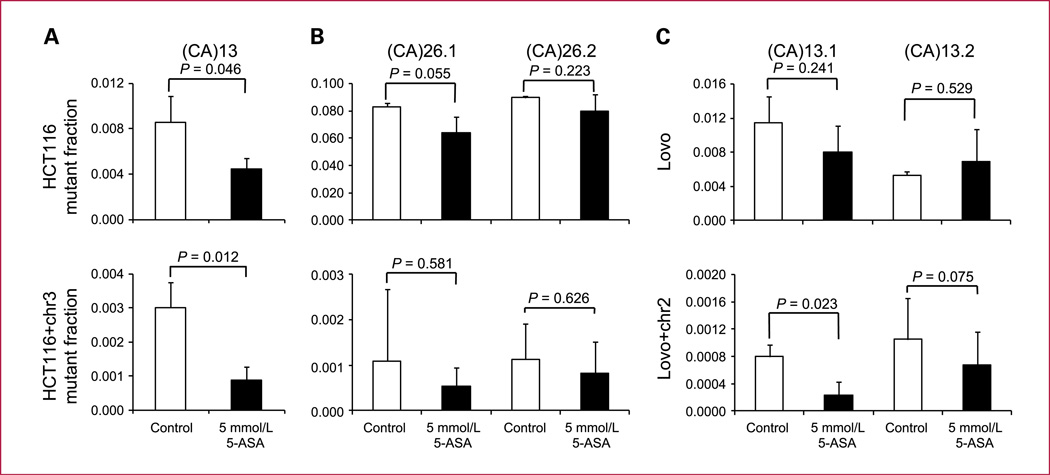
To test the effect of 5-ASA on replication fidelity at poly-(CA) repeats, nonfluorescent HCT116 and HCT116+chr3 cells harboring (CA) 13 (12) or (CA)26 reporter plasmids as well as hMSH2-deficient LoVo and hMSH2-proficient LoVo+chr2 cells harboring the (CA)13 plasmid were sorted into six-well plates. Twenty-four hours later, 5-ASA (5 mmol/L) was added, and after 8 days, the EGFP-positive mutant fraction was analyzed by flow cytometry. 5-ASA treatment resulted in a significant reduction of the mutant fraction in HCT116 and HCT116+chr3 cells harboring a (CA)13 repeat from 8.5 ± 2.3 (×10−3) to 4.4 ± 1.0 (×10−3) and from 3.0 ± 0.8 (×10−3) to 0.9 ± 0.4 (×10−3), respectively (Fig. 2A) confirming previous results (12). This was also found to some extent in one LoVo clone (CA)13.1 but not in LoVo (CA)13.2 (Fig. 2C). Both MMR-proficient clones in LoVo+chr2 also improved. HCT116 (CA)26 clones are hypermutable with a large mutant fraction (8% of cells after 8 days of culture). In these clones as well as in their HCT116+chr3 counterparts, we did not observe a significant reduction of the mutant fraction (Fig. 2B). These results suggest that a reduction of mutations by 5-ASA depends on the stability of poly-(CA) repeats.
Fig. 2.
Effect of 5-ASA on dinucleotide microsatellites. The effect of 5-ASA on replication fidelity within the dinucleotide repeat sequences (CA)13 (A and C) and (CA)26 (B) repeats was tested. Nonfluorescent cells (5 × 103) were sorted into six-well plates and treated with 5 mmol/L 5-ASA for a period of 8 d. The EGFP-positive mutant fraction was analyzed by flow cytometry. Columns, mean of triplicate cultures for each clone; bars, SEM.
Effect of 5-ASA on mononucleotide repeats
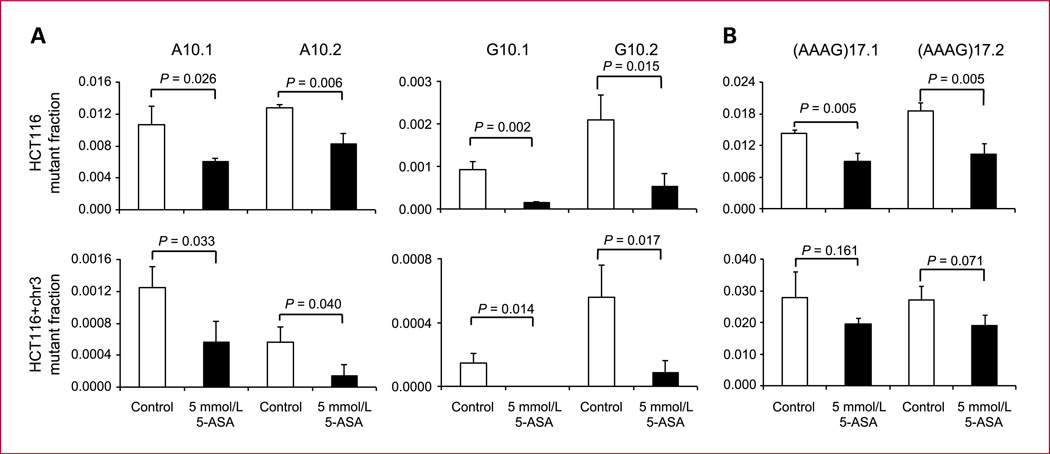
Mononucleotide repeats are abundant in the human genome and are frequently mutated in the coding region of tumor suppressor genes within MSI cancers (15). We investigated the effect of 5-ASA on the replication fidelity within poly-A and poly-G tracts. Nonfluorescent HCT116 and HCT116+chr3 cells containing an A10 or G10 repeat were incubated with 5 mmol/L 5-ASA. In HCT116 cells with an A10 repeat, 5-ASA significantly reduced the mutant fraction from 1.1 ± 0.2 (×10−2) to 0.6 ± 0.05 (×10−2) in clone A10.1 and from 1.3 ± 0.04 (×10−2) to 0.8 ± 0.1 (×10−2) in the A10.2 clone (P < 0.05). In HCT116+chr3 cells with an A10 repeat, 5-ASA reduced the mutant fraction from 1.2 ± 0.3 (×10−3) to 0.6 ± 0.3 (×10−3) in clone A10.1 and from 0.6 ± 0.2 (×10−3) to 0.1 ± 0.1 (×10−3) in clone A10.2 (P < 0.05; Fig. 3A). Similarly, in the G10 repeat harboring HCT116 clones, 5-ASA reduced the mutant fraction from 9.3 ± 1.9 (×10−4) to 1.5 ± 0.8 (×10−4) in G10.1 and from 2.1 ± 0.6 (×10−3) to 0.5 ± 0.3 (×10−3) in the G10.2 clone (P < 0.05). In HCT116+chr3 cells with a G10 repeat, 5-ASA reduced the mutant fraction from 1.5 ± 0.6 (×10−4) to 0.0 ± 0.0 (×10−4) in G10.1 and from 5.6 ± 2.0 (×10−4) to 0.8 ± 0.8 (×10−4) in the G10.2 clone (P < 0.05; Fig. 3B). These results suggest that the treatment with 5-ASA improves the replication fidelity within short poly-A and poly-G tracts.
Fig. 3.
Effect of 5-ASA on mononucleotide and tetranucleotide microsatellites. The effect of 5-ASA on replication fidelity within mononucleotide repeat sequences A10, G10 (A), and (AAAG)17 repeats (B) was tested. Nonfluorescent cells (5 × 103) were sorted into six-well plates and treated with 5 mmol/L 5-ASA for a period of 8 d. The EGFP-positive mutant fraction was analyzed by flow cytometry. Columns, mean of triplicate cultures for each clone; bars, SEM.
Effect of 5-ASA on tetranucleotide repeats
Mutations within tetranucleotide tracts are associated with head, neck, bladder, and respiratory tract cancers (16). We investigated the effect of 5-ASA on the replication fidelity within a poly-(AAAG) repeat. Nonfluorescent HCT116 and HCT116+chr3 cells (5 × 103) containing (AAAG)17 were treated with 5-ASA for 8 days. In HCT116 (AAAG)17.1 and HCT116 (AAAG)17.2, a significant reduction of the mutant fraction from 1.4 ± 0.07 (×10−2) to 0.9 ± 0.2 (×10−2) in (AAAG)17.1 and from 1.9 ± 0.2 (×10−2) to 1.0 ± 0.2 (×10−2) in the (AAAG)17.2 clone was observed (P < 0.05; Fig. 3B, top). However, this effect did not reach significance in MMR-proficient HCT116+chr3 clones (Fig. 3B, bottom). Interestingly the mutant fraction in HCT116+chr3 is similar to HCT116. In fact, both HCT116 and HCT116+chr3 cells are lacking hMSH3, which is necessary to repair insertion/deletion loops within tetranucleotide repeat tracts (17). Similar to (CA)26, the high mutation rate in these cells may interfere with the ability of 5-ASA to sufficiently reduce the mutation rate in tetranucleotide repeats. This effect might be different in normal colon epithelial cells that are hMSH3 proficient.
Effect of 5-ASA on poly-A repeats in TGFBR2 and ACVR2
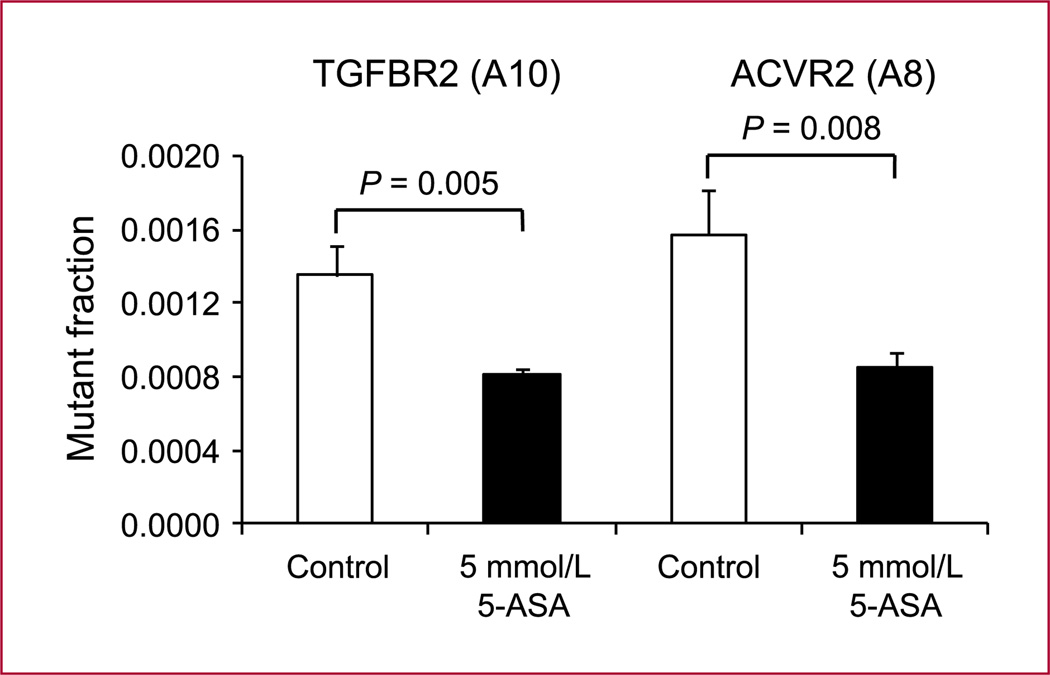
The type II receptor for TGFBR2 harbors an A10 repeat within exon 3. Frameshift mutations within this polyadenine sequence lead to a truncated TGFBR2 protein, which is nonfunctional and interrupts TGF-β signaling (18). ACVR2 contains polyadenine tracts in exons 3 and 10 but only the A8 tract in exon 10 is mutated in ~85% of colorectal cancers with MSI (19). We tested the effect of 5-ASA on the replication fidelity within these exon-coding poly-A tracts (surrounded by flanking regions of exon 3 and exon 10 of TGFBR2 and ACVR2) in HCT116 cells. In TGFBR2 (A10) as well as in ACVR2 (A8), a significant reduction of the mutant fraction from 1.4 ± 0.2 (×10−3) to 0.8 ± 0.04 (×10−3) and from 1.6 ± 0.2 (×10−3) to 0.8 ± 0.1 (×10−3), respectively was observed (P < 0.05; Fig. 4). These results suggest that 5-ASA leads to a reduction of mutational events within poly-A tracts in tumor suppressor genes TGFBR2 and ACVR2.
Fig. 4.
Effects of 5-ASA on poly-A microsatellites in TGFBR2 and ACVR2. 5-ASA was tested on replication fidelity in A10 and A8 repeats within flanking regions of exon 3 and 10 of TGFBR2 and ACVR2, respectively. Nonfluorescent cells (5 × 103) were sorted into six-well plates and treated with 5 mmol/L 5-ASA for a period of 8 d. The EGFP-positive mutant fraction was quantified by flow cytometry. Columns, mean of triplicate cultures for each clone; bars, SEM.
Discussion
In the eukaryotic genome, polymerase slippage during DNA replication within microsatellite sequences leads to frameshift mutations (20). Such events involve the insertion or deletion of repeat units. Several factors influence the stability of such repeat sequences. These factors are the length, composition, repeat unit size of DNA repeats (21), and most likely also the surrounding sequence. Mononucleotide repeats are the most abundant repeat motifs in humans and are frequently mutated within certain types of cancer such as HNPCC (15) but also dinucleotide repeats such as in epidermal growth factor receptor or within the IGF-I gene, which is linked to breast cancer or colitis-associated cancers exhibit frequent mutations (22). Mutations within tetranucleotide repeats are associated with head, neck, bladder, and respiratory tract cancers (16). This study indicates that 5-ASA reduces mutations within mononucleotide, dinucleotide, and tetranucleotide repetitive sequences in vitro irrespective of a functional MMR, a finding that may be used for chemoprevention in HNPCC.
Previously, 5-ASA showed an improvement of replication fidelity within a (CA)13 repeat using a vector-based assay for the detection of frameshifts (9). Here, we confirm this finding and extend the analysis to mononucleotide and tetranucleotide sequences as well as exonic TGFBR2 and ACVR2 microsatellites. TGFBR2 harbors an A10 repeat within exon 3. Frameshift mutations within this polyadenine sequence lead to a truncated, nonfunctional protein (18). ACVR2 contains polyadenine tracts in exons 3 and 10 but only the A8 tract in exon 10 is mutated in ~85% of colorectal cancers with MSI indicating that the flanking sequence of a gene influences frameshift mutations. It is of clinical relevance that 5-ASA leads to a reduction of mutations at these specific sequences. For the current experiments, we used the same 5-ASA concentration (5 mmol/L) that was previously the most active (9) and which compares to nontoxic, intraluminal in vivo 5-ASA concentrations in ulcerative colitis (estimated between 12.6 and 23.7 mmol/L; ref. 23). At this concentration, all cell lines exhibited a mild reduction of cell growth, which has been also observed in other studies and was attributed to the activation of a replication checkpoint (14, 24).
Changes in clones containing the hypermutable (CA)26 and (AAAG)17 sequences did not reach the level of significance. This may be due to the high instability of such repeats or higher variations in the experiments. In addition, HCT116+chr3 cells with the (AAAG)17 repeat reveal a high mutation rate due to the lack of hMSH3, a MMR component that is critical for the repair of tetranucleotide repeats (17). Interestingly, in hMSH2-deficient LoVo cells, the effect of 5-ASA on replication fidelity was less pronounced, although 5-ASA also affected cell proliferation and cell cycle (14). A simple explanation is the slower growth rate of LoVo (compared with HCT116) with less than half cell division cycles in the same period of time and thus fewer opportunities to cause or prevent frameshift mutations. However, it is also possible that the different genetic make up of LoVo and HCT116 cells is responsible for this difference, an interpretation that warrants caution when translating our in vitro data into the in vivo situation.
It has been shown that aspirin increases MMR protein expression and subsequent apoptosis, suggesting that the upregulation of the MMR system might be another chemopreventive mechanism of aspirin and related nonsteroidal antiinflammatory drugs (25). Aspirin also induces genetic selection for microsatellite stability in a subset of MMR-deficient cells and could provide prophylactic therapy for HNPCC patients in which mutations of the hMSH2 and hMLH1 genes are associated with an increased risk for colorectal cancer (26). However, a recent study conducted in patients with HNPCC showed no beneficial effects of aspirin (27). This might be related to the fact that aspirin does not improve replication fidelity within microsatellite sequences as previously shown with an in vitro model (9). It could also be negative in this study due to the germline's nature of the patients defect, or the dose, duration, and timing in the disease process. It would also be interesting to study a possible effect in sporadic colorectal cancer patients with MSI.
Here, we used an EGFP-based frameshift reporter system to analyze and compare the effects of 5-ASA on replication fidelity within different mononucleotide, dinucleotide, and tetranucleotide repeats in MMR-proficient and MMR-deficient colon epithelial cells. 5-ASA increases replication fidelity in mononucleotide, dinucleotide, and tetranucleotide repeats and reduces microsatellite instability in tumor suppressor genes TGFBR2 and ACVR2, a feature that might be beneficial for the prevention of cancer in patients with HNPCC. However, biological effects observed in vitro might not be predictive that the same effects will occur in a more complex in vivo system. Unfortunately, representative in vivo models for HNPCC are lacking. Selective deletion of MSH2 in mice has been associated with increased intestinal tumors (28). However, both the distribution (small bowel predominant) and the mutational spectrum (common adenomatous polyposis coli point mutations) are different from HNPCC tumors. In addition, the mouse TGFBR2 gene does not carry an intronic microsatellite making a direct comparison difficult. Despite these shortcomings, we will study the potential chemopreventive effect of 5-ASA in villin-cre MSH2LoxP/LoxP mice in vivo.
Translational Relevance.
Mesalazine (5-aminosalicylic acid, 5-ASA) is widely used in the treatment of patients with ulcerative colitis. Several studies suggest an activity of 5-ASA in preventing colorectal cancer in the setting of ulcerative colitis. Beside its antiinflammatory properties, 5-ASA improves replication fidelity within a (CA)13 repeat in vitro. However, it has not been shown whether this effect is also achieved within other repetitive sequences such as poly-A or poly-G tracts, which are important mutational targets in certain tumor suppressor genes such as transforming growth factor β receptor II. This communication provides evidence that 5-ASA improves replication fidelity within such mononucleotide repeats in vitro, a feature that might be beneficial for the prevention of cancer in patients with hereditary nonpolyposis colorectal cancer.
Acknowledgments
We thank Drs. Rick Boland, Minoru Koi, Ajay Goel, and Thomas Jascur (Baylor University Medical Center, Dallas) for the HCT116+chr3, LoVo+chr2 cells and for the kind gift of pIREShyg2-EGFP-A10, pIREShyg2-EGFP-G10, and pIREShyg2-EGFP-G16 reporter plasmids and Dr. Martin Willheim and Guenther Hofbauer for cell sorting (Core Unit Zellsortierung, Medical University of Vienna).
Grant Support
Austrian Science Fund (FWF grant P18270) and the Christian Doppler Research Association.
Footnotes
Disclosure of Potential Conflicts of Interest
C. Gasche has research collaborations with Giuliani SpA and Shire Pharmaceuticals. The other authors do not have a conflict of interest to report.
References
- 1.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 2.Croog VJ, Ullman TA, Itzkowitz SH. Chemoprevention of colorectal cancer in ulcerative colitis. Int J Colorectal Dis. 2003;18:392–400. doi: 10.1007/s00384-002-0476-6. [DOI] [PubMed] [Google Scholar]
- 3.Rubin DT, Cruz-Correa MR, Gasche C, et al. Colorectal cancer prevention in inflammatory bowel disease and the role of 5-aminosalicylic acid: a clinical review and update. Inflamm Bowel Dis. 2007;14:265–274. doi: 10.1002/ibd.20297. [DOI] [PubMed] [Google Scholar]
- 4.Velayos FS, Terdiman JP, Walsh JM. Effect of 5-aminosalicylate use on colorectal cancer and dysplasia risk: a systematic review and metaanalysis of observational studies. Am J Gastroenterol. 2005;100:1345–1353. doi: 10.1111/j.1572-0241.2005.41442.x. [DOI] [PubMed] [Google Scholar]
- 5.Velayos FS, Loftus EV, Jr, Jess T, et al. Predictive and protective factors associated with colorectal cancer in ulcerative colitis: a case-control study. Gastroenterology. 2006;130:1941–1949. doi: 10.1053/j.gastro.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Rubin DT, Turner JR. Surveillance of dysplasia in inflammatory bowel disease: the gastroenterologist-pathologist partnership. Clin Gastroenterol Hepatol. 2006;4:1309–1313. doi: 10.1016/j.cgh.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terdiman JP, Steinbuch M, Blumentals WA, Ullman TA, Rubin DT. 5-Aminosalicylic acid therapy and the risk of colorectal cancer among patients with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:367–371. doi: 10.1002/ibd.20074. [DOI] [PubMed] [Google Scholar]
- 8.Jess T, Gamborg M, Munkholm P, Sorensen TI. Overall and cause-specific mortality in ulcerative colitis: meta-analysis of population- based inception cohort studies. Am J Gastroenterol. 2007;102:609–617. doi: 10.1111/j.1572-0241.2006.01000.x. [DOI] [PubMed] [Google Scholar]
- 9.Gasche C, Goel A, Natarajan L, Boland CR. Mesalazine improves replication fidelity in cultured colorectal cells. Cancer Res. 2005;65:3993–3997. doi: 10.1158/0008-5472.CAN-04-3824. [DOI] [PubMed] [Google Scholar]
- 10.Chung H, Young DJ, Lopez CG, et al. Mutation rates of TGFBR2 and ACVR2 coding microsatellites in human cells with defective DNA mismatch repair. PLoS ONE. 2008;3:e3463. doi: 10.1371/journal.pone.0003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koi M, Umar A, Chauhan DP, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. ancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 12.Gasche C, Chang CL, Natarajan L, et al. Identification of frame-shift intermediate mutant cells. Proc Natl Acad Sci U S A. 2003;100:1914–1919. doi: 10.1073/pnas.0437965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasche C, Chang CL, Rhees J, Goel A, Boland CR. Oxidative stress increases frameshift mutations in human colorectal cancer cells. Cancer Res. 2001;61:7444–7448. [PubMed] [Google Scholar]
- 14.Luciani MG, Campregher C, Fortune JM, Kunkel TA, Gasche C. 5-ASA affects cell cycle progression in colorectal cells by reversibly activating a replication checkpoint. Gastroenterology. 2007;132:221–235. doi: 10.1053/j.gastro.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bubb VJ, Curtis LJ, Cunningham C, et al. Microsatellite instability and the role of hMSH2 in sporadic colorectalcancer. Oncogene. 1996;12:2641–2649. [PubMed] [Google Scholar]
- 16.Xu L, Chow J, Bonacum J, et al. Microsatellite instability at AAAG repeat sequences in respiratory tract cancers. Int J Cancer. 2001;91:200–204. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1031>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 17.Haugen AC, Goel A, Yamada K, et al. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 2008;68:8465–8472. doi: 10.1158/0008-5472.CAN-08-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz S, Wang J, Myeroff L, et al. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 19.Jung B, Doctolero RT, Tajima A, et al. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 21.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brentnall TA, Crispin DA, Bronner MP, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. cancer Res. 1996;56:1237–1240. [PubMed] [Google Scholar]
- 23.Staerk LL, Stokholm M, Bukhave K, Rask-Madsen J, Lauritsen K. Disposition of 5-aminosalicylic acid by olsalazine and three mesalazine preparations in patients with ulcerative colitis: comparison of intraluminal colonic concentrations, serum values, and urinary excretion. Gut. 1990;31:1271–1276. doi: 10.1136/gut.31.11.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinacher-Schick A, Schoeneck A, Graeven U, Schwarte-Waldhoff I, Schmiegel W. Mesalazine causes a mitotic arrest and induces caspase- dependent apoptosis in colon carcinoma cells. Carcinogenesis. 2003;24:443–451. doi: 10.1093/carcin/24.3.443. [DOI] [PubMed] [Google Scholar]
- 25.Goel A, Chang DK, Ricciardiello L, Gasche C, Boland CR. A novel mechanism for aspirin-mediated growth inhibition of human colon cancer cells. Clin Cancer Res. 2003;9:383–390. [PubMed] [Google Scholar]
- 26.Ruschoff J, Wallinger S, Dietmaier W, et al. Aspirin suppresses the mutator phenotype associated with hereditary nonpolyposis colorectal cancer by genetic selection. Proc Natl Acad Sci U S A. 1998;95:11301–11306. doi: 10.1073/pnas.95.19.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burn J, Bishop DT, Mecklin JP, et al. Effect of aspirin or resistant starch on colorectal neoplasia in the Lynch syndrome. N Engl J Med. 2008;359:2567–2578. doi: 10.1056/NEJMoa0801297. [DOI] [PubMed] [Google Scholar]
- 28.Kucherlapati MH, Lee K, Nguyen AA, et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 2009 doi: 10.1053/j.gastro.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]