Abstract
Background and objective: Liver regeneration is a complex process regulated by a group of genetic and epigenetic factors. A variety of genetic factors have been reported, whereas few investigations have focused on epigenetic regulation during liver regeneration. In the present study, valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, was used to investigate the effect of HDAC on liver regeneration. Methods: VPA was administered via intraperitoneal injection to 2/3 partially hepatectomized mice to detect hepatocyte proliferation during liver regeneration. The mice were sacrificed, and their liver tissues were harvested at sequential time points from 0 to 168 h after treatment. DNA synthesis was detected via a BrdU assay, and cell proliferation was tested using Ki-67. The expressions of cyclin D1, cyclin E, cyclin dependent kinase 2 (CDK2), and CDK4 were detected by Western blot analysis. Chromatin immunoprecipitation (ChIP) assay was used to examine the recruitment of HDACs to the target promoter regions and the expression of the target gene was detected by Western blot. Results: Immunohistochemical analysis showed that cells positive for BrdU and Ki-67 decreased, and the peak of BrdU was delayed in the VPA-administered mice. Consistently, cyclin D1 expression was also delayed. We identified B-myc as a target gene of HDACs by complementary DNA (cDNA) microarray. The expression of B-myc increased in the VPA-administered mice after hepatectomy (PH). The ChIP assay confirmed the presence of HDACs at the B-myc promoter. Conclusions: HDAC activities are essential for liver regeneration. Inhibiting HDAC activities delays liver regeneration and induces liver cell cycle arrest, thereby causing an anti-proliferative effect on liver regeneration.
Keywords: Liver regeneration, Epigenetic factors, Cell cycle, VPA, B-myc
1. Introduction
The liver is one of few organs in the body capable of regeneration after injury. In a study by Higgins and Anderson (1931), using a 2/3 hepatectomy (PH) model, the remaining rat liver mass recovered its original weight within one week after surgery. The regenerative properties of the liver contribute substantially to the treatment of liver diseases and surgical operations. For example, liver transplantation is considered the most effective feasible therapy for end-stage liver diseases (Fujiyoshi and Ozaki, 2011; Riehle et al., 2011). During shortage of donated organs, living liver donors are one of the most important sources of livers for transplantation (Merion, 2010). Thus, the fast regeneration of liver tissue is of particular importance for both donors and recipients. Currently, the regulation of liver regeneration is a crucial issue in hepatology research. Numerous cytokines and growth factors reportedly participate in liver regeneration. However, most studies have focused on the genetic regulation of liver regeneration and few have focused on its epigenetic control (Zimmermann, 2004).
Recently, investigations have shown that epigenetic control is essential in many biological processes, such as development, tumorigenesis, cell proliferation, apoptosis, and recovery from injury. Studies have demonstrated the participation of epigenetic events in the pathogenesis of many diseases (Cheng and Blumenthal, 2011; Katsuyama and Paro, 2011; Martín-Subero and Esteller, 2011). Epigenetic control is the regulation of gene transcription without any change in the DNA sequence. Among epigenetic events, histone acetylation/deacetylation is one of the most clearly understood genetic modifications. The level of histone acetylation is regulated by two important types of enzymes, histone deacetylases (HDACs) and histone acetyltransferases (HATs), which work in opposition. HATs acetylate histone tails and set the chromosome to a relaxed status. This status is suitable for the binding of transcription factors to the promoter regions of genes, which then initiate transcription. HDACs work in reverse by removing the acetyl group of the histone tail, setting the chromosome to a compact status. Consequently, the DNA is then inaccessible for the binding of transcription factors, and certain gene expression is inhibited. Therefore, HATs can be regarded as activators and HDACs as repressors of gene expression. However, this is not always true as in the case of HDACs that regulate repressor genes, which inhibit target gene expression (Turner, 2000; Nakayama and Takami, 2001).
B-myc is a member of the myc family discovered by Ingvarsson et al. (1988). It is expressed at low levels in mature tissue cells. Unlike most members of the myc family, B-myc has only an encoded exon similar to the C-myc 2# exon in structure. B-myc protein has an N-terminal region similar to that of C-myc (Ingvarsson et al., 1988). Hence, B-myc also plays a regulatory role in genetic transcription (Resar et al., 1993; Facchini and Penn, 1998). Considering B-myc lacks the basic helix-loop-helix-zipper DNA-binding domain possessed by other members of the myc family, it might bond with other regulatory nuclear transcription proteins for joint transcription regulation (Asker et al., 1995; Sakamuro and Prendergast, 1999; Cornwall et al., 2001; Burton et al., 2006). Previous studies have found that the myc box II (MB II) structural domain of B-myc could bond with the GAL4/C-myc chimera to inhibit C-myc (Resar et al., 1993; Gregory et al., 2000; Cornwall et al., 2001), which demonstrates its inhibitory function during cell proliferation. Hence, B-myc may be a negative regulatory factor in proliferation.
In the present research, the focus was on the effect of HDACs in the control of liver regeneration. Valproic acid (VPA), a class I HDAC inhibitor, has been widely used as an antiepileptic agent for nearly 30 years to repress HDAC activity and to investigate the effects of HDACs on liver regeneration. VPA was administered by injecting into 2/3 partially hepatectomized mice. DNA synthesis was detected in hepatocytes, and was accordingly impaired using a BrdU incorporation assay. The expressions of cell cycling proteins such as cyclin D1, cyclin E, cyclin dependent kinase 2 (CDK2), and CDK4 were also monitored during liver regeneration. Furthermore, the B-myc expression level was examined, and revealed enhanced protein levels of B-myc. ChIP (Chromatin immunoprecipitation) assays confirmed that HDAC1 and HDAC2 were recruited to the promoter region of the B-myc gene. Our results suggest that inhibition of HDAC activities causes delayed liver regeneration at least in part, by disrupting their target genes such as B-myc.
2. Materials and methods
2.1. Animals, surgical procedures, and drug treatment
One hundred young adult male C57BL/6 mice (18–22 g, 2–3 months old) were obtained from the Animal Center of Sichuan University, West China Hospital, Chengdu, China, and were bred in the animal facilities at the Ministry of Health, West China Hospital, Sichuan University.
We randomly divided 84 mice into three groups: Group 1, VPA treated only; Group 2, 2/3 partial PH only; and Group 3, VPA treated plus 2/3 PH. The last 15 mice were used to determine the appropriate dosage of VPA (Sigma, Carlsbad, CA, USA).
PH was performed on the 2–3 months old male mice, as previously described (Mitchell and Willenbring, 2008).
For the VPA treated group, the mice were injected intraperitoneally (i.p.) every 12 h with appointed doses of VPA until the animals were sacrificed at 0, 12, 24, 36, 48, 72, 120, or 168 h after treatment.
All the animal experiments were performed according to the guidelines of the Animal Care and Use Committee of Sichuan University.
2.2. BrdU analysis and immunohistochemistry
One hour before sacrifice, the mice were injected i.p. with BrdU (Sigma). At the indicated time, the mice were sacrificed and their liver tissues were harvested. DNA synthesis was monitored by BrdU staining (Dako, Carpinteria, CA, USA). Hepatocyte proliferation was detected using Ki-67 (Thermo Fisher, Waltham, MA, USA). Staining with hematoxylin and eosin (HE) and with immunohistochemical stains was performed as described by Ke et al. (2007). At least 2 000 cells were counted to determine the proportion of hepatocytes that expressed the previously mentioned markers.
2.3. Liver function tests
Liver function tests, including serum albumin (Alb), alanine aminotransferase (ALT), and aspartate aminotransferase (AST) determination, were conducted by the Biochemical Laboratory of Sichuan University, West China Hospital, Chengdu, China.
2.4. Western blot analysis
The protein isolated from the liver tissue was subjected to Western blot analysis as described by Rodríguez et al. (2006). The total proteins were transferred onto a polyvinylidene fluoride membrane after separation by electrophoresis using a 12% sodium dodecyl sulfate polyacrylamide gel. Western blot analyses were performed using anti-acH3 (Abcam, Shatin, HK), anti-total H3 (Cell Signaling Technology, Boston, MA, USA), anti-cyclin D1 (Epitomics, Burlingame, CA, USA), anti-cyclin E (Cell Signaling Technology), anti-CDK2 (Epitomics), anti-CDK4 (Santa Cruz, Buffalo, CA, USA) and B-myc (Epitomics) antibodies. Mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Millipore, Billerica, MA, USA) antibody was used as the internal control.
The immunoblot bands of each sample were densitometrically analyzed using the Quantity One Analysis package (Bio-Rad, Hercules, CA, USA).
2.5. ChIP assay
Fresh mice liver tissues were harvested immediately after the mice were sacrificed. Chromatin was isolated according to the EZ-Zyme chromatin prep kit guidelines (Millipore). The tissue samples were immunoprecipitated using RNA polymerase II (Millipore), anti-acH3 (Abcam), HDAC1 (Abcam), and HDAC2 (Abcam) antibodies, and then reversed cross-linked using an EZ-Magna ChIP G chromatin immunoprecipitation kit (Millipore). Proteins were digested with proteinase K and the recovered DNA was purified and subjected to polymerase chain reaction (PCR) amplification. The primers used for ChIP were as follow. B-myc promoter 1: 5′-ccagctcatgttcacacaggcaaa-3′ for the sense primer, and 5′-tgaaggatagctcgctggtagaga-3′ for the antisense primer; B-myc promoter 2: 5′-atgtagcccagccatggtaatcct-3′ for the sense primer, and 5′-cagcatcatcccacacacctgtaa-3′ for the antisense primer; B-myc promoter 3: 5′-tccatcgtcagaggaacggacaat-3′ for the sense primer, and 5′-cgcccagtgactcttctactttca-3′ for the antisense primer. The DNAs recovered from the chromatin that was not immunoprecipitated (input) and the chromatin that was immunoprecipitated with protein G in the absence of primary antibodies, were used as the controls.
2.6. Statistical analysis
All averaged data are presented as mean±standard error of the mean (SEM), and all statistical analyses were performed using the PASW Statistics 17.0 statistical package. Statistical significance was assessed using a t-test. The differences were considered significant at P<0.05.
3. Results
3.1. VPA suppresses HDAC activity in the liver
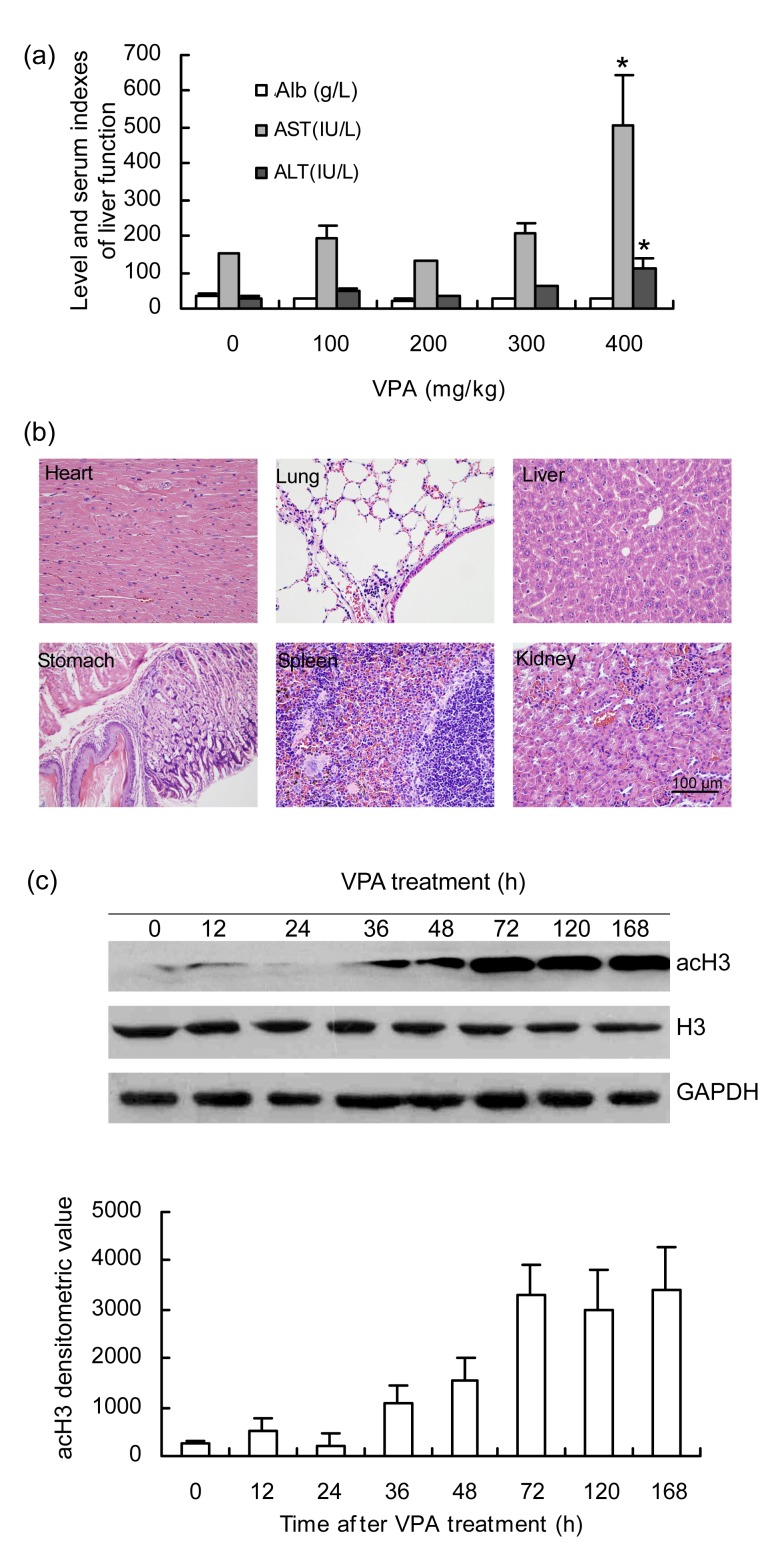
To detect an appropriate dosage, different VPA doses were tested on the mice (intraperitoneal, administration of VPA from 100 to 400 mg/kg twice daily). The liver function tests showed that ALT and AST levels were within the normal range under a twice-daily dosage of 100 to 300 mg/kg VPA. Alb did not change significantly (Fig. 1a), and HE staining of the vital organs showed no significant pathologic changes (Fig. 1b). Histologically, the heart, liver, spleen, lung, kidneys, and stomach were normal and in good physiologic status. Reversible changes were detected only in the liver. Slight cell swelling or fatty change was observed in some cases. A small number of inflammatory cells, mainly lymphocytes, were observed locally in the pulmonary, interstitial, and gastric mucosa. Necrosis, apoptosis, or hemorrhage was not detected. When the dosage was increased to 400 mg/kg twice daily, nearly all the mice manifested tremors and convulsions after injection. More than 50% died after 2 d of VPA administration. The last surviving mice treated with 400 mg/kg VPA twice daily showed abnormal liver function as indicated by high levels of ALT and AST (Fig. 1a). Therefore, a dosage of 300 mg/kg twice daily was administered for further study. The mice were sacrificed at 0, 12, 24, 36, 48, 72, 120, or 168 h, and their liver tissues were harvested. To test the effect of VPA on HDAC activity, the global levels of acH3 were detected by Western blot and compared with total H3 at each particular time point after treatment. Increased levels of acH3 protein were detected in the VPA-treated mice after 36 h, and reached a peak at 72 h. Expression remained high until 168 h of treatment (Fig. 1c). Hence, the twice-daily administration of 300 mg/kg of VPA represses HDAC function and significantly enhances acH3 expression under normal liver function.
Fig. 1.
Effects on mice pathophysiology after treatment with VPA by intraperitoneal injection
N=3 for each group treated with VPA under 100 to 300 mg/kg, twice daily (bid); N=6 for the group under the dosage of 400 mg/kg, bid. (a) Mice liver function detected after 2 d of VPA administration under the dosage of 100 to 400 mg/kg, i.p. bid (* P<0.05 when compared with normal conditions). (b) Histology of vital organs stained with HE after VPA treatment (300 mg/kg, i.p. bid) for 2 d. (c) Western blot analysis of histone acetylation state detected with antibody acH3 at 0, 12, 24, 36, 48, 72, 120, or 168 h after VPA treatment (300 mg/kg, i.p. bid). The levels of total H3 were also detected at the same time points. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. The lower panel shows a densitometry histogram of acH3
3.2. VPA scarcely suppresses hepatocyte proliferation in intact livers
Under VPA administration, the mice were sacrificed and their liver tissues were harvested at 0, 12, 24, 36, 48, 72, 120, or 168 h post-treatment. Histologically, the liver tissues were normal and integrated. In the HE-stained liver paraffin sections, the lobules, portal triads, and central veins were intact. The sinusoids were clear and without angiectasis. The hepatocytes were polygonal and/or round with a large amount of eosinophilic cytoplasm. Necrosis was not observed, but slight swelling and a small amount of fatty change were detected in some cases. A small number of inflammatory cells were observed in some regions (Fig. 2a). BrdU and Ki-67 staining showed slight changes under the VPA treatment (Fig. 2).
Fig. 2.
Effect of VPA on liver histology and hepatocyte proliferation
N=4 for every time point of each group. (a) Histological and immunohistochemical staining of mice liver. Upper panel: hepatic histological specimen stained by HE at 0, 48, 120, or 168 h after VPA administration; middle panel: DNA synthesis detected by BrdU stain at the same time points; lower panel: proliferation of liver cells detected by immunohistochemical staining of mice liver treated by VPA using anti-Ki-67 antibodies at the same time points. Nuclear staining with BrdU and Ki-67 was considered positive. A positive rate was counted after VPA injection of BrdU (b) and Ki-67 (c) at 0, 12, 24, 36, 48, 72, 120, or 168 h
In the current study, VPA did not suppress hepatocyte proliferation despite its inhibitory effect on HDAC under physiologic states.
3.3. VPA rapidly enhances liver histone acetylation after 2/3 PH
To investigate the influence of hepatocytic histone acetylation under VPA administration during liver regeneration, the mice were first administered with 300 mg/kg VPA. Afterwards, a standard 2/3 PH was performed, and VPA was administered continually until the mice were sacrificed. More than 90% of the mice were alive and appeared normal until sacrificed. The subjects were examined at various times after operation until 168 h. Results of total H3 appeared to be similar in the PH and PH plus VPA groups. A significant increase in acH3 was detected immediately after 2/3 PH. The simple 2/3 PH mice showed markedly lower levels of liver acH3 (Fig. 3). Our data suggest that VPA can rapidly enhance histone acetylation during liver regeneration after 2/3 PH.
Fig. 3.
Histone acetylation status influenced by VPA in the liver regeneration process
N=4 for each group at each time point. Histone acetylation was detected with anti-acH3 antibody and compared with total H3 at 0, 12, 24, 36, 72, 120, or 168 h after 2/3 PH treatment with and without VPA (300 mg/kg, i.p. bid). (a) Western blot analysis of acH3; (b) Densitometric analysis of acH3. GAPDH was used as a loading control. * P<0.05,** P<0.001, when the VPA(PH group is compared with the PH group at the same time point
3.4. VPA delays early liver regeneration after 2/3 PH
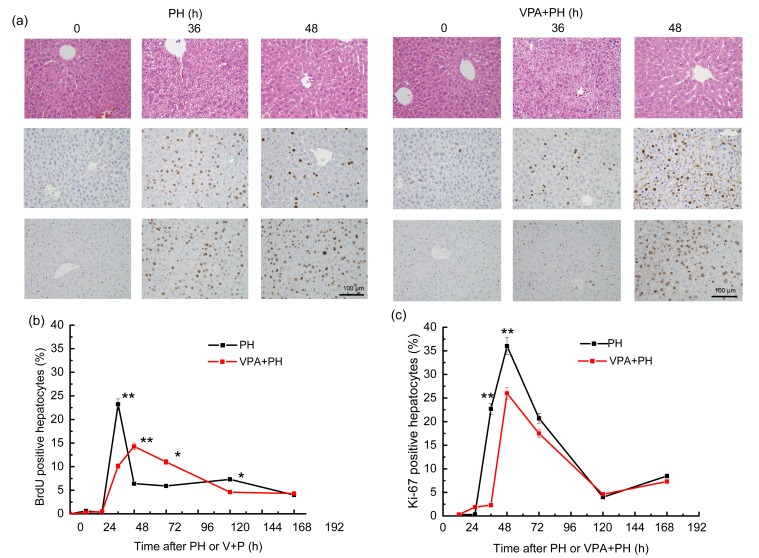
The mice were randomly divided into two groups. One group was administered with VPA until sacrificed, and 2/3 PH was performed after the initial VPA injection. The other group received only PH. No obvious difference was observed between the two groups in terms of survival ratio or histologic conditions (Fig. 4a). To test the effects of VPA on hepatocyte proliferation during liver regeneration after 2/3 PH, liver tissues were harvested at 0, 12, 24, 36, 48, 72, 120, or 168 h after treatment to examine DNA synthesis using BrdU staining. Only a small number of cells were BrdU-positive at 0 h. In the PH group, more BrdU-positive cells were observed at 12 h and the numbers reached a peak at 36 h after the operation, and then declined to a near normal level at 168 h after 48 h post-surgery. Hepatectomy stimulated the hepatocytes in the G0 phase to immediately enter the G1 and DNA synthesis phases under normal conditions. However, in the VPA+PH group, DNA synthesis was significantly delayed and suppressed. The peak of the BrdU-positive cells was delayed to 48 h. The ratio of positive cells also declined significantly (Figs. 4a and 4b). VPA suppresses DNA synthesis during liver regeneration after 2/3 PH. Ki-67 expression was subsequently determined in these two groups. The results indicate that during the regeneration process after PH, Ki-67 is expressed from 24 h, reaching its peak at 48 h. The ratio of positive cells was at a high level from 36 to 72 h. Most of the hepatocytes were shown to be in the G2 and M phases after DNA synthesis at 36 h. VPA decreased Ki-67 expression (Figs. 4a and 4c). The data for BrdU and Ki-67 staining demonstrate that VPA suppresses liver proliferation. So the results indicated that VPA can impair liver regeneration by repressing hepatocyte proliferation.
Fig. 4.
Histology and hepatocyte proliferation influenced by VPA in the liver regeneration process
N=4 for each group. (a) Upper panel: hepatic histological specimen stained with HE at 0, 36, and 48 h after 2/3 PH with or without VPA administration; middle panel: DNA synthesis detected using BrdU stain at the same time points; lower panel: proliferation of liver detected by immunohistochemical staining of mice liver treated with VPA using anti-Ki-67 antibodies at the same time points. Nuclear staining with BrdU and Ki-67 was considered positive. A positive rate was counted for the two groups after treatment of BrdU (b) and Ki-67 (c) at 0, 12, 24, 36, 48, 72, 120, or 168 h. * P<0.05,** P<0.001 when VPA(PH group is compared with the PH group at the same time point
3.5. VPA suppresses cyclins in the early stage of liver regeneration
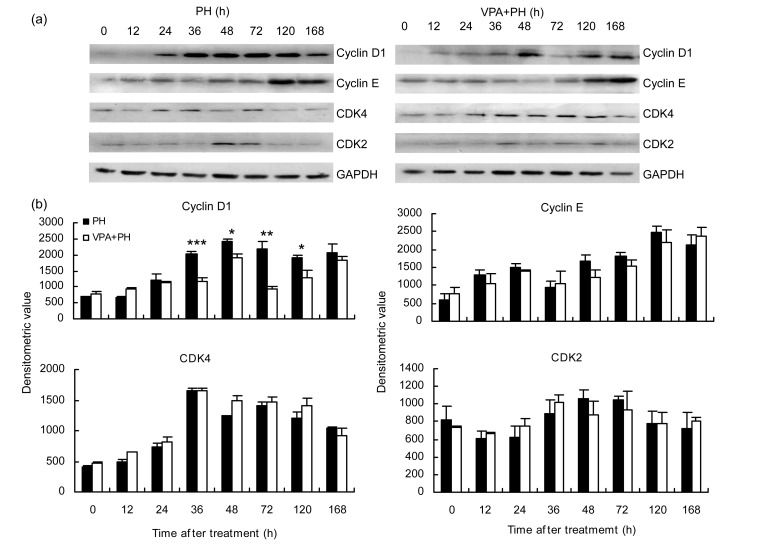
Because DNA replication was impaired by VPA during liver regeneration, and cell cycle-related kinases are necessary for cell proliferation, we then investigated the effects of VPA on the cell cycle. Cyclin D1, cyclin E, CDK4, and CDK2 were tested at sequential time points in the two groups by Western blot analysis. Cyclin D1 was the only enzyme significantly altered between the two groups. Cyclin D1 was expressed from 24 h and increased continuously after 2/3 PH. After reaching its peak at 36 h, cyclin D1 declined until 168 h. Under VPA+PH, cyclin D1 expression was postponed until 48 h, and maintained at a lower level than that following PH (Fig. 5). Regardless of the CDK4 levels, suppression of cyclin D1 expression by VPA would cause a decrease in cyclin D1/CDK4 complex expression and cell cycle arrest at the G1 phase. DNA synthesis in the hepatocytes then decreased.
Fig. 5.
Effect of VPA on the cell cycle in the process of liver regeneration
N=3 for each. (a) Expressions of cell cyclin proteins in the two groups of PH and VPA+PH at sequential time points were detected by Western blot with antibody cyclin D1, cyclin E, CDK4 and CDK2 at 0, 12, 24, 36, 48, 72, 120, or 168 h in the PH and VPA+PH groups. (b) Corresponding densitometric analyses of immunodetection of certain antibodies. GAPDH was performed as a loading control. * P<0.05, ** P<0.01, *** P<0.001 when the VPA(PH group is compared with the PH group at the same time point
3.6. VPA upregulates B-myc expression to arrest hepatocyte proliferation
The target genes of HDAC during liver regeneration were screened using a cDNA microarray. The hepatocyte proliferation inhibitor B-myc was identified as the target gene (Table A1). The expression of B-myc during liver regeneration was verified by Western blot. The results show that B-myc decreased markedly after PH, and remained at a lower level compared with its expression in intact liver. However, the VPA+PH group demonstrated a different trend. B-myc increased rapidly after 12 h and a distinct discrepancy in PH was observed at the same time points. The B-myc expression level still greatly outstripped the normal until 48 h. After 48 h the expression gradually decreased. According to this result, compared with the PH group, VPA enhanced B-myc expression from 12 to 48 h after PH (Fig. 6a).
Fig. 6.
Effect of VPA on expression of B-myc in the process of liver regeneration after PH
(a) Time-course of the expression of B-myc in the two groups, PH and VPA+PH, at sequential time points was detected and analyzed by Western blot (n=3 for each). GAPDH was performed as a loading control. * P<0.05 when the VPA+PH group is compared with the PH group at the same time point. (b) Recruitment of HDAC1, HDAC2, acH3, and RNA polymerase ΙΙ to the indicated regions of the B-myc promoters detected using ChIP on chromatin isolated from the mice liver at 48 h after PH or VPA+PH treatment (input and no antibody (NA) used as controls)
3.7. Inhibition of HDAC1/2 by VPA suppresses B-myc transcription
HDACs are epigenetic factors which can regulate transcription of target genes by changing the chromatin architecture. Based on the increased B-myc expression under VPA treatment, which suppressed liver regeneration, a ChIP assay was performed to test whether HDACs are recruited to the promoter regions of the B-myc gene. Liver tissues treated or untreated with VPA were harvested at 48 h after 2/3 PH. Antibodies specific for HDAC1, HDAC2, acH3, and RNA polymerase II, and primers for multiple regions of B-myc promoters were used. The recruitment of HDAC1 and HDAC2 to a chromatin region of the B-myc promoter was detected in PH, and was reduced markedly in the VPA+PH treatment. The recruitment of RNA polymerase II to the specific region of the B-myc promoter was detected clearly in the VPA+PH treatment, but was rarely observed in the PH treatment. This pattern was consistent with the lower level of B-myc transcripts during natural liver regeneration (Fig. 6b). Together, these results further validate the conclusion that inhibition of HDACs by VPA increases B-myc transcription during liver regeneration after PH.
4. Discussion
With the progress of epigenetic studies, epigenetic regulation has been shown to play an important role in an increasing number of fields (Katsuyama and Paro, 2011; Martín-Subero and Esteller, 2011). Histone HDAC is one of the two types of enzymes that regulate histone acetylation (Turner, 2000; Nakayama and Takami, 2001; Thiaqalinqam et al., 2003). HDAC1 and HDAC2 exist extensively in tissues and organs and play an important role in tissue development and regeneration (Thiaqalinqam et al., 2003; Shen et al., 2008). Among the multitude of HDAC inhibitors, VPA is normally used in in vivo experiments to inhibit the activity of HDAC1/2 (Gurvich et al., 2004; Michaelis et al., 2004).
The results of our current study indicated that intraperitoneal VPA injection in mice remarkably improved the acetylation of H3 in liver cells under normal liver function, but did not inhibit hepatocyte proliferation in intact mouse liver tissues. In contrast, 24 h after intraperitoneal VPA injection in mice, the proliferation indices (the positivity rates of BrdU and Ki-67) indicated that proliferative cells increased slightly, but the proliferation level remained low. The reason may be related to the slight damage caused by drug injection. Although the mice had good living conditions and their liver functions were in the normal range, the morphologic features of their liver tissue indicated that the liver cells were subjected to slight reversible injury, such as cellular swelling and fatty degeneration. Such reversible injuries might stimulate the proliferation of several liver cells.
VPA clearly inhibited the regeneration of mice liver after 2/3 PH. VPA delayed the rapid proliferation of liver cells during the early phase of liver regeneration. The results of BrdU and Ki-67 staining indicated that VPA inhibited and delayed DNA synthesis, and arrested liver cells in the G1/S phase. This result was consistent with the report by Takai et al. (2004). As VPA arrested the proliferation of liver cells in the G1 stage, the cyclic proteins cyclin D1 and cyclin E in the G1/S phase and their CDK partners were examined further. The results indicated that VPA delayed and down-regulated cyclin D1 expression during liver regeneration. During mitosis, cyclin D1 bonds with CDK4 to phosphorylate Rb and activate the transcription factor E2F (Simile et al., 2004). It facilitates gene transcription and the transition of cells from phase G1 to S, and is the most important cyclic protein in this phase (Satyanarayana and Kaldis, 2009). However, some studies have shown that cyclin D1 only shortens the G1 phase instead of facilitating cell proliferation when a mitogen is absent, similar to mitogen-activated protein kinase (MAPK) kinase 1 (MEK1) (Cheng et al., 1998; Sherr and Roberts, 1999; Sherr, 2000). The absence of cyclin D1 prevents cells from entering the S phase (Sherr, 1995; Awad and Gruppuso, 2000; Nelsen et al., 2001; Takai et al., 2004; Patil et al., 2009). During liver regeneration, the transition of liver cells from the G1 to the S phase is a key event initiating regeneration, in which cyclin D1 plays an important role (Fausto, 2000). VPA may trigger the delay and inhibition of cyclin D1 expression, contributing to the inhibition of liver cell proliferation, as proven in VPA in vitro studies (Michaelis et al., 2004). VPA has been found to inhibit the proliferation of various tumor cell lines in vitro, including blood and nervous system tumors and several solid cancers, such as breast and gastric cancers (Takai et al., 2004; Mongan and Gudas, 2005; Kaiser et al., 2006; Bartolini et al., 2008). Inhibition of the proliferation of various cancer cells by VPA is expressed as retardation of the G1/S phase and inhibition of cyclin D1 expression.
VPA inhibition of HDAC1/2 function triggers the super-acetylation of histone in liver cells. The acetylation of histone promotes the transcription of relevant genes. Hence, VPA may activate the expression of some factors that inhibit proliferation during liver regeneration, contributing to a delay in liver regeneration. The liver tissue of liver-specific Hdac1/2 gene knockout mice 36 h after 2/3 PH and that of wild mice were compared at the same time points and under the same conditions. Microarray detection (Table A1) was conducted to screen the target gene B-myc. To verify the function of B-myc in VPA inhibition of liver regeneration after PH, the expression of B-myc was examined at different time points after the surgery by comparing the VPA+PH group and the PH group. The change in these two groups showed that B-myc expression in the experimental group continued to be higher than that in the control group within 12–48 h after PH. This result corresponded well with the changes in proliferation indices and the expression of cyclin D1. Furthermore, the ChIP detection result showed that HDAC1/2 bound at the transcription initiation area of B-myc and VPA weakened this connection. This finding indicated that HDAC1/2 regulates B-myc gene expression in liver regeneration. VPA inhibited HDAC1/2, reducing the bonding between HDAC1/2 and the B-myc gene, and boosted B-myc. Higher B-myc expression participated in reducing the regenerative capacities of the liver. In practice, HDAC1/2 cannot bond to the promoter directly. It needs to form a protein complex with a certain recruiting protein. Different complexes perform diverse biological functions (Turner, 2000; Nakayama and Takami, 2001; Shen et al., 2008). The functional difference between HDAC1 and HDAC2 has been reported to be an important factor that determines the difference in the proliferation of hepatocytes (Wang et al., 2008a; Wang et al., 2008b). HDAC1 interacted with CCAAT-enhancer binding protein alpha (C/EBPα) in old mice. The HDAC1-C/EBPα complex blocked E2F bonding on certain gene promoters causing inhibition of expression of genes, such as Forkhead box M1 (FoxM1B), which is required for liver regeneration. Hence, the proper process of liver regeneration was impaired. However, the HDAC1-C/EBPβ complex promoted liver regeneration by inhibiting the expression of C/EBPα in young mice (Wang et al., 2008a; Wang et al., 2008b). According to our results, we propose that different epigenetic factors control a number of diverse genes. HDAC1/2 interacts with diverse recruiting proteins, regulating several genes, including B-myc, and plays an important part in the process of liver regeneration in cooperation with other epigenetic factors. As for the detailed mechanism of liver proliferation regulated by B-myc, such as the interaction between the HDAC1/2 complex and the B-myc promoter, and the recruiting proteins of the HDAC1/2 complex, we shall carry out further studies on gene knockout mice.
In summary, the epigenetic factors, HDAC1 and HDAC2, are essential for liver regeneration. VPA inhibits HDAC activities, reducing cyclin D1, preventing liver cells from entering the DNA synthesis phase, increasing B-myc expression, and at least partially, participating in the regulation of liver regeneration.
Acknowledgments
We would like to thank Prof. Q. Richard LU (Department of Developmental Biology and Kent Waldrep Foundation Center for Basic Neuroscience Research on Nerve Growth and Regeneration, University of Texas Southwestern Medical Center, Dallas, Texas, USA) for giving us the liver-specific Hdac1/2 gene knockout mice. We also thank Dr. Yu-jun SHI, Dr. Li-jia CHEN, Dr. Ji BAO and Miss Fei CHEN (Key Laboratory of Transplant Engineering and Immunology of Ministry of Health, West China Hospital, Sichuan University, Chengdu, China) for their help during the research.
APPENDIX
Table A1.
Differentially expressed genes of liver-specific Hdac1/2 gene knockout mice in the liver regeneration process
| SEQ-ID | P value | Fold change | Regulation | Gene name | Gene-ID | Chromosome |
| BC056436 | 0.017731 | 6.108081 | Up | B-myc | 107771 | chr2 |
| BC039953 | 0.001419 | 3.057304 | Up | Src | 20779 | chr2 |
| AK142435 | 0.018637 | 2.662801 | Up | Camk2d | 108058 | chr3 |
| BC132185 | 0.046963 | 2.31571 | Up | Tgfa | 21802 | chr6 |
| AK164302 | 0.019604 | 1.88951 | Up | Pten | 19211 | chr19 |
| AK032770 | 0.0117 | 1.776138 | Up | Apbb2 | 11787 | chr5 |
| AK171909 | 0.031938 | 1.772888 | Up | Stk11 | 20869 | chr10 |
| BC037707 | 0.033992 | 1.735755 | Up | Sept8 | 20362 | chr11 |
| AK036317 | 0.023272 | 1.665878 | Up | Raf1 | 110157 | chr6 |
| BC013718 | 0.045114 | 1.565469 | Up | Ddit3 | 13198 | chr10 |
| AK031757 | 0.04134 | 1.530431 | Up | Sept9 | 53860 | chr11 |
| AK171368 | 0.023827 | 1.505595 | Up | Sept11 | 52398 | chr5 |
| BC065160 | 0.009677 | 1.502571 | Up | Bin1 | 30948 | chr18 |
Analysis results of significantly up-regulated genes participating in the cell cycle, detected by cDNA microarray in the liver tissue of liver-specific Hdac1/2 gene knockout mice during liver regeneration. SEQ_ID was the sequence identifier of the microarray probe. Fold change refers to the absolute ratio of normalized intensities between the knockout mice and the wild mice. Column ID shows the gene ID in NCBI
Footnotes
Project (Nos. 30971118 and 31000601) supported by the National Natural Science Foundation of China
References
- 1.Asker C, Magnusson KP, Piccoli SP, Andersson K, Klein G, Cole MD, Wiman KG. Mouse and rat B-myc share amino acid sequence homology with the C-myc transcriptional activator domain and contain a B-myc specific carboxyl terminal region. Oncogene. 1995;11(10):1963–1969. [PubMed] [Google Scholar]
- 2.Awad MM, Gruppuso PA. Cell cycle control during liver development in the rat: evidence indicating a role for cyclin D1 posttranscriptional regulation 1. Cell Growth Differ. 2000;11(6):325–334. [PubMed] [Google Scholar]
- 3.Bartolini G, Orlandi M, Papi A, Ammar K, Tonelli R, Franzoni M, Pession A, Rocchi P, Ferreri AM. Growth inhibition and proapoptotic activity induction by IIF and valproic acid on RA-resistant leukemia cells. Anticancer Res. 2008;28(1A):283–288. [PubMed] [Google Scholar]
- 4.Burton RA, Mattila S, Taparowsky EJ, Post CB. B-myc: N-terminal recognition of myc binding proteins. Biochemistry. 2006;45(32):9857–9865. doi: 10.1021/bi060379n. [DOI] [PubMed] [Google Scholar]
- 5.Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1) PNAS. 1998;95(3):1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng X, Blumenthal RM. Introduction—epiphanies in epigenetics. Prog Mol Biol Transl Sci. 2011;101:1–21. doi: 10.1016/B978-0-12-387685-0.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornwall GA, Collis R, Xiao Q, Hsia N, Hann SR. B-myc, a proximal caput epididymal protein, is dependent on androgens and testicular factors for expression. Biol Reprod. 2001;64(6):1600–1607. doi: 10.1095/biolreprod64.6.1600. [DOI] [PubMed] [Google Scholar]
- 8.Facchini LM, Penn LZ. The molecular role of myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12(9):633–651. [PubMed] [Google Scholar]
- 9.Fausto N. Liver regeneration. J Hepatol. 2000;32(1 Suppl.):19–31. doi: 10.1016/S0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 10.Fujiyoshi M, Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J Hepatobilliary Pancreat Sci. 2011;18(1):13–22. doi: 10.1007/s00534-010-0304-2. [DOI] [PubMed] [Google Scholar]
- 11.Gregory MA, Xiao Q, Cornwall GA, Lutterbach B, Hann SR. B-myc is preferentially expressed in hormonally-controlled tissues and inhibits cellular proliferation. Oncogene. 2000;19(42):4886–4895. doi: 10.1038/sj.onc.1203851. [DOI] [PubMed] [Google Scholar]
- 12.Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64(3):1079–1086. doi: 10.1158/0008-5472.CAN-03-0799. [DOI] [PubMed] [Google Scholar]
- 13.Higgins GM, Anderson RM. Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 14.Ingvarsson S, Asker C, Acelson H, Klein G, Sumegi J. Structure and expression of B-myc, a new member of the myc gene family. Mol Cell Biol. 1988;8(8):3168–3174. doi: 10.1128/MCB.8.8.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser M, Zavrski I, Sterz J, Jakob C, Fleissner C, Kloetzel PM, Sezer O, Heider U. The effects of the histone deacetylase inhibitor valproic acid on cell cycle, growth suppression and apoptosis in multiple myeloma. Haematologica. 2006;91(2):248–251. [PubMed] [Google Scholar]
- 16.Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS Lett. 2011;585(11):1617–1624. doi: 10.1016/j.febslet.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Ke Q, Erbolat , Zhang HY, Bu H, Li S, Shi DN, Yang GH, Chen HJ, Wei B. Clinicopathologic features of pleomorphic hyalinizing angiectatic tumor of soft parts. Chin Med J. 2007;120(10):876–881. [PubMed] [Google Scholar]
- 18.Martin-Subero JI, Esteller M. Profiling epigenetic alterations in disease. Adv Exp Med Biol. 2011;711:162–177. doi: 10.1007/978-1-4419-8216-2_12. [DOI] [PubMed] [Google Scholar]
- 19.Merion RM. Current status and future of liver transplantation. Semin Liver Dis. 2010;30(4):411–421. doi: 10.1055/s-0030-1267541. [DOI] [PubMed] [Google Scholar]
- 20.Michaelis M, Michaelis UR, Fleming I, Suhan T, Cinatl J, Blaheta RA, Hoffmann K, Kotchetkov R, Busse R, Nau H, et al. Valproic acid inhibits angiogenesis in vitro and in vivo. Mol Pharmacol. 2004;65(3):520–527. doi: 10.1124/mol.65.3.520. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 22.Mongan NP, Gudas LJ. Valproic acid, in combination with all-trans retinoic acid and 5-aza-2′-deoxycytidine, restores expression of silenced RARβ2 in breast cancer cells. Mol Cancer Ther. 2005;4(3):477–486. doi: 10.1158/1535-7163.MCT-04-0079. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama T, Takami Y. Participation of histones and histone-modifying enzymes in cell functions through alterations in chromatin structure. J Biochem. 2001;129(4):491–499. doi: 10.1093/oxfordjournals.jbchem.a002882. [DOI] [PubMed] [Google Scholar]
- 24.Nelsen CJ, Rickheim DG, Timchenko NA, Stanley MW, Albrecht JH. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res. 2001;61(23):8564–8568. [PubMed] [Google Scholar]
- 25.Patil MA, Lee SA, Macias E, Lam ET, Xu CR, Jones KD, Ho C, Rodriguez-Puebla M, Chen X. Role of cyclin D1 as a mediator of c-Met- and β-catenin-induced hepatocarcinogenesis. Cancer Res. 2009;69(1):253–261. doi: 10.1158/0008-5472.CAN-08-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resar LM, Dolde C, Barret JF, Dang CV. B-myc inhibits neoplastic transformation and transcriptional activation by c-myc. Mol Cell Biol. 1993;13(2):1130–1136. doi: 10.1128/MCB.13.2.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riehle KJ, Dan YY, Campbell JS, Fausto N. New concepts in liver regeneration. J Gastroenterol Hepatol. 2011;26(7):1218. doi: 10.1111/j.1440-1746.2010.06539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG, Martínez-Chantar ML, Viña J, Viña JR, Mato JM, et al. Id2 leaves the chromatin of the E2F4-p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398(3):431–437. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamuro D, Prendergast G. New myc-interacting proteins: a second myc network emerges. Oncogene. 1999;18(19):2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 30.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28(33):2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 31.Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJM, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11(9):1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20(5):187–190. doi: 10.1016/S0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 33.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–3695. [PubMed] [Google Scholar]
- 34.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 35.Simile MM, de Miglio MR, Muroni MR, Frau M, Asara G, Serra S, Muntoni MD, Seddaiu MA, Daino L, Feo F, et al. Down-regulation of c-myc and Cyclin D1 genes by antisense oligodeoxy nucleotides inhibits the expression of E2F1 and in vitro growth of HepG2 and Morris 5123 liver cancer cells. Carcinogenesis. 2004;25(3):333–341. doi: 10.1093/carcin/bgh014. [DOI] [PubMed] [Google Scholar]
- 36.Takai N, Desmond JC, Kumagai T, Gui D, Said JW, Whittaker S, Miyakawa I, Koeffler HP. Histone deacetylase inhibitors have a profound antigrowth activity in endometrial cancer cells. Clin Cancer Res. 2004;10(3):1141–1149. doi: 10.1158/1078-0432.CCR-03-0100. [DOI] [PubMed] [Google Scholar]
- 37.Thiaqalinqam S, Cheng KH, Lee HJ, Mineva N, Thiaqalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983(1):84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 38.Turner BM. Histone acetylation and epigenetic code. Bioessays. 2000;22(9):836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 39.Wang GL, Salisbury E, Shi XR, Timchenko L, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPα in the inhibition of liver proliferation in old mice. J Biol Chem. 2008;283(38):26169–26178. doi: 10.1074/jbc.M803544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang GL, Salisbury E, Shi XR, Timchenko L, Medrano EE, Timchenko NA. HDAC1 promotes liver proliferation in young mice via interactions with C/EBPβ. J Biol Chem. 2008;283(38):26179–26187. doi: 10.1074/jbc.M803545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann A. Regulation of liver regeneration. Nephrol Dial Transplant. 2004;19(Suppl. 4):iv6–iv10. doi: 10.1093/ndt/gfh1034. [DOI] [PubMed] [Google Scholar]