Abstract
Trans-acting genetic variants play a substantial, albeit poorly characterized, role in the heritable determination of gene expression. Using paired purified primary monocytes and B-cells we identify novel, predominantly cell-specific, cis- and trans-eQTL (expression quantitative trait loci). These include multi-locus trans-associations to LYZ in monocytes and to KLF4 in B-cells. Additionally, we observe B-cell specific trans-association of rs11171739 at 12q13.2, a known autoimmune disease locus, to IP6K2 (pB-cell=5.8×10−15), PRIC285 (pB-cell=3.0×10−10) and an upstream region of CDKN1A (pB-cell=2×10−52; pmonocyte=1.8×10−4), suggesting roles for cell cycle regulation and PPARγ signaling in disease pathogenesis. We also find specific HLA alleles forming trans-association with the expression of AOAH and ARHGAP24 in monocytes but not in B-cells. In summary, we demonstrate that mapping gene expression in defined primary cell populations identifies new cell-specific trans-regulated networks and provides insights into the genetic basis of disease susceptibility.
INTRODUCTION
Defining the genetic determinants of gene expression is crucial to understanding the biological and medical significance of genetic variation. This has particular relevance in the drive to identify functional variants underlying observed disease associations from genome-wide association studies (GWAS)1. It is increasingly apparent that functional activity of many genetic polymorphisms is dependent upon context, requiring the study of relevant cell or tissue types in a particular biological state2-4. This context-specificity means that whilst lymphoblastoid cell lines (LCLs) and other tissues have provided important insights, they may fail to capture the in vivo activity of particular variants in disease relevant tissues5,6. Recent cell and tissue specific studies highlight the importance of context in the identification of expression associated genetic variants3,4,7-10. In umbilical cord-derived cultured cells, up to 80% of regulatory variants act in a cell-type specific manner3, whilst comparison of skin, fat and LCLs identify only 30% of eQTLs to be common between tissues4. The basis for this specificity remains unresolved, but may relate to variation at tissue specific distal enhancers as opposed to conserved promoter elements3. Analyses performed on non-cultured primary tissue have typically used sources with a heterogeneous cell composition, such as peripheral blood mononuclear cells (PBMCs)3,11 or fat4. Whilst this provides general insights into tissue specific eQTLs, highly cell-type specific eQTLs may be missed due to signal saturation from other cell types where the eQTL is absent. This is especially pertinent in the elucidation of trans-acting eQTLs, where tissue specificity appears to be of increased relevance12.
Here we sought to determine physiologically active cell type-specific eQTLs of high relevance to immunity and inflammation in paired samples of monocytes and B-cells, freshly purified by positive selection. Our analysis highlights both the extent of eQTL cellular specificity, especially for trans-acting variants, and the underlying inherent complexity of eQTL action. We observe multiple examples of genes with eQTL in both cell types but to different loci, and of eQTL showing opposing cell-type dependent directional effects. Mapping genetic determinants of gene expression in these immune cell types is shown to be highly informative for reported GWAS hits, notably involving immune, infectious and inflammatory disease.
RESULTS
Defining eQTLs in purified B-cell and monocyte populations
B-cells are lymphocytes with crucial roles in adaptive and humoral immunity whilst monocytes form an innate myeloid derived cell population that initiates an inflammatory, cytokine mediated response upon microorganism invasion. Their divergent functions and origins ensure these cell populations form highly informative primary tissue for insight into immune and inflammatory diseases. Furthermore, whereas multiple LCL eQTL analyses have been performed, as yet there are no large studies focused on B-cells, the cells immortalized to derive LCLs. To investigate eQTLs in these primary cell types we used positive selection, a method demonstrated to result in superior cell purity for microarray analysis13 to separate CD19+ B-cells and CD14+ monocytes from PBMCs prepared using the whole blood of 288 healthy European volunteers (Online methods). Purity of samples was confirmed with flow cytometry and was 90-95% for B-cells and approaching 99% for monocytes. Genome-wide gene expression profiling and genotyping was performed using HumanHT-12 v4 BeadChips (Illumina) and HumanOmniExpress-12v1.0 BeadChips (Illumina). Following processing and quality control we performed eQTL mapping at 651210 markers for each of 283 individuals.
Cell-specific cis-eQTL are common, complex and directional in effects
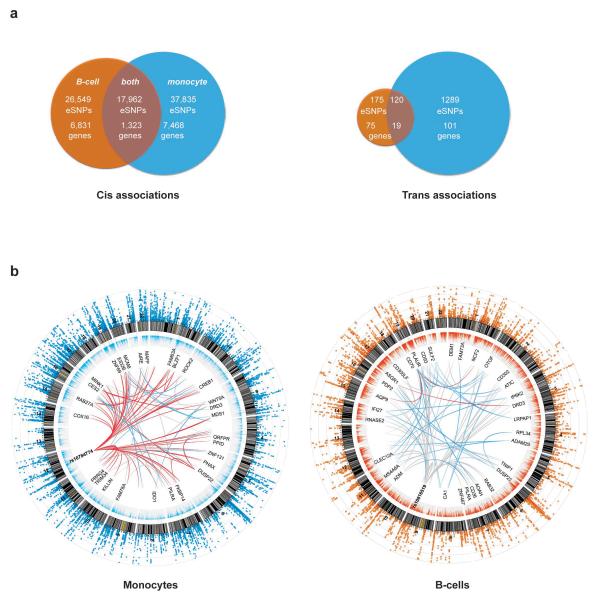
Identification of locally acting eQTL (referred to here as cis-acting) was performed by testing SNPs that fell within a 2.5Mb interval either side of the probe for association with expression in each cell type using linear and Spearman rank models. In this large, highly purified paired sample set we found little difference between the significance values using either approach - however, only eQTL that reached a permuted p<1×10−3 in both analyses were carried forward. We identified 82,346 eQTL (SNP-probe interactions, referred to hereafter as eSNPs) at permuted p<0.001, 32.2% of which were unique to B-cells, 45.9% to monocytes and 21.8% shared between cell types (Figure 1, Supplementary Figure 1, Supplementary Tables 1-2). This corresponded to 7468, 6831 and 1323 genes (8441, 7589 and 1466 probes) in which there was at least one cis-eSNP with permuted p<0.001 specific to B-cells, monocytes and shared by both cell types, respectively (a full description of associations at different significance thresholds is given in Supplementary Table 3). Comparative analysis of this dataset demonstrated a high concordance of eSNPs identified here that were shared between cell types and those previously identified in primary cell eQTL analysis10,14. This degree of commonality was not so clearly demonstrable upon analysis of LCLs15, possibly reflective of intrinsic differences between primary and cultured cells (Supplementary Figure 2). Cell specific eSNPs may exist secondary to cell-specific function of genetic variants, or alternatively when transcript expression is restricted to one cell type only. In order to identify eSNPs formed due to cell specific functional variants we defined genes expressed at similar levels across cell types but which demonstrated robust cell-specific effects. Even after excluding genes whose mean log2 expression across the cohort differed in magnitude by >0.5 between cell types, we identified 70149 cis-eSNPs in almost identical proportions by cell type. In general, whilst cell-specific eQTLs were observed more frequently, they tended to have smaller effect sizes than those shared between cells (median r2 5.1%, 5.5% and 9.8% for B-cell-specific, monocyte-specific and shared respectively) with 54, 107 and 351 eSNPs explaining more than 50% of the variance in expression of 14, 37 and 61 genes respectively. Using further RNA purified from randomly selected individuals within the cohort we were able to replicate the cellular specificity for selected genes by real-time PCR (n=14-29 homozygous individuals per gene) (Supplementary Figure 3). When RNA from crude PBMCs from the same individuals was analysed however, the signal was frequently absent (Supplementary Figure 3b,d,i), supporting the importance of cell purification in the identification of subtle primary cell-specific eQTL.
Figure 1. Shared and cell–specific cis and trans associations in B-cells and monocytes.
(a) Venn diagrams illustrating the number of eSNPs unique to each cell subset and shared between datasets in cis (left panel) and in trans. B-cell data depicted in orange, monocyte in blue. (b) Circos plots for monocyte dataset (left panel) and B-cell dataset. From outside rim inwards: The outermost rim depicts a Manhattan plot for eQTLs from the respective dataset, the second rim depicts relative expression of genes, the third rim depicts an arbitrary selection of genes (constrained due to space) with significant trans-eQTLs (p<1×10−11) and the innermost network depicts ‘spokes’ between nodal eSNPs and their trans-regulated genes. Such nodal eSNPs include rs10784774 which marks a monocyte specific master-regulatory region while rs10816519 identifies a B-cell specific master regulatory region. Red spokes: > 10 eSNPs from this locus map to the trans-eQTLs, blue spokes: >1 and <10, grey spokes: 1 eSNP.
Consistent with other reported eQTL analyses, we find the effect size and statistical significance of an eQTL varies as a function of distance from transcription start site (TSS) (Supplementary Figure 4). Notably, the density distribution of eSNPs demonstrating cell-specific effects on expression was more dispersed around the TSS in comparison to that of shared eQTL. This supports the premise that cell specific eQTL are enriched in more distant enhancer elements involved in cell-specific expression3.
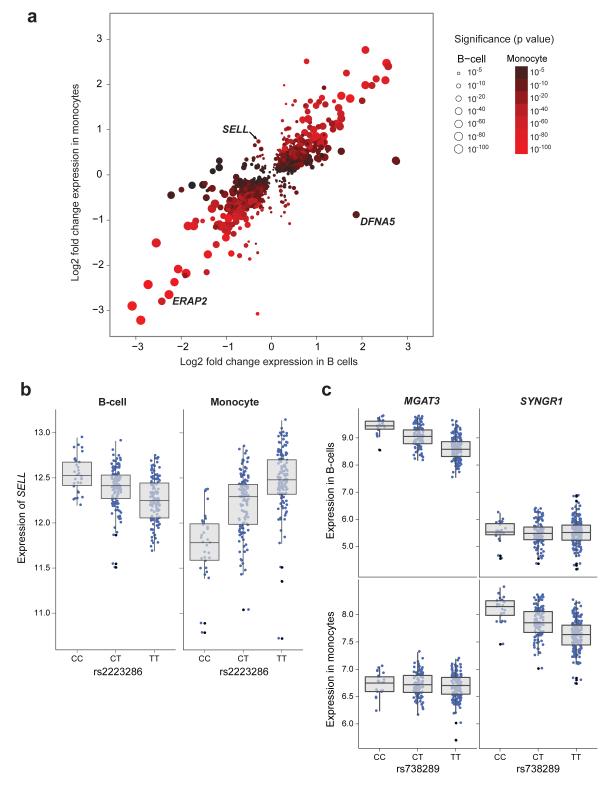
Previously reported eQTL shared between cell types have the same directional effect3. Whilst in general this holds true for our analysis of primary monocytes and B-cells, we observe several eQTL with cell type-dependent directional effects: the same eSNP being associated with opposing directional effects. We identified 197 eSNPs in 35 genes demonstrating significant ‘directional eQTL’ in B-cells and monocytes (permuted p<0.001 both cell types, total opposing directional effect >0.5 in terms of magnitude of mean log2 expression difference between major and minor alleles) (Figure 2a, Supplementary Table 4). To further investigate the significance of these directional findings we estimated the differences in the slopes using z-scores and demonstrated that for 31 of the 35 genes the differences were highly significant (Bonferroni corrected p<1×10−38) (Supplementary Table 4), thus strongly suggesting these were not chance observations. The most significant directional eQTL were noted to the gene DFNA5, previously implicated in familial deafness16 and a target of promoter methylation in gastric cancer17. Other notable genes with directional effect include MCOLN2, encoding a cation channel involved in type I interferon responses18, and SELL (also known as L-selectin, CD62L). The latter is of particular interest given the role of this cell surface receptor in monocyte recruitment to lymphoid tissues during inflammation19 and previous association of this region with amyotrophic lateral sclerosis20. We did not observe the reported association with KIFAP3 expression in LCLs20 but note the disease associated variants show a cell directional effect with the neighboring gene SELL. Real-time PCR quantification of DFNA5 and SELL on further RNA purified from randomly selected homozygous individuals (DFNA5 n=16; SELL n=20-28) confirmed these directional effects were not a product of array-mediated artifact (Supplementary Figure 3h,i).
Figure 2. eSNPs shared between cell-types may lead to opposing directional effects on gene expression or associate with expression of different genes in a cell-specific manner.
(a) Using eSNPs shared between cell types the most significant eSNP per probe is plotted with the fold change in expression this eSNP causes in homozygous form between major and minor alleles (fold-change monocytes y- axis, B-cells x-axis). Whilst the majority of eSNPs shared between datasets cause the same directional change, there are examples of eSNPs that cause opposing directional changes in expression dependent upon cell-type. Only one eSNP is plotted per probe and only eSNPs with examples of >2 individuals homozygous in the minor allele with permuted p<0.001 in both B-cells and monocyte datasets are annotated. (b) rs2223286 is associated with profound directional effects in the expression of SELL dependent upon genotype, with the minor C allele associated with increased expression of SELL in B-cells and reduced expression of SELL in monocytes (pB-cell=4.6×10−11, pmonocyte=1.1×10−22). (c) rs738289 is an example of an eSNP that forms eQTL to differing genes dependent upon cell type. In B-cells this eSNP is strongly associated with the expression of MGAT3 (pB-cell= 9.8×10−26) with no association to SYNGR1 expression; whilst in monocytes this eSNP is significantly associated with the expression of SYNGR1 (pmonocyte = 1.2×10−17) with no association to MGAT3 expression.
The extent to which a particular genetic variant may demonstrate diverging effects according to cell type is unclear. Here we find many variants show multiple, cell specific actions, with 6.4% of cell specific eSNPs additionally forming eQTL to different genes in the alternative cell type (Supplementary Figure 5a). For example rs738289 and linked eSNPs in the first intron of MGAT3, a gene encoding a glycosyltransferase21, showed association with MGAT3 in B-cells (p=9.8×10−26); by contrast in monocytes this eSNP was solely associated with the neighboring gene SYNGR1 encoding expression of vesicle associated protein (p=1.2×10−17) (Figure 2c). Just as cell type may define the gene a particular variant regulates, we observe certain genes whose expression is modulated by different variants in a cell specific manner. Such examples include TSPAN3 (Supplementary Figure 5b) and AKAP7 (not shown), in keeping with the effect of regulatory genetic variants being dependent on the environment defined by the cell type.
Identification of multiple cell-specific trans-associated eQTL
Although a considerable element of heritable gene expression is proposed to act in trans, trans-eQTL have proved difficult to define in the cell populations studied to date. It is probable that trans-acting variants are involved in a more multifactorial process than those acting in cis, reflecting contributions both of prevailing environmental cues and the cellular context. We explored this question in each of our primary cell populations and mapped trans associated eSNPs using a conservative significance threshold of 1×10−11, based on a Bonferonni correction given the number of SNPs analysed. For autosomal genes and SNPs, we identified 1704 eSNPs involving 75 genes specific to B-cells, 101 to monocytes and 19 shared (Figure 1, Supplementary Table 2). It is increasingly apparent that particular variants act as master regulators of transcription as trans-eQTL12,14. Many of these eSNPs form cis-eQTL to genes that probably define the nodal gene of these subsets (Supplementary Table 2, Supplementary Figure 6) and we observe two highly cell specific examples in the form of KLF4 (Supplementary Figures 7,8) and LYZ (discussed below). We found that only 7% of trans-eSNPs are shared between cell types, implying trans-eQTLs show a greater degree of cell specificity than cis-eQTLs. Given that B-cells constitute ~5% of PBMCs and monocytes 10-15%13, cell specific traits will often be undetectable in eQTL analysis of a heterogeneous population of PBMCs due to saturation of cell specific signals by expression signatures from other cell types.
LYZ as a monocyte-specific master regulator of a large gene set
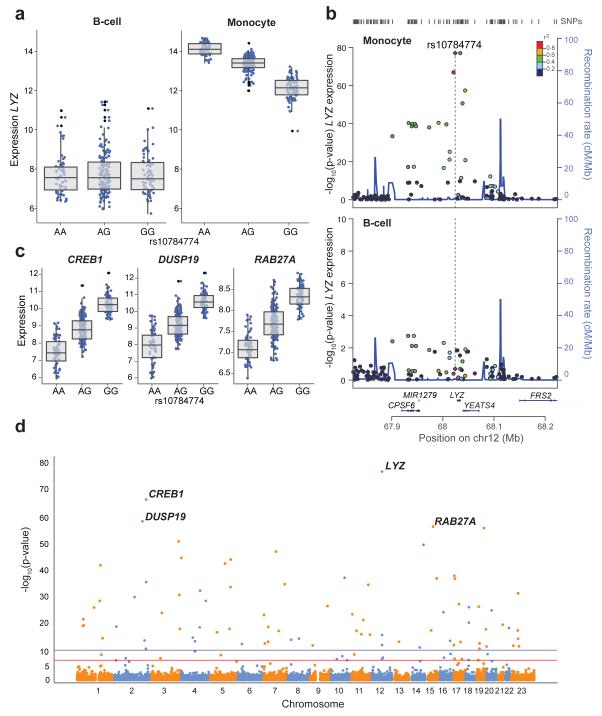
Monocytes have evolved to scavenge and present antigens through a highly conserved endosomal pathway. This is assisted via the release of hydrolyzing enzymes from secretory granules. Lysozyme forms a key component of monocyte secretory granules and degrades bacterial cell wall derived peptidoglycan. We find evidence of a novel master regulatory region involving cis- and trans-eQTL at the 12q15 locus for rs10784774 and associated eSNPs spanning LYZ, encoding lysozyme (Figure 3). This locus forms a monocyte specific cis-eQTL (Supplementary Figure 9) to a probe mapping to the 3′ UTR of LYZ (pmonocyte=8.9×10−78) and forms a trans eQTL to a remarkable 62 annotated genes (72 probes) throughout the genome at a p<1×10−11, the most significant association being to the gene CREB1 (pmonocyte=2.03×10−67) and many others at a lower significance (Figure 3, Supplementary Figure 10 and Supplementary Table 5). Interrogation of a previously published primary cell eQTL analyses of negatively selected monocytes from whole blood10 and whole blood RNA from a mixed disease and control cohort14 replicates this strong association between 12q15 and expression of both LYZ and CREB1 (CREB1, p=5.1×10−22 (rs11177644) monocytes10; p=3.2×10−67 (rs2168029) whole blood14). The multi-faceted transcriptional properties of CREB1 in immune cells22 ensure it is a highly plausible candidate to regulate expression of many downstream genes, consistent with recent reports of other transcription factors underlying multiple eQTL to one locus12. Of note, LYZ expression was most strongly correlated to that of CREB1 (r2=0.82, p=5.7×10−107) and CREB1 expression was highly correlated with the majority of trans-associated genes to this locus (Supplementary Table 5). This would support a pathway whereby cis-modulation of LYZ results in differential expression of CREB1, possibly through downstream effects via altered lysozyme secretion, with resultant modulation of a network of trans-associated genes.
Figure 3. rs10784774 marks a monocyte specific cis-eSNP to LYZ and forms a monocyte specific master regulator of multiple genes including CREB1.
(a & b) rs10784774 is significantly associated with the expression of a probe mapping to the 3′UTR of LYZ, encoding lysozyme, in monocytes only (p 78 B-cell N.S., pmonocyte=8.9×10−78). (c) rs10784774 additionally forms an eSNP to 72 probes mapping across the genome, the most significantly associated being in the genes CREB1(p = 2.0×10−67), DUSP19 (p = 2.2×10−59), and RAB27A (p = 2.0×10−57). (d) Manhattan plot demonstrating chromosomal location of all genes with probes mapping to rs10784774. The blue line represents 1.0×10−11, red line 5.0×10−8.
We noted a weaker cis-eQTL for rs10784774 with YEATS4 expression in both cell types (pmonocyte=1.26×10−13, pB-cell=1.55×10−7). Although YEATS4 is a transcription factor23 and therefore could potentially account for the observed trans-eQTL, the exclusivity of the trans-eQTL to monocytes would argue against this. Furthermore, correlation analyses between monocyte YEATS4, CREB1 and LYZ expression and that of other probes demonstrates the strongest coefficient of correlation to expression of LYZ and CREB1 for all trans associated genes to this locus (Supplementary Table 5).
Imputation demonstrates that rs10784774 forms the 5′ SNP in a haplotype spanning LYZ of 4 SNPs in almost complete LD (Supplementary Figure 11). Analysis of ENCODE ChIP-seq data24 demonstrates a p300 binding site overlapping rs10784774 and further sites overlapping the promoter (Supplementary Figure 12). To investigate the allelic relationship between p300 and LYZ expression, genotype conditioned correlation analysis between basal expression of p300 and LYZ was performed. We observed a strong inverse correlation between expression of p300 and LYZ expression in individuals homozygous with the ancestral A allele at rs10784774 but no association with expression in the homozygous G allele (AA: r2 0.41, p=4.4×10−8; AG: r2 0.22, p=1.4×10−9; GG: r2 0.02, p=0.11), supportive of allele-specific regulation of LYZ via p300.
Trans-eQTL for the disease-associated 12q13.2 locus with cell cycle regulators
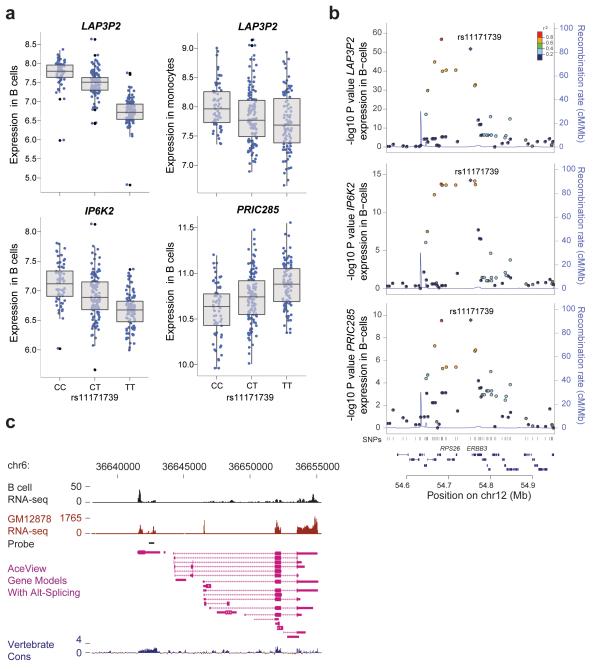
The generation of self-reactive auto-antibodies is a characteristic feature of autoimmune disease. To prevent this, B-cells expressing self-reactive antibody are removed prior to maturation to antibody secreting plasma cells. Here we show SNPs at 12q13.2, a locus showing reproducible association with multiple autoimmune diseases including type I diabetes (T1D)25-28 regulate the expression of genes implicated in cell-cycle control in trans in B-cells only. Previous studies have demonstrated rs11171739 at 12q13.2 forms a cis-eQTL involving RPS26 in liver8, LCLs15 and leukocytes11. Differential RPS26 expression however appears unable to explain susceptibility to T1D at this locus29 and the causal genes are unresolved. We find a highly significant trans-eQTL in B-cells with IP6K2 (encoding inositol hexakisphosphate kinase-2) (pB-cell=5.8×10−15) (Figure 4a). IP6K2 is required for p53-mediated apoptosis30 and is able to regulate the accumulation of p21/CIP1, a protein involved in cell division, apoptosis of pathogenic memory B-cells31 and is a known susceptibility gene for SLE32. Intriguingly, a further trans-association was found for a transcript mapping 5′ to the actual gene encoding p21/CIP1 (CDKN1A) (pB-cell=2×10−52, pmonocyte=1.8×10−4). This region is denoted as the pseudogene LAP3P2 but we find using RNA-seq it is a region of high transcriptional activity in primary B-cells (Figure 4c). rs11171739 has also been associated with type 2 diabetes (T2D)33 and we note on further analysis of this region a trans eQTL between this SNP and PRIC285 (pB-cell=3.0×10−10) a transcriptional coactivator involved in PPARγ signaling. Given the role of PPARγ agonists in T2D management this association may be of notable biological relevance34.
Figure 4. rs11171739, a SNP at 12q13.2 with strong autoimmune association forms B-cell specific trans-eQTL to 3 separate genes.
(a) rs11171739 specifically associates with the B-cell trans expression of the genes LAP3P2 (pB-cell=2.0×10−52, pmonocyte=1.7×10−4), IP6K2 (pB-cell =5.8×10−15) and PRIC285 (pB-cell=3.0×10−10). (b) B-cell regional association plots of these genes demonstrating their expression by SNP marker across chromosome 12q13.2, a known autoimmune risk locus. (c) LAP3P2 lies ~1kb 5′ to CDKN1A and is denoted as a pseudogene. We observe a high degree of transcriptional activity in our RNA-seq data from CD19+ primary B-cells, and this is also seen in LCLs (GM12878)24. This region is highly conserved amongst vertebrates.
Disease associated HLA alleles underlie trans-associations to the MHC
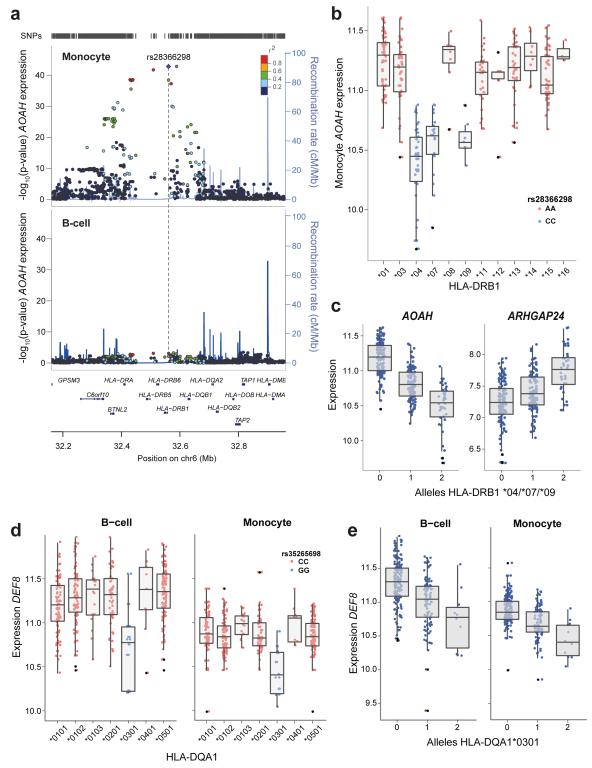
HLA genes play a critical role in the processing and presentation of antigens, they are highly polymorphic and show robust association with disease but the functional basis of this remains unresolved. Here we describe several striking associations to the MHC region (p<1×10−11), segregating by HLA allele, which may provide insights into the basis of associations with ulcerative colitis, rheumatoid arthritis and T1D. We describe a trans-association of AOAH, encoding acyloxyacyl hydrolase lipase that degrades bacterially derived lipopolysaccharide35, to the MHC class II region. An association between the MHC and AOAH has recently been described in a study of whole blood RNA from a mixed population of disease and control cohorts (peak eSNP rs2395185, 7.0×10−38)14. We replicate this finding at this SNP (rs2395185 pmonocyte=5.7×10−39), with the most strongly associated eSNP in our dataset being rs28366298 (pmonocyte=1.6×10−43). Notably, we find only very weak association of this region in B-cells (pB-cell>1×10−3) (Figure 5a), suggesting much of the signal in whole blood studies is monocyte derived, and providing underlying potential mechanistic insight. To further define this trans-association we inferred the underlying HLA alleles to 2 and 4 digit resolution in all individuals through genotype imputation36,37. This enables us to refine the association of AOAH expression and the MHC to the presence or absence of HLA class II alleles DRB1*04, *07 and *09 (p<2.2×10−16, 2 way ANOVA) which define HLA-DRB4 (Figure 5b, Supplementary Figure 13). Specifically, individuals homozygous for any of these alleles express 0.6 fold (95% C.I. 0.57-0.67) the level of AOAH as individuals homozygous for all other alleles at DRB1. Interestingly we observe a second monocyte-specific trans-association for DRB1*04/*07/*09 to ARHGAP24 (peak eSNP rs28366298 pmonocyte=4.59×10−14). ARHGAP24 encodes a negative regulator of Rho-GTPase activating protein implicated in cell migration38. Converse to AOAH, we observe that individuals homozygous for DRB1*04/*07/*09 alleles express 1.4 fold (95% C.I. 1.30-1.54) the level of ARHGAP24 than all other alleles. These alleles are associated with autoimmune disease susceptibility, as is HLA-DRB4 (defining the DR53 antigen)39,40. Our study shows that specifically monocytes from individuals with HLA-DRB4 have significantly reduced AOAH expression, a protein with anti-inflammatory properties, in tandem with increased expression of ARHGAP24.
Figure 5. Imputation of HLA status resolves cell-specific trans-associated gene expression to carriage of specific classical HLA alleles.
(a) Regional association plots from monocyte and B-cell data demonstrating the monocyte specific trans association of the chromosome 7 gene AOAH, to the class II MHC gene HLA-DRB1. There is no association in B-cell dataset, whilst the peak eSNP from monocyte dataset is rs28366298 (pmonocyte=1.6×10−43). (b) Imputation of class II alleles to 2 digit resolution demonstrates that the C allele of rs28366298 is specific to DRB1 *04, *07 and *09 alleles and these alleles are associated with reduced AOAH expression. For clarity only homozygotes are plotted with two AOAH expression values per individual (corresponding to each DRB allele) are displayed. (c) The number of DRB1 *04/*07/*09 alleles carried by an individual is significantly associated with reduced expression of AOAH (pB-cell <2.2×10−16, one-way ANOVA) and increased expression of ARHGAP24 (pmonocyte=3.0×10−14, one-way ANOVA). (d) Imputation of DQA1 status to 4 digits demonstrates that the G allele of rs35265698 is significantly associated with reduced expression of DEF8 in both B-cells and monocytes (pB-cell=6.2×10−13, pmonocyte=9.0×10−17). This allele is unique to DQA1*0301 as illustrated. For clarity only homozygotes of rs35265698 are plotted with two DEF8 expression values per individual (corresponding to each DQA allele) displayed. (e) Higher numbers of DQA1*0301 alleles possessed by an individual is significantly associated with reduced DEF8 expression (pB-cell=6.2×10−13, pmonocyte <2.2×10−16)
Common to both cell types, we observed a trans-eQTL to DEF8 (rs2760985 pmonocyte=9×10−17 pB-cell=6.2×10−13) (Figure 5c), a chromosome 16 encoded gene expressed in peripheral blood mononuclear cells of unknown function41. This association resolves to presence of the DQA1*0301 allele. These novel trans-associations to class II MHC alleles represent the first observations of differential expression of genes defined in trans by the presence of specific HLA alleles. Whilst it is unclear whether these molecular signatures are causative or arising from the disease risk associated HLA alleles, these trans-associations provide original insight into the mechanistic basis of the MHC and disease susceptibility.
Finally, although HLA allele carriage is associated with haplotype specific expression patterns across the MHC42, the high density of SNPs across this region, especially within classical HLA genes, results in exclusion of many probes. For HLA-C, an eQTL has been reported for a SNP rs926494243 located 35kb upstream of the gene associated with HIV-1 viral load and AIDS progression43,44. Our array data suggests a significant cis-eQTL, common to both cell types, but is potentially confounded by SNPs in the probe sequence. To overcome this we performed quantitative RT-PCR across the cohort using primers to regions with minimal SNP density and discrete from the probe binding site. This demonstrated a significant eQTL for rs9264942 (p=7.4×10−9) in PBMCs consistent with previous findings43 but the most significant association for HLA-C mapped to rs10484554 (p=5.2×10−16) (Supplementary Figure 14a), a GWAS SNP showing highly significant association with AIDS progression45 and psoriasis46,47. HLA imputation analysis shows the cis-eQTL can be defined in terms of presence of HLA alleles C*0602 and C*1203 (Supplementary Figure 14b,c). Further investigation is required to resolve the role of HLA-C and genetic variation at this locus in terms of disease susceptibility. C*0602 has been strongly implicated in susceptibility to psoriasis48, while current evidence shows that the association with HIV control and progression is more complex, with HLA-B mediated peptide presentation playing a key role49.
Informativeness of cell-specific eQTL analysis for fine mapping and functional characterisation of GWAS
Understanding the functional impact of genetic markers of susceptibility to disease is of central importance in determining mechanisms of pathogenesis1,50. Whilst disease susceptibility loci can be associated with differential gene expression, just as with eQTL in general, this effect may be cell-specific. It is also probable many disease-associated eQTL only manifest under physiological conditions of mixed cell populations permitting appropriate cellular interactions and antigen exposure. We hypothesized that analysis of primary B-cells and monocytes freshly isolated from peripheral blood may identify such eQTL with the attractive promise of delineating functional activity to the innate or adaptive arms of the immune response. We investigated eQTLs involving SNP markers identified as a disease risk alleles at genome-wide significance (p < 5×10−8) for 548 common traits listed in the Catalog of Published Genome-Wide Association Studies (10th September 2011). We find that 49.4% of traits (17.3% of reported GWAS SNPs) are associated with one or more significant cis-eQTL based on the listed GWAS markers or identified proxy SNPs from the 1000 Genomes Project (CEU cohort, r2 > 0.8). A complete integrated dataset indexed by trait is provided (Supplementary Table 6). Overall, we find that 730 genes show significant cis-eQTL related to GWAS, of which 33.5% are B-cell-specific, 45.5% are monocyte-specific and 21% are found in both cell types.
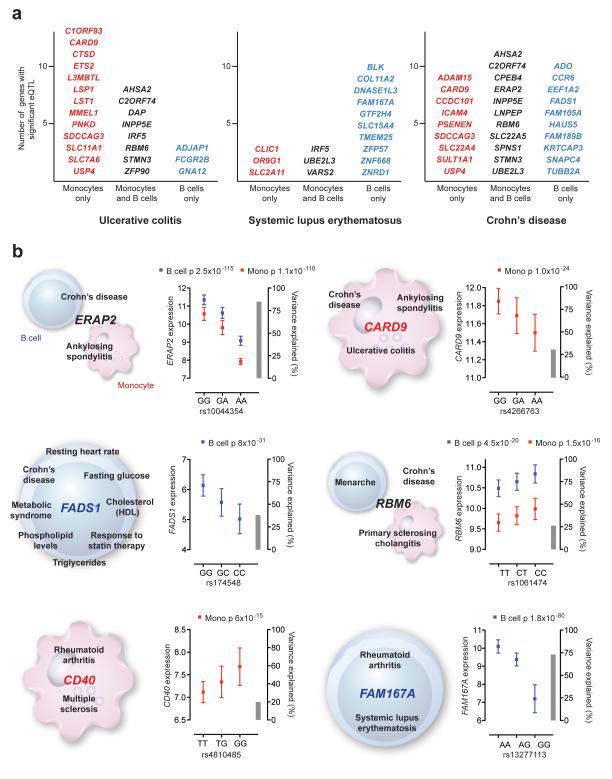
It remains a possibility that GWAS SNPs have divergent effects between cell types. In support of this we find 4.6% of reported GWAS SNPs have cis-eQTL to differing genes in a cell type dependent manner. For example, rs10781500, a SNP associated with ulcerative colitis51 and ankylosing spondylitis52 shows contrasting cis-eQTL for three genes at 9q34.3 dependent on cell type. In B-cells the SNP associates with expression of the small nuclear RNA activating complex polypeptide gene SNAPC4 (p=3.1×10−5) while in monocytes the association is with SDCCAG3 (p=1.9×10−12) and CARD9 (p=1.0×10−24); SDCCAG3 is a poorly characterised gene encoding a molecule implicated in presentation of TNF receptor 1 on the cell surface53 while CARD9 encodes a caspase recruitment domain-containing signaling protein with a regulatory role in innate immunity and apoptosis54. Our analysis of highly purified primary cell populations also highlights that certain traits demonstrate a preponderance of cell specific eQTL, as illustrated by systemic lupus erythematosus (SLE) for B-cells, while others traits show a predominance of monocyte-specific eQTL, as seen in ulcerative colitis (UC) (Figure 6). These diseases have contrasting and complex etiologies involving adaptive and innate immunity, the role of B-cells and autoantibody production being paramount in SLE, while in UC innate immunity involving dysregulated mucosal immunity and monocyte lineage cells are critically important. Our analysis defines a catalog of genes showing cell-specific or shared cis-eQTL for genetic variants implicated in many common diseases (Figure 6) (Supplementary Figure 15, Supplementary Table 6), facilitating the fine mapping and functional characterisation of specific genes and variants within GWAS disease intervals.
Figure 6. Cell specific cis-eQTL involving SNP markers associated with disease.
Genes showing significant cis-eQTL that involve SNPs reported at genome-wide significance (p < 5×10−8) in the Catalog of Published Genome-Wide Association Studies (www.genome.gov/GWA studies) (accessed 10th September 2011) or proxy SNPs identified for these disease markers from the 1000 Genomes Project (CEU cohort, r2 > 0.8) are shown. (a) List of genes with shared cis-eQTL/GWAS SNPs for UC, SLE and CD grouped by cell type and excluding HLA genes. Specific examples are described further in Supplementary Information. (b) Examples of genes showing eQTL involving SNPs associated with multiple GWAS traits.
DISCUSSION
In this study we illustrate that eQTL analysis of expression data obtained from highly purified primary cells enriches eQTL identification and reveals underlying cellular specific complexity not appreciable from mixed tissue. It is clear that whilst many eQTL with large effect sizes are shared across cells, the majority of eQTL in primary tissue are cell-specific. The degree to which a single variant can associate with the expression of different genes in monocytes and B-cells is particularly striking. It is probable that analysis of further divergent primary cell populations will increase the number of examples of such cell-specific eQTL. The delineation of cell-directional eQTLs is also of importance; in the case of those observed with SELL, which encodes a membrane protein involved in cellular recruitment to areas of inflammation, the intriguing possibility is raised that a polymorphism may influence the predominant cell subset recruited.
This study reveals trans-eQTLs possess an even higher degree of cellular autonomy than cis, reflecting them manifesting as products of upstream gene expression and cellular cross-talk. Notably, we identify novel master-regulatory regions specific to monocytes via LYZ and B-cells via the expression of KLF4. The functional implications with respect to infectious disease susceptibility of the multi-locus LYZ eQTL remain to be explored, but the existence of several genes including ERAP2, encoding an aminopeptidase involved in the loading of class I MHC molecules55 as well as RAB27A, encoding a GTPase critical to the membranous tethering of secretory lysosomes56, would suggest this eQTL may impact upon antigen presentation. The identification of B-cell specific trans-eQTLs to rs11717139, a locus with reproducible association to autoimmune diseases, in genes with putative roles in cell-cycle and apoptosis provides novel candidate genes for analysis in autoimmune disease susceptibility. B-cells are intrinsically prone to apoptosis providing protection against the generation of self-reactive antibodies. Regulation of genes involved in apoptosis would therefore be predicted to alter this process and predispose to autoimmunity57. It is notable that these genes do not have strong cis-eQTLs which otherwise might be expected to be disease associated.
Finally, we demonstrate for the first time the possession of specific HLA alleles regulates gene expression in trans in a cell specific manner. The monocyte specific trans association of AOAH and ARHGAP24 expression to the possession of DRB1*04 , *07 and *09 alleles is particularly intriguing. Only these DRB1 alleles are associated with expression of HLA-DRB4, encoding the DR53 superantigen58. The reduced expression of AOAH, an enzyme that hydrolyses the potent innate immune stimulant lipopolysaccharide, in monocytes might be anticipated to pre-dispose towards inflammation. Whether the further association of ARHGAP24, encoding a protein involved in actin remodelling59, a requirement for monocyte spreading and locomotion60 is secondary to reduced AOAH expression will require further investigation. This study demonstrates that healthy individuals possess cell specific expression signatures attributable to HLA allele carriage, reflecting the overwhelming importance HLA status plays upon everyday immune interactions.
Comparative analysis of highly pure cell subsets has the potential to unearth added intricacy in the nature of eQTLs, highlighting that the majority act in a cell specific manner and many eQTLs will likely remain undiscovered if analysis focuses on heterogeneous tissues. This study adds strong evidence to support the concept that a large number of eQTL function in a highly context specific manner in vivo and provide a plausible explanation for a degree of disease susceptibility.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to all the volunteers who participated in this study together with members of the Knight laboratory and Adrian Hill (University of Oxford) for their support. We thank Adam Auton for his assistance in the identification of probes containing SNPs with European MAF >0.01% using the 1000 Genome dataset and to John Broxholme for bioinformatic support and mapping of all probes to the reference sequence. We thank Greg Gibson (Georgia Institute of Technology), Adrian Hill and colleagues for critical reading of the manuscript and helpful suggestions. This work was supported by the Wellcome Trust (Grants 074318 [J.C.K.], 088891 [B.P.F.], and 075491/Z/04 [core facilities Wellcome Trust Centre for Human Genetics]), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC Grant agreement no. 281824 (J.C.K.) and the NIHR Oxford Biomedical Research Centre.
Footnotes
URLs. Catalog of Published Genome-Wide Association Studies (www.genome.gov/GWA studies)
European Genome-Phenome Archive (EGA) (www.ebi.ac.uk/ega/)
The R Project for Statistical Computing (http://www.r-project.org/)
Array Express (http://www.ebi.ac.uk/arrayexpress/)
ACCESSION NUMBERS: Gene expression data is available through ArrayExpress under accession number E-MTAB-945. Genotyping data has been deposited at the European Genome-Phenome Archive (EGA) and is available on request.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerrits A, et al. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–9. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 8.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwan T, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Idaghdour Y, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42:62–7. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–564. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons PA, et al. Microarray analysis of human leucocyte subsets: the advantages of positive selection and rapid purification. BMC Genomics. 2007;8:64. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehrmann RS, et al. Trans-eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 16.Van Laer L, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–7. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- 17.Akino K, et al. Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer. Cancer Sci. 2007;98:88–95. doi: 10.1111/j.1349-7006.2006.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Manivannan A, Crane I, Dawson R, Liversidge J. Critical but divergent roles for CD62L and CD44 in directing blood monocyte trafficking in vivo during inflammation. Blood. 2008;112:1166–74. doi: 10.1182/blood-2007-06-098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landers JE, et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106:9004–9. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benson V, Grobarova V, Richter J, Fiserova A. Glycosylation regulates NK cell-mediated effector function through PI3K pathway. Int Immunol. 2010;22:167–77. doi: 10.1093/intimm/dxp123. [DOI] [PubMed] [Google Scholar]
- 22.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010;185:6413–9. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer U, et al. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum Mol Genet. 1997;6:1817–22. doi: 10.1093/hmg/6.11.1817. [DOI] [PubMed] [Google Scholar]
- 24.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hakonarson H, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–6. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 26.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plagnol V, Smyth DJ, Todd JA, Clayton DG. Statistical independence of the colocalized association signals for type 1 diabetes and RPS26 gene expression on chromosome 12q13. Biostatistics. 2009;10:327–34. doi: 10.1093/biostatistics/kxn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koldobskiy MA, et al. p53-mediated apoptosis requires inositol hexakisphosphate kinase 2. Proc Natl Acad Sci U S A. 2010;107:20947–51. doi: 10.1073/pnas.1015671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawson BR, et al. Deficiency of the cyclin kinase inhibitor p21(WAF-1/CIP-1) promotes apoptosis of activated/memory T cells and inhibits spontaneous systemic autoimmunity. J Exp Med. 2004;199:547–57. doi: 10.1084/jem.20031685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim K, et al. A regulatory SNP at position -899 in CDKN1A is associated with systemic lupus erythematosus and lupus nephritis. Genes Immun. 2009;10:482–6. doi: 10.1038/gene.2009.5. [DOI] [PubMed] [Google Scholar]
- 33.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomaru T, et al. Isolation and characterization of a transcriptional cofactor and its novel isoform that bind the deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. Endocrinology. 2006;147:377–88. doi: 10.1210/en.2005-0450. [DOI] [PubMed] [Google Scholar]
- 35.Hagen FS, et al. Expression and characterization of recombinant human acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides. Biochemistry. 1991;30:8415–23. doi: 10.1021/bi00098a020. [DOI] [PubMed] [Google Scholar]
- 36.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–72. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavelin I, Geiger B. Characterization of a novel GTPase-activating protein associated with focal adhesions and the actin cytoskeleton. J Biol Chem. 2005;280:7178–85. doi: 10.1074/jbc.M411990200. [DOI] [PubMed] [Google Scholar]
- 39.Rioux JD, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 41.Hotfilder M, Baxendale S, Cross MA, Sablitzky F. Def-2, -3, -6 and -8, novel mouse genes differentially expressed in the haemopoietic system. Br J Haematol. 1999;106:335–44. doi: 10.1046/j.1365-2141.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- 42.Vandiedonck C, et al. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 2011;21:1042–54. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV 1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas R, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–4. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Limou S, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho PY, et al. Investigating the role of the HLA-Cw*06 and HLA-DRB1 genes in susceptibility to psoriatic arthritis: comparison with psoriasis and undifferentiated inflammatory arthritis. Ann Rheum Dis. 2008;67:677–82. doi: 10.1136/ard.2007.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–4. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–7. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neznanov N, Neznanova L, Angres B, Gudkov AV. Serologically defined colon cancer antigen 3 is necessary for the presentation of TNF receptor 1 on cell surface. DNA Cell Biol. 2005;24:777–85. doi: 10.1089/dna.2005.24.777. [DOI] [PubMed] [Google Scholar]
- 54.Ruland J. CARD9 signaling in the innate immune response. Ann N Y Acad Sci. 2008;1143:35–44. doi: 10.1196/annals.1443.024. [DOI] [PubMed] [Google Scholar]
- 55.Andres AM, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elstak ED, et al. The munc13-4-rab27 complex is specifically required for tethering secretory lysosomes at the plasma membrane. Blood. 2011;118:1570–8. doi: 10.1182/blood-2011-02-339523. [DOI] [PubMed] [Google Scholar]
- 57.Gray M, Miles K, Salter D, Gray D, Savill J. Apoptotic cells protect mice from autoimmune inflammation by the induction of regulatory B cells. Proc Natl Acad Sci U S A. 2007;104:14080–5. doi: 10.1073/pnas.0700326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knowles RW, et al. Complexity of the supertypic HLA-DRw53 specificity: two distinct epitopes differentially expressed on one or all of the DR beta-chains depending on the HLA-DR allotype. J Immunol. 1986;137:2618–26. [PubMed] [Google Scholar]
- 59.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8:803–14. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 60.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–45. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 61.Tedder TF, Isaacs CM. Isolation of cDNAs encoding the CD19 antigen of human and mouse B lymphocytes. A new member of the immunoglobulin superfamily. J Immunol. 1989;143:712–7. [PubMed] [Google Scholar]
- 62.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 65.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 67.Pruim R, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.