Abstract
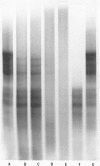
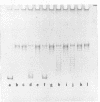
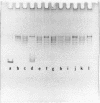
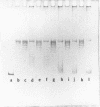
We have formed complexes of relaxed closed circular Col E1 DNA with various combinations of histones, and examined the effects of treating the complexes with nicking-closing enzyme. Germond et al (1) have shown that when a mixture of the four core histones of the nucleosome (HIA, H2B, H3 and H4) is used in such an experiment, the subsequently isolated DNA is supercoiled. We find that the arginine-rich histone pair, H3 and H4, is sufficient to induce the supercoiling observed in this experiment. Both H3 and H4 are required, and in the absence of either, no other histones are effective. H3 and and H4 are as efficient, per unit weight, as a mixture of the four histones in inducing supercoils. We also show that there is a large difference between the DNA bending energy needed to form a nucleosome and that needed to form one turn of normal superhelical DNA. These two processes are energetically quite distinct and probably separable. We estimate the free energy of interaction between DNA-bound histone pairs, and find that one or two such interactions would generate enough energy to fold the DNA into a nucleosome.
Full text
PDF





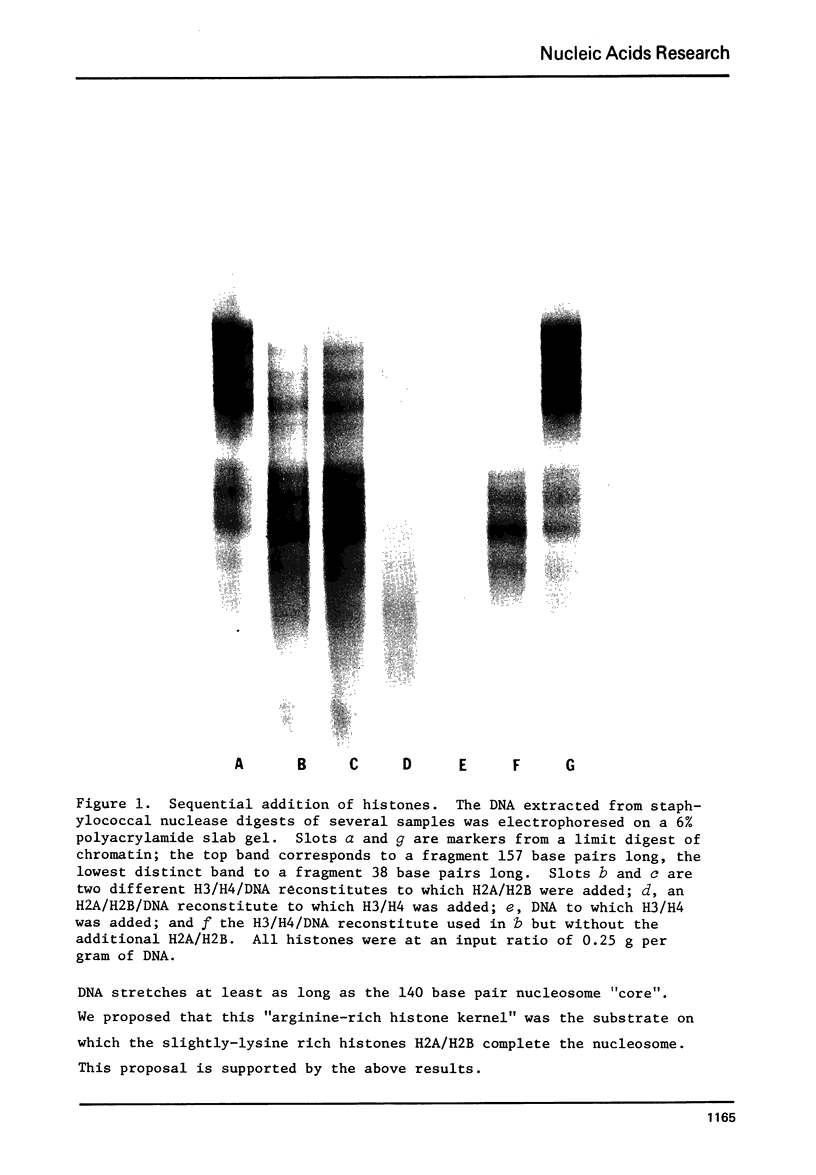





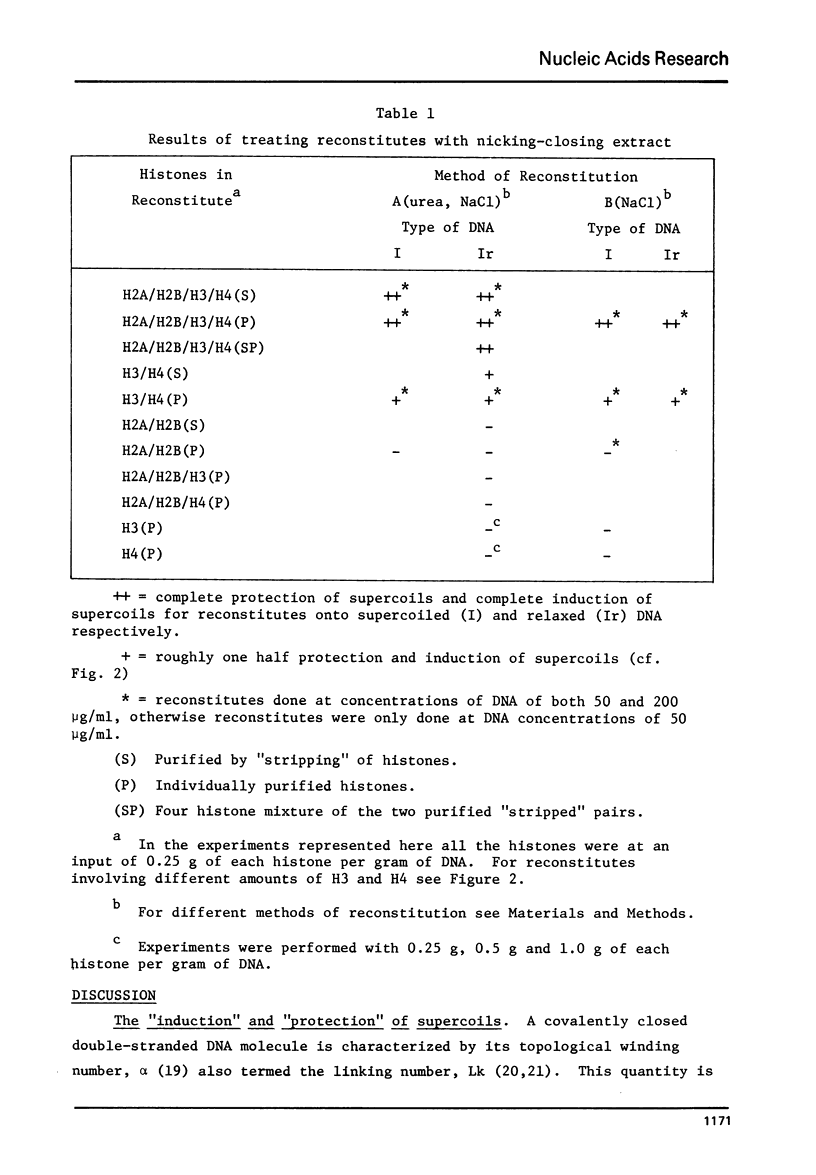










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baase W. A., Wang J. C. An omega protein from Drosophila melanogaster. Biochemistry. 1974 Oct 8;13(21):4299–4303. doi: 10.1021/bi00718a009. [DOI] [PubMed] [Google Scholar]
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Bradbury E. M., Butler-Browne G. S., Carpenter B. G., Stephens R. M. Physical studies of chromatin. The recombination of histones with DNA. Eur J Biochem. 1976 Feb 2;62(1):21–31. doi: 10.1111/j.1432-1033.1976.tb10093.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Cary P. D., Crane-Robinson C., Rattle H. W., Boublik M., Sautière P. Conformations and interactions of histone H2A (F2A2, ALK). Biochemistry. 1975 May 6;14(9):1876–1885. doi: 10.1021/bi00680a012. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Rattle H. W. Simple computer-aided approach for the analyses of the nuclear-magnetic-resonance spectra of histones. Fractions F1, Fsa1, F2B, cleaved halves of F2B and F2B-DNA. Eur J Biochem. 1972 May 23;27(2):270–281. doi: 10.1111/j.1432-1033.1972.tb01836.x. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Janin J. Principles of protein-protein recognition. Nature. 1975 Aug 28;256(5520):705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi C., Pignatti P. F., Croissant O., Yaniv M. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J Virol. 1975 Jan;17(1):204–211. doi: 10.1128/jvi.17.1.204-211.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. A histone cross-complexing pattern. Biochemistry. 1974 Nov 19;13(24):4992–4997. doi: 10.1021/bi00721a019. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubiger D., Hearst J. E. Effect of superhelical structure on the secondary structure of DNA rings. Biopolymers. 1967;5(8):691–696. doi: 10.1002/bip.1967.360050803. [DOI] [PubMed] [Google Scholar]
- Godfrey J. E., Eisenberg H. The flexibility of low molecular weight double-stranded DNA as a function of length. I. Light scattering measurements and the estimation of persistence lengths from light scattering, sedimentation and viscosity. Biophys Chem. 1976 Sep;5(3):301–318. doi: 10.1016/0301-4622(76)80042-7. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr Sedimentation coefficient of polyoma virus DNA. Biopolymers. 1967;5(10):1009–1019. doi: 10.1002/bip.1967.360051012. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Jolly D., Eisenberg H. Photon correlation spectroscopy, total intensity light scattering with laser radiation, and hydrodynamic studies of a well fractionated DNA sample. Biopolymers. 1976 Jan;15(1):61–95. doi: 10.1002/bip.1976.360150107. [DOI] [PubMed] [Google Scholar]
- Keller W. Characterization of purified DNA-relaxing enzyme from human tissue culture cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2550–2554. doi: 10.1073/pnas.72.7.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Page M. I., Jencks W. P. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Tatchell K., Van Holde K. E., Richards B. M. Low-angle neutron scattering from chromatin subunit particles. Nucleic Acids Res. 1975 Nov;2(11):2163–2176. doi: 10.1093/nar/2.11.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Morgan A. R. Partial purification of "omega" protein from calf thymus. Biochemistry. 1975 Nov 18;14(23):5205–5209. doi: 10.1021/bi00694a029. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Roark D. E., Geoghegan T. E., Keller G. H. A two-subunit histone complex from calf thymus. Biochem Biophys Res Commun. 1974 Jul 24;59(2):542–547. doi: 10.1016/s0006-291x(74)80014-8. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Allfrey V. G. A method for the purification of histone fraction F3 by affinity chromatography. Arch Biochem Biophys. 1973 Jan;154(1):185–191. doi: 10.1016/0003-9861(73)90047-7. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y., Tomizawa J. I. Replication of colicin E1 plasmid DNA in cell extracts. Proc Natl Acad Sci U S A. 1974 Mar;71(3):802–806. doi: 10.1073/pnas.71.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Herman T. M., Kovacic R. T., Beaudreau G. S., Van Holde K. E. Analysis of subunit organization in chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1976 Feb;73(2):505–509. doi: 10.1073/pnas.73.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Gray H. B., Jr, Vinograd J. Sedimentation velocity behavior of closed circular SV40 DNA as a function of superhelix density, ionic strength, counterion and temperature. J Mol Biol. 1971 Nov 28;62(1):21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P., Grossman L. I., Vinograd J. Isolation and partial characterisation of the relaxation protein from nuclei of cultured mouse and human cells. Eur J Biochem. 1975 Jun 16;55(1):79–93. doi: 10.1111/j.1432-1033.1975.tb02140.x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- van der Westhuyzen D. R., Böhm E. L., von Holt C. Fractionation of chicken erythrocyte whole histone into the six main components by gel exclusion chromatography. Biochim Biophys Acta. 1974 Aug 8;359(2):341–345. doi: 10.1016/0005-2795(74)90233-5. [DOI] [PubMed] [Google Scholar]