Abstract
Plasmodium falciparum is the agent of malignant malaria, one of mankind's most severe maladies. The parasite exhibits antigenic polymorphisms that have been postulated to be ancient. We have proposed that the extant world populations of P. falciparum have derived from one single parasite, a cenancestor, within the last 5,000–50,000 years. This inference derives from the virtual or complete absence of synonymous nucleotide polymorphisms at genes not involved in immune or drug responses. Seeking to conciliate this claim with extensive antigenic polymorphism, we first note that allele substitutions or polymorphisms can arise very rapidly, even in a single generation, in large populations subject to strong natural selection. Second, new alleles can arise not only by single-nucleotide mutations, but also by duplication/deletion of short simple-repeat DNA sequences, a process several orders of magnitude faster than single-nucleotide mutation. We analyze three antigenic genes known to be extremely polymorphic: Csp, Msp-1, and Msp-2. We identify regions consisting of tandem or proximally repetitive short DNA sequences, including some previously unnoticed. We conclude that the antigenic polymorphisms are consistent with the recent origin of the world populations of P. falciparum inferred from the analysis of nonantigenic genes.
The Malaria Plague and Control Efforts
The World Health Organization estimates that there are 300–500 million clinical cases of malaria per year, more than 1 million children die in sub-Saharan Africa, and more than 2 billion people are at risk throughout the world (1). Plasmodium falciparum is the agent of malignant malaria, the most fatal version of the disease. Malaria has been an elusive target for medical intervention. Epidemiological control efforts were first directed against the Anopheles mosquito vectors, which soon evolved resistance to massively applied insecticides. Current efforts against the mosquito vectors seek to produce transgenic mosquitoes that are unable to transmit Plasmodium, followed by massive release of the transformed vectors in endemic regions.
Greater efforts yet are invested in the development of protective vaccines or remedial drugs directed against the parasite. These exertions are handicapped, however, by the parasite's rapid evolution of drug resistance and antigens. Underlying this evolution is a wealth of genetic variation that arises rapidly by rearrangement of modular repeating elements that generate ever newly protected phenotypes.
The merozoite form of the Plasmodium parasite found in the human bloodstream is haploid. A fraction of these haploids differentiate into gametocytes, which are taken up in the mosquito's blood meal. Gametes fuse in the mosquito midgut to form transient diploids, which then undergo meiosis to yield haploid infectious forms, called sporozoites. Protective immunity against P. falciparum was demonstrated in the 1970s by immunization of human patients with irradiated sporozoites (2). Parasite genes that code for antigenic determinants subsequently have been isolated and characterized. Notable among the genes intensively investigated and chosen for vaccine development are those encoding surface proteins of the sporozoite (Csp, coding for the circumsporozoite protein) and the merozoite (Msp-1 and Msp-2, coding for the merozoite surface proteins 1 and 2). The success of efforts to develop an effective malaria vaccine is contingent on determining the extent of diversity of these genes and on identifying the mechanisms by which this variation is generated and persists in populations of P. falciparum.
Assessment of DNA sequence variation in P. falciparum has been based almost exclusively on examination of genes coding for antigenic determinants, where amino acid polymorphisms (nonsynonymous nucleotide polymorphisms) are common and likely to be affected by natural selection. Numerous studies have indicated that Csp, Msp-1, Msp-2, and other antigenic genes are polymorphic and that their multiple allelic forms differ in their ability to abrogate recognition by the host's immune response (3–7). These observations have been interpreted as instantiation of widespread polymorphism throughout the genome. Yet, we have investigated allelic variation in a diverse set of gene loci and found a complete absence of silent site polymorphism (8) and have proposed a recent derivation (within thousands of years) of the extant P. falciparum world populations from a single propagule.
It seems paradoxical that P. falciparum antigenic genes would be so highly polymorphic, because these genes must have shared the recent allelic homogenization caused by the population bottleneck we have inferred. Indeed, some authors have hypothesized that the polymorphisms of genes encoding P. falciparum surface proteins are very old, even older than the species itself.
We shall argue herein that the antigenic gene polymorphisms of P. falciparum are consistent with the conclusion drawn from the analysis of synonymous DNA sites, that the current world populations of the parasite are of recent origin, derived from a single strain within the last several thousand years. We will review our previous analysis of Csp (8) and then we will examine the Msp-1 and Msp-2 polymorphisms.
Evolutionary Association of P. falciparum with the Hominid Lineage
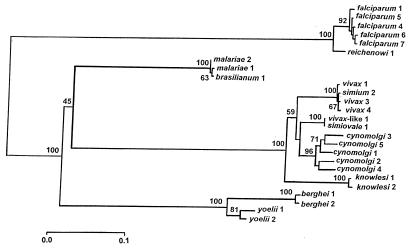
Fig. 1 is a phylogenetic tree of Plasmodium species derived from Csp gene sequences (ref. 9; for very similar trees based on other genes see refs. 10–12). Estimates of divergence times are shown in Table 1.
Figure 1.
Phylogeny of 12 Plasmodium species inferred from Csp gene sequences. P. falciparum, malariae, and vivax are human parasites; berghei and yoelii are rodent, and all others are primate parasites. The numbers refer to different strains. Bootstrap values above branches assess the reliability of the branch clusters; values above 70 are considered statistically reliable. Reprinted with permission from ref. 12.
Table 1.
Time (in million years) of divergence between Plasmodium species, based on genetic distances at two gene loci (see Figs. 1 and 2; adapted from refs. 9 and 10)
| Plasmodium | rRNA | Csp |
|---|---|---|
| falciparum vs. reichenowi | 11.2 ± 2.5 | 8.9 ± 0.4 |
| vivax vs. monkey* | 20.9 ± 3.8 | 25.2 ± 2.1 |
| vivax vs. malariae | 75.7 ± 8.8 | 103.5 ± 0.6 |
| falciparum vs. vivax/malariae | 75.7 ± 8.8 | 165.4 ± 1.6 |
brasilianum not included.
It is apparent that the three human parasites, P. falciparum, Plasmodium malariae, and Plasmodium vivax are very remotely related to each other, so that the evolutionary divergence of these three human parasites greatly predates the origin of the hominids. Plasmodium ovale, a fourth human parasite, is also remotely related to the other three (13). These results are consistent with the diversity of physiological and epidemiological characteristics of these four Plasmodium species (14, 15).
P. falciparum is more closely related to Plasmodium reichenowi, the chimpanzee parasite, than to any other Plasmodium species. The time of divergence between these two Plasmodium species is estimated at 8–12 million years ago, which is roughly consistent with the time of divergence between the two host species, human and chimpanzee. A parsimonious interpretation of this state of affairs is that P. falciparum is an ancient human parasite, associated with our ancestors since the divergence of the hominids from the great apes. Fig. 1 shows that P. malariae, a human parasite, is genetically indistinguishable from Plasmodium brasilianum, a parasite of New World monkeys; similarly, human P. vivax is genetically indistinguishable from Plasmodium simium, also a parasite of New World monkeys. It follows that lateral transfer between hosts has occurred in recent times, at least in these two cases. Whether the transfer has been from humans to monkeys or vice versa is a moot question (for discussion, see ref. 12).
Malaria's Eve: Recent Origin of P. falciparum World Populations
Silent (i.e., synonymous) nucleotide polymorphisms are appropriate for estimating the age of genes, because silent nucleotide polymorphisms are often adaptively neutral (or very nearly so). Thus, silent nucleotide polymorphisms reflect the mutation rate and the time elapsed since their divergence from a common ancestor. Table 2 summarizes data for 10 genes (8). The gene sequences analyzed derive from isolates of P. falciparum geographically representative of the global malaria endemic regions (see table 1 in ref. 8; and, for the Csp gene, ref. 16). A scarcity of synonymous polymorphisms also has been observed in a separate study of 10 P. falciparum genes, most of them antigenic (17).
Table 2.
Polymorphisms in 10 loci of P. falciparum
| Gene | Chromosome location | Length, bp | Sequences in the sample, ni | Nonsynonymous polymorphisms, Dn | Synonymous polymorphisms, Ds | Synonymous sites analyzed
|
|
|---|---|---|---|---|---|---|---|
| 4-fold, nili | 2-fold, nimi | ||||||
| Dhfr | 4 | 609 | 32 | 4 | 0 | 2,144 | 4,128 |
| Ts | 4 | 1,215 | 10 | 0 | 0 | 1,250 | 2,640 |
| Dhps | 8 | 1,269 | 12 | 5 | 0 | 1,536 | 2,724 |
| Mdr1 | 5 | 4,758 | 3 | 1 | 0 | 1,350 | 2,088 |
| Rap1 | — | 2,349 | 9 | 8 | 0 | 1,092 | 1,668 |
| Calm | 14 | 441 | 7 | 0 | 0 | 364 | 602 |
| G6pd | 14 | 2,205 | 3 | 9 | 0 | 726 | 1,404 |
| Hsp86 | 7 | 2,241 | 2 | 0 | 0 | 532 | 910 |
| Tpi | — | 597 | 2 | 0 | 0 | 180 | 262 |
| Csp 5′end | 3 | 387 | 25 | 7 | 0 | 688 | 2,010 |
| Csp 3′end | 3 | 378 | 25 | 17 | 0 | 1,050 | 1,625 |
| Total | — | — | — | 51 | 0 | 10,912 | 20,061 |
Modified from ref. 12.
As we have expounded elsewhere (8, 11–12), five possible hypotheses may account for the absence of silent polymorphisms in P. falciparum: (i) persistent low effective population size, (ii) low rates of spontaneous mutation, (iii) strong selective constraints on silent variation, (iv) one or more recent selective sweeps affecting the genome as a whole, and (v) a demographic sweep, i.e., a recent population bottleneck, so that extant world populations of P. falciparum would have recently derived from a single ancestral strain. We have concluded that only the fifth hypothesis is consistent with the observations and have used coalescent theory to estimate the age of the ancestral strain or “cenancestor” (Table 3).
Table 3.
Estimated times to the cenancestor of the world populations of P. falciparum
| Estimated mutation
rate ×
10−9
|
t95 | t50 | |
|---|---|---|---|
| μa | μb | ||
| 7.12 | 2.22 | 24,511 | 5,670 |
| 3.03 | 0.95 | 57,481 | 13,296 |
Adapted from refs. 8 and 11. t95 and t50 are the upper boundaries of the confidence intervals. Thus, in the first row the cenancestor lived less than 24,511 years ago with a 95% probability, and less than 5,670 years ago with a 50% probability. μa and μb are the estimated neutral mutation rates of 4-fold and 2-fold degenerate codons, respectively.
The issue arises of how to account for a recent demographic sweep in P. falciparum. One possible hypothesis is that P. falciparum has become a human parasite in recent times, by lateral transfer from some other host species (18). This hypothesis is contrary to available evidence (9, 10). An alternative explanation is that human parasitism by P. falciparum has long been highly restricted geographically and has dispersed throughout the Old World continents only within the last several thousand years, perhaps within the last 10,000 years, after the Neolithic revolution (19–21). Three possible scenarios may have led to this historically recent dispersion: (i) changes in human societies, (ii) genetic changes in the host-parasite-vector association that have altered their compatibility, and (iii) climatic changes that entailed demographic changes (migration, density, etc.) in the human host, the mosquito vectors, and/or the parasite.
One factor that may have impacted the widespread distribution of P. falciparum in human populations from a limited original focus, probably in tropical Africa, may have been changes in human living patterns, particularly the development of agricultural societies and urban centers that increased human population density (20–26). Genetic changes that have increased the affinity within the parasite-vector-host system also seem to be a viable explanation for a recent expansion. Coluzzi (20, 21) has cogently argued that the worldwide distribution of P. falciparum is recent and has come about, in part, as a consequence of a recent dramatic rise in vectorial capacity caused by repeated speciation events in Africa of the most anthropophilic members of the species complexes of the Anopheles gambiae and Anopheles funestus mosquito vectors. The biological processes implied by this account may have, in turn, been associated with, and even depended on the onset of agricultural societies in Africa and climatic changes, specifically the gradual increase in ambient temperatures after the Würm glaciation, so that about 6,000 years ago climatic conditions in the Mediterranean region and the Middle East made possible the spread of P. falciparum and its vectors beyond tropical Africa (20, 21, 24, 25).
Sherman (26) has noticed the late introduction and low incidence of falciparum malaria in the Mediterranean region, which postdates historical times. Hippocrates (460–370 B.C.) describes quartan and tertian fevers, but there is no mention of severe malignant tertian fevers, which suggests that P. falciparum infections did not yet occur in classical Greece, as recently as 2,400 years ago. The late introduction of falciparum malaria into the Mediterranean region and the Middle East has been attributed to the low vectorial efficiency of the indigenous anopheline mosquitoes (20, 21). Once the demographic and climate conditions became suitable for the propagation of P. falciparum, natural selection would have facilitated the evolution of Anopheles species that were highly anthropophilic and effective falciparum vectors (20, 21, 24).
The selective sweep hypothesis (iv) is, in a way, a special case of the demographic sweep hypothesis (v); i.e., a particular strain may have spread throughout the world and replaced all other strains impelled by natural selection. Natural selection can account for the absence of synonymous variation at any one of the 10 loci shown in Table 2, if the particular gene itself (or a gene with which it is linked) has been subject to a recent worldwide selective sweep, without sufficient time for the accumulation of new synonymous mutations. However, the 10 genes are located on, at least, six different chromosomes (Table 2), and thus six independent selective sweeps would need to have occurred more or less concurrently, which seems prima facie unlikely. A selective sweep simultaneously affecting all chromosomes could happen if one accepts the hypothesis that the population structure of P. falciparum is predominantly clonal, rather than sexual (see refs. 10 and 12). This hypothesis is controversial, although we have argued that it may indeed be the case, the capacity for sexual reproduction of the parasite notwithstanding (12, 16).
The Recent Origin of P. falciparum Populations Vis-à-Vis Antigenic Polymorphisms
The absence of synonymous polymorphisms in most P. falciparum genes must be made congruous with the substantial levels of polymorphism observed in such antigenic genes as Csp, Msp-1, and Msp-2. We propose that nucleotide polymorphism arises in antigenic genes promoted by natural selection acting on two different “mutation” processes. First, the familiar process of single-site nucleotide mutation generates amino acid replacements that give rise to polymorphisms at antigenic sites subject to diversifying selection. Second, there is intragenic recombination that generates variation at a rapid rate in repetitive segments (often occurring in tandem) of antigenic genes. The variation generated by intragenic recombination is also subject to diversifying natural selection because it contributes to the parasite's ability to evade the immune response of the human host. We will show that some of the reported nucleotide variation between antigenic alleles is an artifact stemming from misalignment of gene sequences that are of different lengths as a consequence of unequal numbers of repetitions generated by intragenic recombination.
The CSP
The Csp gene is comprised of two terminal regions that are not repetitive (5′ NR and 3′ NR), which embrace a central region (CR) made up of a variable number (mostly, between 40 and 50) of tandemly arranged 12-nt-long repeats. As shown in Table 2, there are no silent polymorphisms in the 5′NR and 3′NR regions of the gene, which is part of the evidence supporting a recent origin of P. falciparum populations.
The repetitive amino acid sequences encoded within the CR are remarkably conserved (only two amino acid motifs are known in P. falciparum, NANP and NVDP; Table 4), but there is a fair deal of synonymous nucleotide polymorphism among the repeats (Table 5). We have introduced the concept of the repeat allotype (RAT) to refer to variant nucleotide sequences that encode a single amino acid motif (16). Among the known Csp gene sequences of P. falciparum, there are 10 RATs that encode the NANP motif and four RATs that encode the NVDP motif, with an average of about 10 RATs per gene sequence (range 9–11; see Table 6). Table 4 displays the arrangement of the two amino acid motifs in 25 gene sequences of P. falciparum and one of P. reichenowi. The alignment of the RATs can be found in Rich et al. (ref. 16; see also ref. 12). The only known sequence of Csp in P. reichenowi is somewhat shorter than those of falciparum (35 rather than about 45 repeats per sequence, on average), but has a similar number of distinct RATs (10, the same as the falciparum average) and three rather than only two amino acid motifs, two of them identical to those of falciparum.
Table 4.
Composition of the CR of the Csp gene
| Sequence | Repeat motifs | Number of repeats
|
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| M15505 | 1212111111111111111211111111111111111111111111 | 43 | 3 | 0 |
| M83173 | 1212111111111111111211111111111111111111111111 | 43 | 3 | 0 |
| M83149 | 12121211111111111111111111111111111111111111 | 41 | 3 | 0 |
| M83150 | 12121111111111111112111111111111111111111111111 | 44 | 3 | 0 |
| M83156 | 121211111111111111111111111111111111111111111111111 | 49 | 2 | 0 |
| M83158 | 1212121211111111111111111111111111111111111111 | 42 | 4 | 0 |
| M83161 | 1212121111111111111111111211111111111111111 | 39 | 4 | 0 |
| M83163 | 12121111111111111112111111111111111111111111111 | 43 | 3 | 0 |
| M83164 | 12121111111111111112111111111111111111111111111 | 46 | 3 | 0 |
| M83165 | 1212121111111111111111111111111111111111111111 | 43 | 3 | 0 |
| M83166 | 1212121211111111111111111111111111111111111111 | 42 | 4 | 0 |
| M83167 | 12121211111111111111111111111111111111111111111 | 46 | 3 | 0 |
| M83168 | 1212121211111111111111111111111111111111111111 | 42 | 4 | 0 |
| M83169 | 12121211111111111111111111111111111111111111 | 41 | 3 | 0 |
| M83170 | 1212121211111111111111111111111111111111111111 | 42 | 4 | 0 |
| M83174 | 1212121111111111211111111111111111111111111 | 39 | 4 | 0 |
| M19752 | 12121211111111111111111111111111111111111111 | 41 | 3 | 0 |
| M83172 | 121212111111111111111121111111111111111111 | 38 | 4 | 0 |
| K02194 | 12121211111111111111121111111111111111111 | 37 | 4 | 0 |
| M57499 | 12121212111111111111111111111111111111111111 | 40 | 4 | 0 |
| U20969 | 1212121111111111111112111111111111111111 | 36 | 4 | 0 |
| M83886 | 121212111111111111111112111111111111111111 | 38 | 4 | 0 |
| M22982 | 12121211111111111111111112111111111111111111 | 40 | 4 | 0 |
| X15363 | 12121211111111111111111112111111111111111111 | 40 | 4 | 0 |
| M57498 | 12121211111111111111121111111111111111111 | 37 | 4 | 0 |
| P. reichenowi | 12121212131213131311111111111111111 | 26 | 5 | 4 |
The repeat motifs NANP, NVDP, and NVNP are represented by 1, 2, and 3, respectively. Adapted from ref. 12.
Table 5.
Amino acid and nucleotide sequence of the RATs and their incidence
| RAT | Motif
|
Falciparum
|
reichenowi
|
|||
|---|---|---|---|---|---|---|
| Amino acid | Nucleotide | % | Number | % | Number | |
| A | NANP | aatgcaaaccca | 55.1 | 566 | 38.5 | 10 |
| B | NANP | ........t..t | 16.1 | 165 | 30.8 | 8 |
| C | NANP | ........t... | 7.6 | 78 | — | — |
| D | NANP | .....c..t..a | 6.2 | 64 | 3.8 | 1 |
| E | NANP | ..........c | 6.2 | 64 | — | — |
| F | NANP | ..c........c | 5.1 | 52 | — | — |
| G | NANP | .....c...... | 3.1 | 32 | 7.7 | 2 |
| H | NANP | ...........t | 0.3 | 3 | — | — |
| I | NANP | ..c......... | 0.2 | 2 | 3.8 | 1 |
| J | NANP | .....c....c | 0.1 | 1 | — | — |
| Z | NANP | ........t..c | — | — | 15.4 | 4 |
| M | NVDP | ....t.g.t... | 52.3 | 46 | 20.0 | 1 |
| N | NVDP | ....t.g.t..c | 31.8 | 28 | 40.0 | 2 |
| O | NVDP | ..c.t.g.t..t | 14.8 | 13 | — | — |
| P | NVDP | ....t.g.t..t | 1.1 | 1 | 20.0 | 2 |
| X | NVNP | ....t...t..c | — | — | 100.0 | 4 |
Table 6.
Number of RATs in the Csp gene sequences of P. falciparum and in P. reichenowi
| Number of sequences | Different RATs per
sequence
|
Total RATs per sequence
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 10 | 11 | 35 | 40 | 41 | 42 | 43 | 44 | 46 | 47 | 49 | 51 | ||
| P. falciparum | 25 | 12 | 8 | 5 | 1 | 2 | 2 | 2 | 6 | 8 | 1 | 2 | 1 | |
| P. reichenowi | 1 | 1 | 1 | |||||||||||
Nearly all of the synonymous site differences observed in the CR are between RATs that exist within any single allele. This is a strong indication that while RAT diversity may have an ancient origin, it has been maintained within individual alleles and therefore can withstand even the most constricted bottleneck. For example, all 25 Csp CR alleles contain at least one copy of each of the most common RATs (A, B, C, D, E, and F, which amount to 96% of all NANP repeats; and M and N, which amount to 84% of all NVDP repeats). If any one of these alleles were the sole survivor following a bottleneck, it alone would possess nearly all of the diversity currently known for the species; intragenic recombination between the RATs originally present in one allele can generate size polymorphisms in the resulting alleles. The process of bottleneck reduction, ensued by generation of new variations through intragenic recombination, may have occurred numerous times in the evolution of the species, and may continue to do so, given the nature of the parasite lifestyle and its propensity for being confronted by population bottlenecks, for example, upon colonization of new geographic regions or during seasonal epidemic relapses.
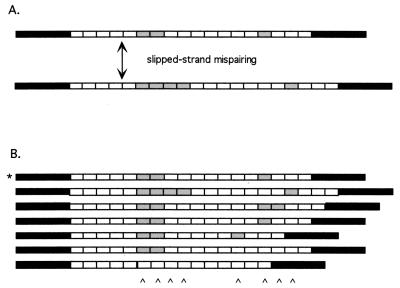
We have proposed that most of the variation in antigenic genes is attributable to duplication and/or deletion of the repeated segments within the genes, which is simply an instance of the general slipped-strand process for generating length variation in repetitive DNA regions (Fig. 2). This process occurs by several mechanisms, each of which is well understood at the molecular level and may involve either intrahelical or interhelical exchange of DNA (27). Intragenic recombination often is associated with the evolution of minisatellite or microsatellite DNA loci, such as those recently described in P. falciparum (28, 29). However, intragenic recombination also has been implicated in generating variability within coding regions in a variety of eukaryotes; including the Drosophila yolk protein gene and the human α2-globin gene, to cite just two examples (30, 31).
Figure 2.
A model of RAT evolution. Black boxes represent flanking single-copy regions; gray and white boxes represent different RATs. (A) A single slippage event yields a duplication of two gray RATs. (B) Six new alleles, derived from a single ancestor (indicated by *) after several cell generations. Slippage produces deletions as well as duplications. Karats at the bottom mark artifactual substitutions appearing when the alleles are aligned. Reprinted with permission from ref. 12.
New RATs can arise by one of two processes: (i) replacement or silent substitutions in a codon, and (ii) the slippage mechanism that leads to RAT proliferation. The two amino acid motifs and the different RAT types have arisen by the first process. The variation in the number of RATs arises by the second process. The second process occurs with a frequency several orders of magnitude greater than the first process (32).
How much of the variation now present in the Csp CR region of falciparum may have arisen by the second process? Notice that only two amino acid motifs are present in the whole set of 25 Csp sequences and that both motifs are present in every one of the sequences (Tables 4 and 5). Thus, there is no evidence that any replacement substitution has occurred in the recent evolution of P. falciparum.
Cryptic Repeats in the Msp-1 Polymorphism
The Msp-1 gene codes for MSP-1 (also referred to as MSA-1, P195, and otherwise), which is a large 185- to 215-kDa protein precursor that is proteolytically cleaved into several membrane protein constituents. The known alleles of Msp-1 belong to one or the other of two allelic classes (group I and group II). There is considerable nucleotide substitution and length variation between the two classes but much less variation within each class (6, 33). The two classes are commonly designated by the strains in which they were originally identified: K1 (group I) and MAD20 (group II).
Tanabe et al. (6) partitioned MSP-1 into 17 blocks, based on the degree of amino acid polymorphism (Table 7). They classified seven blocks (blocks 2, 4, 6, 8, 10, 14, and 16) as highly variable; five blocks (blocks 7, 9, 11, 13, and 15) as semiconserved, and five blocks (blocks 1, 3, 5, 12, and 17, which include the two terminal segments) as conserved. The “highly variable” (as well as the “semiconserved”) amino acid polymorphisms occur only when comparisons are made between the two allele groups, whereas amino acid, as well as synonymous, nucleotide polymorphisms are very low within each allele group. An exception is block 2, which encodes a set of repetitive tripeptides and thus is subject to the same intragenic recombination described above for Csp, as a mechanism for generating polymorphism (in block 4 there is considerable nonsynonymous polymorphism among group I alleles). Table 7 gives the nucleotide diversity (π) for synonymous and nonsynonymous substitutions for each of the 17 blocks, both within and between groups (see Fig. 3). The most extensive amino acid polymorphism between the two allele groups occurs in block 8, which has been assumed to have no simple repeats, but that we will show below to be composed of tandem and proximal repeats (see Fig. 4).
Table 7.
Nucleotide diversity (π) within and between group I and II alleles of the P. falciparum Msp-1 genes
| Block | Length, codons | Synonymous
|
Nonsynonymous
|
||||
|---|---|---|---|---|---|---|---|
| Group I | Group II | Group I + group II | Group I | Group II | Group I + group II | ||
| 1 | 55 | 0.019 | 0.021 | 0.017 | 0.017 | 0.010 | 0.013 |
| 2 | 55 | 0.106 | 0.185 | 0.150 | 0.449 | 0.497 | 0.553 |
| 3 | 202 | 0.038 | 0.006 | 0.042 | 0.018 | 0.000 | 0.023 |
| 4 | 31 | 0.031 | 0.000 | 0.020 | 0.307 | 0.000 | 0.215 |
| 5 | 35 | 0.000 | 0.000 | 0.070 | 0.000 | 0.000 | 0.026 |
| 6 | 227 | 0.000 | 0.000 | 0.282 | 0.004 | 0.001 | 0.300 |
| 7 | 73 | 0.000 | 0.000 | 0.361 | 0.003 | 0.000 | 0.072 |
| 8 | 95 | 0.000 | 0.000 | 0.338 | 0.000 | 0.003 | 0.711 |
| 9 | 107 | 0.000 | 0.023 | 0.409 | 0.005 | 0.043 | 0.126 |
| 10 | 126 | 0.008 | 0.000 | 0.448 | 0.011 | 0.000 | 0.394 |
| 11 | 35 | 0.000 | 0.000 | 0.128 | 0.000 | 0.000 | 0.068 |
| 12 | 79 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 13 | 84 | 0.000 | 0.042 | 0.040 | 0.005 | 0.007 | 0.052 |
| 14 | 60 | 0.000 | 0.018 | 0.212 | 0.002 | 0.005 | 0.371 |
| 15 | 89 | 0.000 | 0.000 | 0.216 | 0.001 | 0.003 | 0.089 |
| 16 | 217 | 0.002 | 0.032 | 0.277 | 0.005 | 0.027 | 0.185 |
| 17 | 99 | 0.002 | 0.019 | 0.007 | 0.010 | 0.027 | 0.016 |
Blocks are as defined by Tanabe et al. (6). Some block lengths vary between group I and II alleles; the value given is the average length of group I and II alleles.
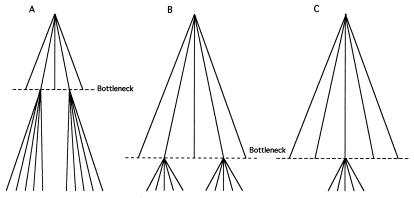
Figure 3.
Three possible models of the evolution of Msp-1 group I and group II alleles. (A) After an ancient bottleneck, only two alleles survive; these two alleles each give rise to new alleles over time. We expect the two allele groups to be very heterogeneous within groups, and more so between groups, with respect to both synonymous and nonsynonymous substitutions. (B) After a recent bottleneck, only two alleles survive, each of which give rise to new alleles. Alleles within a group are fairly similar to each other but alleles from different groups are very heterogeneous throughout the length of the gene, with respect to synonymous and nonsynonymous substitutions. (C) After a recent bottleneck, only one allele survives that gives rise to new alleles over time. Alleles within and between groups are similar, except for occasional (mostly synonymous) substitutions and for differences generated by intragenic recombination, evidenced by the presence of repeats. A and B are inconsistent with the data in Table 6.
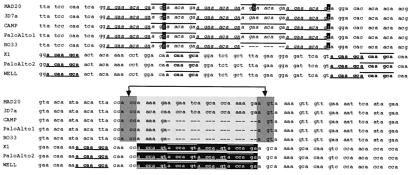
Figure 4.
Partial alignment of MspI (block 8) group I and II alleles. Alternating odd and even occurrence of a repeat is indicated by underline and overbar, respectively. Region R2a consists of five tandem repeats of a 9-bp sequence (agaaacaga, in italics) highlighted in the five group II alleles (Upper); one copy is missing in the RO33 allele. Regions R1a and R1b consist of two repeats, measuring 7-bp (acaagca, in boldface; repeated five times) and 6-bp (accagt, shown in inverted text; repeated four times) found in group I alleles. The five 7-bp repeats (except for two) are separated by several codons, whereas the 6-bp repeats occur in tandem. There are no repeat sequences shared between groups I and II; however, the 6-bp repeat in group I alleles clearly derives from a deletion of the intervening lightly shaded portion of group II alleles, followed by duplication of the resulting accagt motif (junction indicated by arrows). In this regard, the Camp, Palo Alto-1, and RO33 alleles are intermediate between MAD20/3D7 and K1/Palo Alto-2/Wellcome alleles.
The dimorphism observed among group I and II alleles within block 2 has been shown to result by processes analogous to those within the Csp central repeat region (34, 35). The occurrence of repetitive DNA within other blocks has not been described to date. However, we have identified repeats within several of the most polymorphic Msp-1 blocks; in particular, blocks 4, 8, and 14, which heretofore were assumed to be NR. We focus on the repeats detected within block 8, identified by Tanabe et al. (6) as showing the lowest amino acid similarity between groups (10%; π = 0.711 in Table 7). We have identified three group-specific repeats within this block, two in group I alleles (R1a and R1b), and one in group II alleles (R2a). R2a is a 9-bp repeat tandemly replicated five times in all group II alleles (the five uppermost alleles in Fig. 3). R1a is a 7-bp repeat replicated five times, and R1b is a 6-bp repeat replicated four times in all group I alleles. The occurrence of repeats within this very short stretch of DNA is a highly significant departure from chance (12). We have searched the recently completed genomic sequences of P. falciparum chromosomes 2 and 3. The nucleotide sequences of repeats R1a, R1b, and R2a appear 25, 116, and 11 times, respectively, within the 947 kb of chromosome 2. Within the 1,060 kb of chromosome 3, the R1a, R1b, and R2a are present 39, 52, and seven times, respectively. None of the three nucleotide repeats ever appears in tandem on either chromosome 2 or 3. The average distance between each occurrence on these chromosomes is >20 kb, corroborating that their repeated occurrence in the short 147-bp segment of Msp-1 block 8 is a strong departure from random expectation. The Msp-1 gene is located on chromosome 9, which has not yet been assembled as a complete nucleotide sequence; but the distribution of these nucleotide repeats is not likely to differ markedly between chromosomes by chance alone.
We have made two interesting observations while searching chromosome 2 and 3 sequences for the presence of these nucleotide repeats. First, five of the 11 R2a repeats found on chromosome 2 are located within a 558-bp region corresponding to a predicted secreted antigen that appears similar to the glutamic acid-rich protein gene. Second, 67 of the 116 R1a nucleotide repeats on chromosome 2 occur as the 3′ terminus of a 39-nt repeat within the pfEMP member of the var gene family, which is an important component of P. falciparum antigenic variation. The observation of highly significant repeats within regions of the Msp-1 gene previously thought not to be repetitive makes it clear that the extensive between-group nucleotide diversity between the two allelic groups is attributable to the same kinds of repeat variation and rapid divergence known in other antigenic determinants.
Msp-2 Polymorphism
The Msp-2 gene codes for MSP-2 (or MSA-2), a glycoprotein anchored, like MSP-1, in the merozoite membrane, but 45 kDa in size, and thus much smaller than MSP-1. The Msp-2 of P. falciparum shows much greater variability in length, amino acid content, and number of repeats than Csp, but the pattern of allele polymorphism in Msp-2 is consistent with the hypothesis that it has rapidly arisen by intragenic recombination.
Similar to CSP, MSP-2 is characterized by conserved N and C termini, with 43 and 74 residues, respectively (7). Bracketed within these segments, is the highly variable repeat region. Two allelic families have been identified and named after the isolates in which they were first identified. The FC27 family is characterized by at least one copy of a 32-aa sequence and a variable number of repeats, 12 aa in length. The 3D7/Camp family contains tandem amino acid repeats measuring 4–10 aa in length (36).
The 3D7/Camp alleles are more variable in length and sequence of repeat types than those of the FC27 family (37). Fenton et al. (38) have proposed a model to explain the origin of repeat diversity within the 3D7/Camp family of alleles. They divided the 3D7/Camp family into distinct allelic subclasses, which included types A1 and A3, distinguished by amino acid repeats of different length. For example, A1 alleles possess 4-aa motifs, whereas a repeating 8-aa motif occurs in A3. Fenton et al. (38) have shown that the allelic subclasses within the 3D7/Camp family are derived from a common ancestral nucleotide sequence and that the diversity arises from duplication and deletion of repeat subunits.
Recently, Dubbeld et al. (39) have cloned and sequenced the Msp-2 gene of P. reichenowi, which is a “unique mosaic of P. falciparum allelic forms and species-specific elements.” We have used the methods described by Fenton et al. (38) to determine whether the Msp-2 of P. reichenowi provides insight into the ancestry of the FC27 and 3D7/Camp families. Fig. 5 shows the amino acid sequence alignment of two P. falciparum MSP-2s with the P. reichenowi MSP-2. The P. falciparum alleles from the 3D7 and OKS isolates are representative of the 3D7/Camp and FC27 families, respectively. The two P. falciparum alleles are identical at nucleotide sites encoding the N and C termini, but exhibit little similarity, even at the amino acid level, in the intervening repeat region.
Figure 5.
Amino acid alignment of P. falciparum (3D7 and OKS) and P. reichenowi Msp-2 alleles. Open boxes demarcate the conserved N and C termini. The inferred RHRs are shaded in black (RHR1), dark gray (RHR2), and light gray (RHR3). The nucleotide alignments for the inferred repeats of these regions are shown in Figs. 6 and 7.
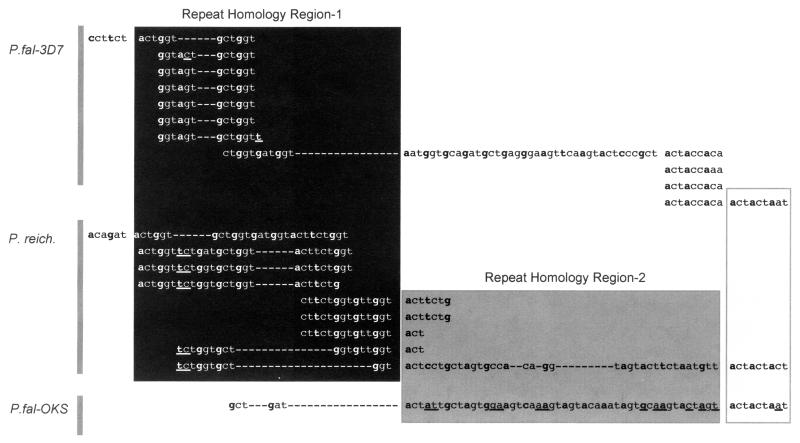
A closer look at the nucleotides within the central portion of the gene manifests the homology of three distinct regions, which we define as repeat homology regions (RHRs). RHR1 shows common ancestry between the P. reichenowi Msp-2 and the 3D7 Msp-2 alleles (Fig. 6, black shading). Diversity within this region results from proliferation of a ggtgct hexamer (38). This hexamer is ancestral to both the 3D7/Camp and the P. reichenowi Msp-2 allelic repeats within this region. Although the conservation of these codons is clear among these two alleles, it appears that they have been lost altogether in the FC27-like alleles (represented by OKS in Figs. 5–7). However, the region adjacent to RHR1 in the P. reichenowi Msp-2 sequence is similar to the first 21 aa of the 32-aa repeat found within the FC27 family, and this sequence is the basis for the inferred RHR2 (Fig. 5, dark gray shading). The last 9 nt of RHR2 also manifest the homology between all three sequences, including the short stretch following the (actaccaa)4 repeat in 3D7. Note also the overlap between repeating nucleotides of P. reichenowi Msp-2 in both RHR1 and RHR2.
Figure 6.
Partial nucleotide alignments of three Msp-2 gene sequences to manifest the repeats and the homologies between P. falciparum 3D7 and P. reichenowi (RHR1, black shading) and between P. falciparum OKS and P. reichenowi (RHR2, dark shading). Sequences read left to right and down where homologous repeats are present. The open box at the 3′ end of RHR2 shows a region of high similarity among all three alleles. Bold letters indicate the first nucleotide of each codon. Differences between aligned sequences are highlighted by underline. The alignment of repeats follows the convention of Fenton et al. (38), so that repeats within and between sequences are aligned to show their homology.
Figure 7.
Nucleotide alignment of two Msp-2 gene sequences to manifest the repeats within RHR3 of P. falciparum OKS. This repeat region is not present in P. falciparum 3D7 or P. reichenowi. The repeat region of OKS continues contiguously from first to second to third to fourth row, left to right.
A third RHR is located further downstream and shows the relationship between the 12-aa repeats of OKS and P. reichenowi Msp-2 (Fig. 7). The repeat region in OKS is surrounded on either side by a 10-bp sequence (tacagaaagt), which occurs as only a single 5′ copy in the P. reichenowi Msp-2 allele. Despite the lengthy repeat insertion in the OKS sequence, the homology of OKS and the P. reichenowi Msp-2 in the region downstream of this repeat is apparent. And so it appears that the repeats were generated sometime after the split between P. falciparum and P. reichenowi.
Analysis of the single P. reichenowi sequence allows us to approximate the ancestral sequence of the two P. falciparum Msp-2 allele families. Indeed, the comparison of the three RHRs discloses that whereas the precursor sequences for the various repeats probably were derived from the common P. falciparum–P. reichenowi ancestral species, the extant diversity among the Msp-2 alleles has derived since the divergence of the two species. The distinctive dimorphism of the two P. falciparum alleles results from proliferation of repeats in two different regions of the molecule. Presumably because the overall MSP-2 molecule is constrained in size, the proliferation of repeats leads consequently to loss of nucleotides along the gene regions; i.e., the 3D7/Camp repeat precursors were lost in FC27 alleles, and the FC27 repeat precursors were lost in the 3D7 alleles.
As noted for Csp, the repetitive DNA sequences found within the Msp-2 (and Msp-1) genes, as well as those in other P. falciparum antigenic determinants, are subject to much higher rates of mutation than NR sequences found within the same locus. Indeed, the paucity of silent substitutions within the NR regions indicates that intragenic recombination has generated repeat diversity in relatively short periods of time. Empirical estimates of mutation rates among repetitive DNA sequences, such as satellite DNA, are as high as 10-2 mutations/per generation and therefore several orders of magnitude greater than rates for point mutations (32). The high mutation rates, coupled with strong selection for immune evasion, yield an extremely accelerated evolutionary rate for P. falciparum antigens.
Antigenic Polymorphism, Intragenic Recombination, and Population Structure
Homologous comparisons among allelic variants of antigenic genes manifest that most of the variation is attributable to the rapid mutational processes associated with intragenic recombination. The increased rate of evolution among these genes reconciles the recent origin of extant P. falciparum populations with the abundance of antigenic diversity observed globally and locally. We have noted that nucleotide diversification can result from either intrahelical or interhelical events. An example of intrahelical recombination is that of mitotic, slipped-strand mismatch repair, which is considered to be the principal source of variation in repetitive units such as satellite DNA (Fig. 5). Interhelical recombination derives from the classical process of meiotic crossing over and recombination within or between loci on homologous chromosomes.
Both of these processes occur in P. falciparum. Kerr et al. (40) have shown that meiotic, interhelical recombination occurs between mixed Msp-2 genotype parasites passaged in laboratory animals. This process constitutes the basis for generating linkage maps of P. falciparum chromosomes (28). But we have shown that, despite the abundant intragenic recombination within Csp CR, there is an apparent absence of recombination between the 5′ and 3′ NR regions, suggesting that the duplication and deletion of RATs occur by mitotic processes such as the slipped-strand process modeled in Fig. 5 (16). This process also has been implicated as the cause of repeat variation in Msp-2 (38).
The debate over the relevance of sexual recombination between P. falciparum types may remain unsettled for some time. It is becoming increasingly clear that the population structure of P. falciparum may not be uniform throughout the species, but depends on local factors related to parasite, vector, and host biology (5, 41–43). An accurate determination of these factors is contingent on careful analysis of parasite genotypes and appropriate determination of homologous comparisons.
Acknowledgments
We are grateful to Benjamin Rosenthal and F. Ellis McKenzie for thoughtful insights and comments.
Abbreviations
- CSP
circumsporozoite protein
- MSP-1 and MSP-2
merozoite surface proteins 1 and 2, respectively
- CR
central region
- NR
not repetitive
- RAT
repeat allotype
- RHR
repeat homology region
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.World Health Organization. Tropical Disease Report, Twelfth Programme Report. Geneva: World Health Organization; 1995. [Google Scholar]
- 2.Clyde D F, McCarthy V C, Miller R M, Hornick R B. Am J Med Sci. 1973;266:398–403. doi: 10.1097/00000441-197312000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Ukhayakumar V, Shi Y-P, Kumar S, Jue D L, Wohlhueter R M, Lal A A. Infect Immun. 1994;62:1410–1413. doi: 10.1128/iai.62.4.1410-1413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zevering Y, Khamboonruang C, Good M F. Eur J Immunol. 1994;24:1418–1425. doi: 10.1002/eji.1830240627. [DOI] [PubMed] [Google Scholar]
- 5.Babiker H, Walliker D. Parasitol Today. 1997;13:262–267. doi: 10.1016/s0169-4758(97)01075-2. [DOI] [PubMed] [Google Scholar]
- 6.Tanabe K, Mackay M, Goman M, Scaife J G. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 7.Smythe J A, Coppel R L, Kay K P, Martin R K, Oduola A M J, Kemp D J, Anders R F. Proc Natl Acad Sci USA. 1991;88:1751–1755. doi: 10.1073/pnas.88.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rich S M, Licht M C, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1998;95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escalante A A, Barrio E, Ayala F J. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- 10.Escalante A A, Ayala F J. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayala F J, Escalante A A, Lal A A, Rich S M. In: Malaria: Parasite Biology, Pathogenesis, and Protection. Sherman I W, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 285–300. [Google Scholar]
- 12.Ayala F J, Escalante A A, Rich S M. Parassitologia. 1999;41:55–68. [PubMed] [Google Scholar]
- 13.Qari S H, Shi Y P, Pieniazek N J, Collins W E, Lal A A. Mol Phylogenet Evol. 1996;6:157–165. doi: 10.1006/mpev.1996.0068. [DOI] [PubMed] [Google Scholar]
- 14.Coatney R G, Collins W E, Warren M, Contacos P G. The Primate Malarias. Washington, DC: U.S. Government Printing Office; 1971. [Google Scholar]
- 15.López-Antuñano F, Schumunis F A. In: Parasitic Protozoa. 2nd Ed. Kreier J P, editor. Vol. 5. New York: Academic; 1993. pp. 135–265. [Google Scholar]
- 16.Rich S M, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escalante A A, Lal A A, Ayala F J. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waters A P, Higgins D G, McCutchan T F. Proc Natl Acad Sci USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coluzzi M. Parassitologia. 1994;36:223–227. [PubMed] [Google Scholar]
- 20.Coluzzi M. Evoluzione Biologica i Grandi Problemi della Biologia. Rome: Accademia dei Lincei; 1997. pp. 263–285. [Google Scholar]
- 21.Coluzzi M. Parassitologia. 1999;41:277–283. [PubMed] [Google Scholar]
- 22.Livingstone F B. Am Anthropol. 1958;60:533–562. [Google Scholar]
- 23.Wiesenfeld S L. Science. 1967;157:1134–1140. doi: 10.1126/science.157.3793.1134. [DOI] [PubMed] [Google Scholar]
- 24.de Zulueta J. Parassitologia. 1973;15:1–15. [PubMed] [Google Scholar]
- 25.de Zulueta J. Parassitologia. 1994;36:7–15. [PubMed] [Google Scholar]
- 26.Sherman I W. In: Malaria: Parasite Biology, Pathogenesis, and Protection. Sherman I W, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 3–10. [Google Scholar]
- 27.Levinson G, Gutman G A. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 28.Su X, Wellems T E. Genomics. 1996;33:430–444. doi: 10.1006/geno.1996.0218. [DOI] [PubMed] [Google Scholar]
- 29.Anderson T J C, Su X Z, Bockarie M, Lagog M, Day K P. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- 30.Oron-Karni V, Filon D, Rund D, Oppenheim A. Hum Mol Genet. 1997;6:881–885. doi: 10.1093/hmg/6.6.881. [DOI] [PubMed] [Google Scholar]
- 31.Ho K F, Craddock E M, Piano F, Kambysellis M P. J Mol Evol. 1996;43:116–124. doi: 10.1007/BF02337356. [DOI] [PubMed] [Google Scholar]
- 32.Schug M D, Hutter C M, Noor M A, Aquadro C F. Genetica. 1998;102–103:359–367. [PubMed] [Google Scholar]
- 33.Hughes A L. Mol Biol Evol. 1992;9:381–393. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]
- 34.Frontali C. Genetica. 1994;94:91–100. doi: 10.1007/BF01443424. [DOI] [PubMed] [Google Scholar]
- 35.Frontali C, Pizzi E. Acta Leiden. 1991;60:69–81. [PubMed] [Google Scholar]
- 36.Felger I, Tavul L, Kabintik S, Marshall V, Genton B, Alpers M, Beck H P. Exp Parasitol. 1994;79:106–116. doi: 10.1006/expr.1994.1070. [DOI] [PubMed] [Google Scholar]
- 37.Felger I, Marshal V M, Reeder J C, Hunt J A, Mgone C S, Beck H P. J Mol Evol. 1997;45:154–160. doi: 10.1007/pl00006215. [DOI] [PubMed] [Google Scholar]
- 38.Fenton B, Clark J T, Khan C M A, Robinson J V, Walliker D, Ridley R, Scaife J G, McBride J S. Mol Cell Biol. 1991;11:963–974. doi: 10.1128/mcb.11.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubbeld M A, Kocken C H, Thomas A W. Mol Biochem Parasitol. 1998;92:187–192. doi: 10.1016/s0166-6851(97)00244-2. [DOI] [PubMed] [Google Scholar]
- 40.Kerr P J, Ranford-Cartwright L C, Walliker D. Mol Biochem Parasitol. 1994;66:241–248. doi: 10.1016/0166-6851(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 41.Paul R E L, Packer M J, Walmsley M, Lagog M, Ranford-Cartwright L C, Paru R, Day K P. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 42.Conway D J, Roper C, Oduola A M J, Arnot D E, Kremsner P G, Grobusch M P, Curtis C R, Greenwood B M. Proc Natl Acad Sci USA. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakihama N, Kimura M, Hirayama K, Kanda T, Na-Bangchang K, Jongwutiwes S, Conway D, Tanabe K. Gene. 1999;230:47–54. doi: 10.1016/s0378-1119(99)00069-4. [DOI] [PubMed] [Google Scholar]