Abstract
In a medium containing 10mM Tris, pH 8, 10 mM MG++, 50 mM K+ and 10 mM NH4, the binding of an E. coli RNA polymerase holoenzyme unwinds the DNA helix by about 240 degrees at 37 degrees C. In this medium the total unwinding of the DNA increases linearly with the molar ratio of polymerase to DNA. The number of binding sites at which unwinding can occur is very large. If the K+ concentration is increased at 200 mM, the enzyme binds to only a limited number of sites, and the bound and free enzyme molecules do not exchange at an appreciable rate. The unwinding angle of the DNA per bound enzyme in this high salt medium is measured to be 140 degrees at 37 degrees C. The total unwinding angle for a fixed number of bound polymerase molecules per DNA is strongly temperature dependent, and decreases with decreasing temperature.
Full text
PDF



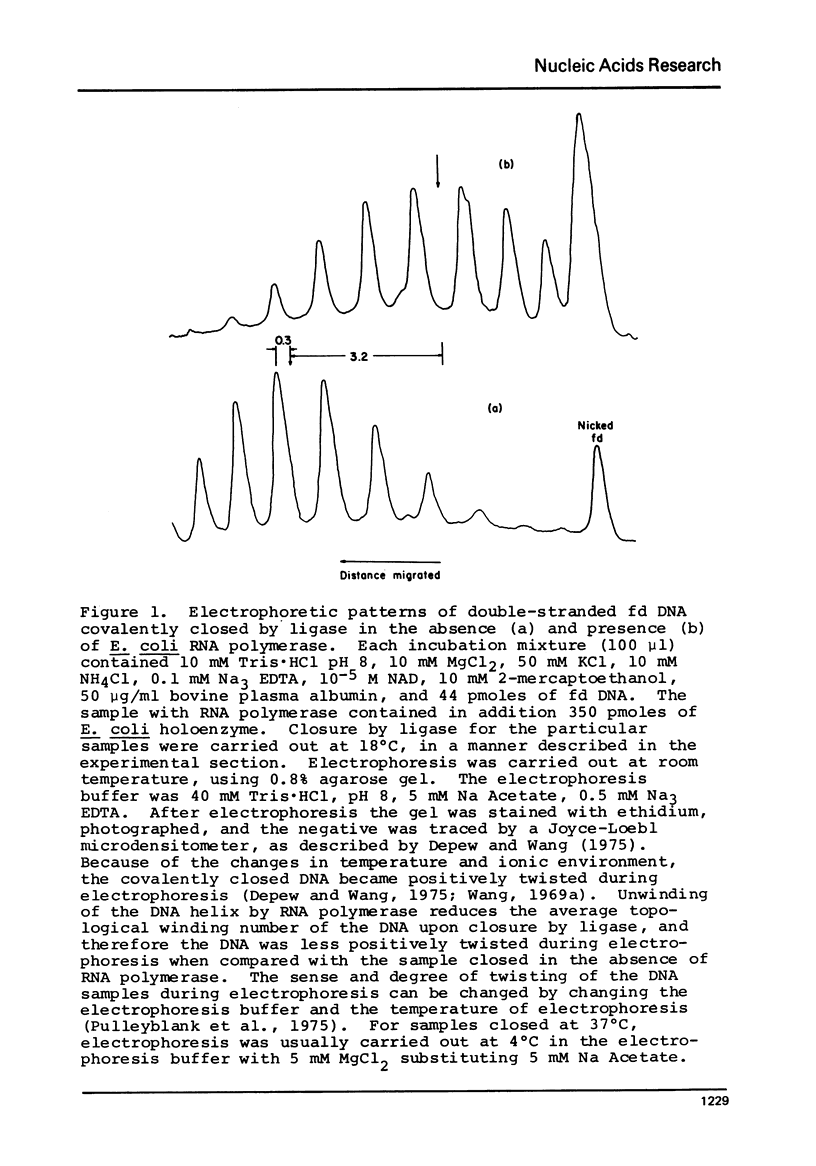

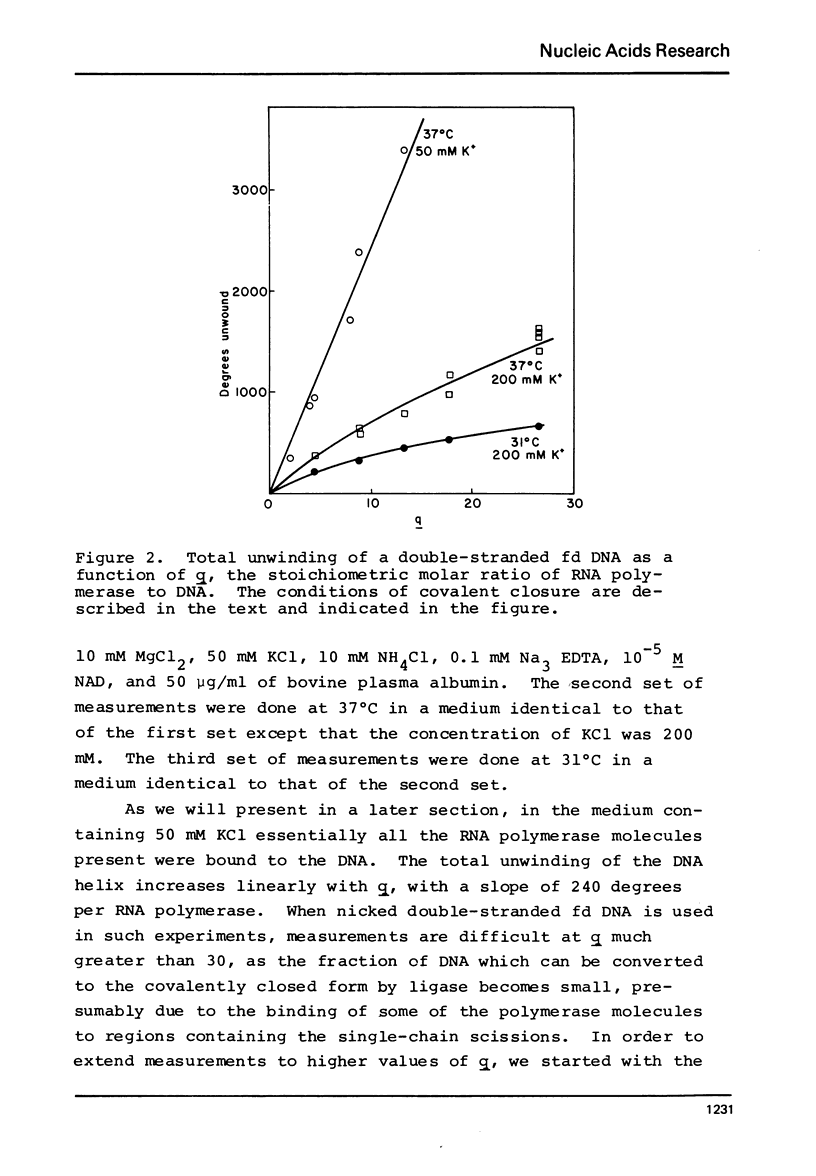


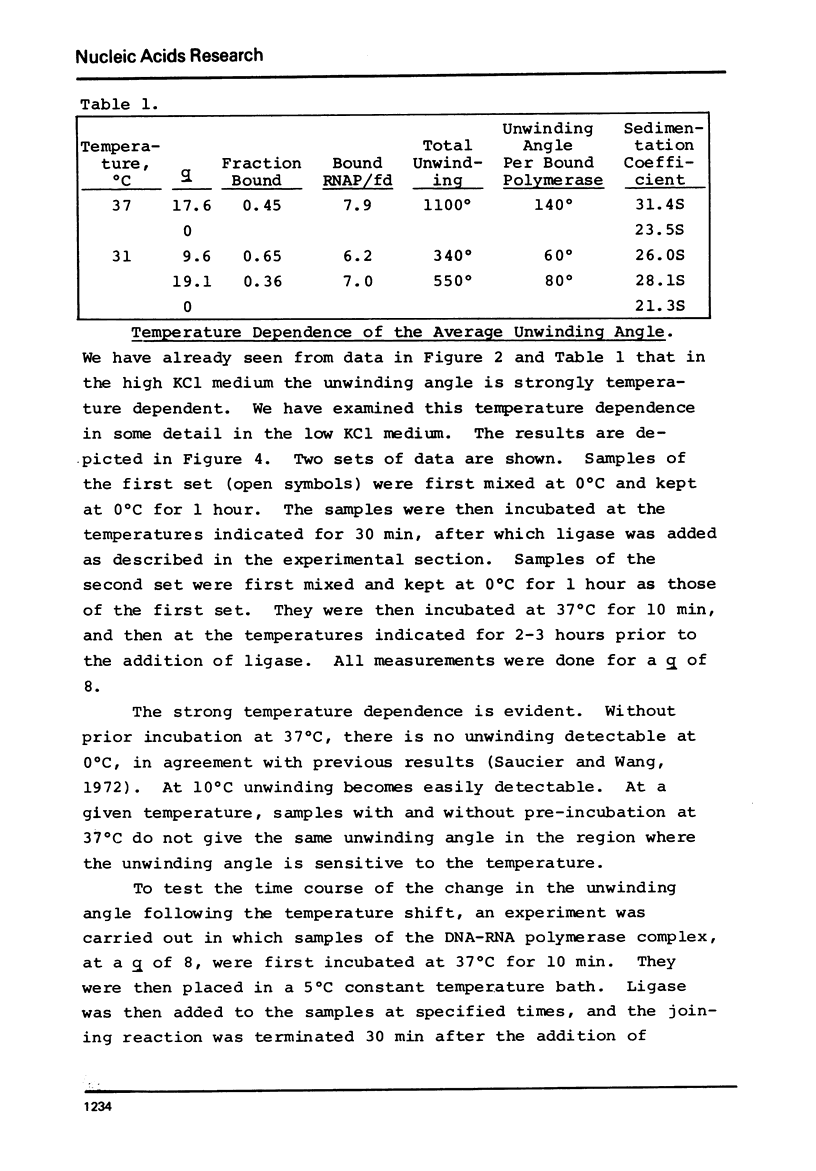
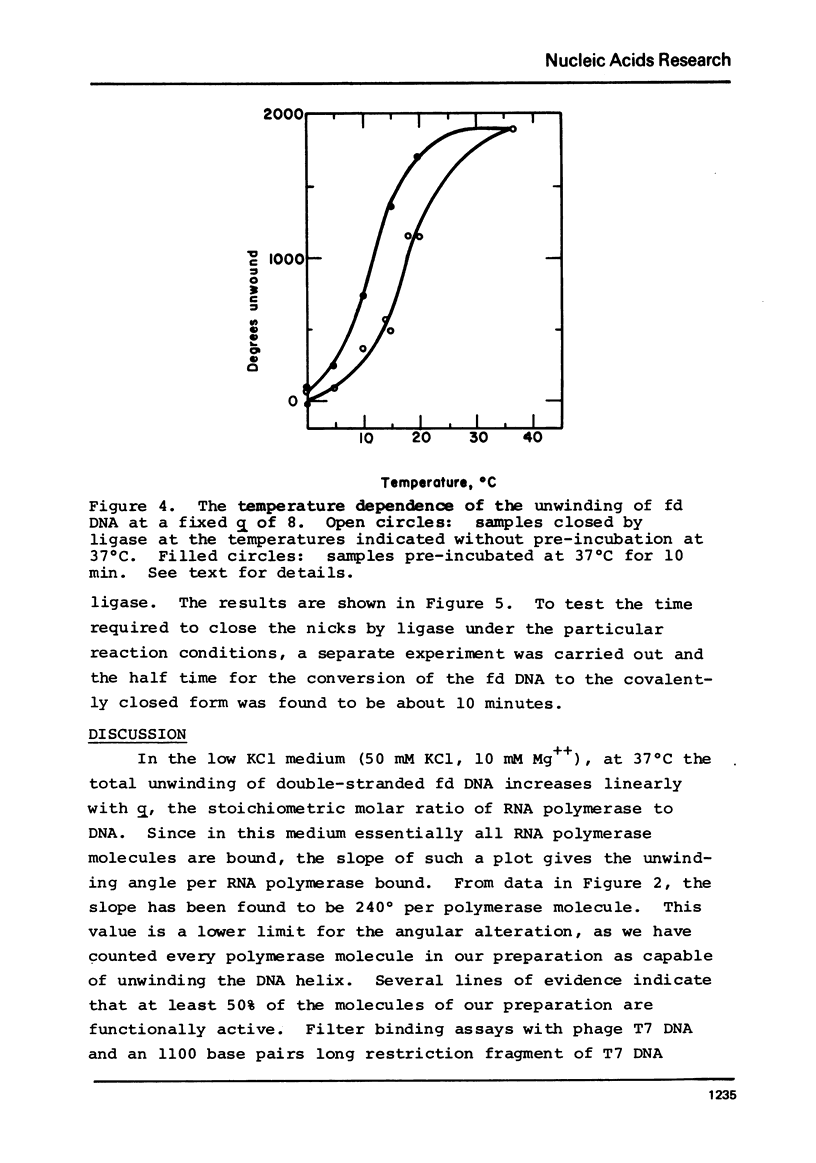





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W. Host specificity of DNA produced by Escherichia coli. 9. Host-controlled modification of bacteriophage fd. J Mol Biol. 1966 Oct;20(3):483–496. doi: 10.1016/0022-2836(66)90004-0. [DOI] [PubMed] [Google Scholar]
- Berg D., Chamberlin M. Physical studies on ribonucleic acid polymerase from Escherichia coli B. Biochemistry. 1970 Dec 22;9(26):5055–5064. doi: 10.1021/bi00828a003. [DOI] [PubMed] [Google Scholar]
- Berkowitz S. A., Day L. A. Molecular weight of single-stranded fd bacteriophage DNA. High speed equilibrium sedimentation and light scattering measurements. Biochemistry. 1974 Nov 5;13(23):4825–4831. doi: 10.1021/bi00720a022. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. The selectivity of transcription. Annu Rev Biochem. 1974;43(0):721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- Dausse J. P., Sentenac A., Fromageot P. Interaction of RNA polymerase from Escherichia coli with DNA. Effect of temperature and ionic strength on selection of T7 DNA early promoters. Eur J Biochem. 1976 Jun 1;65(2):387–393. doi: 10.1111/j.1432-1033.1976.tb10352.x. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubert J. M., Hirschbein L. Reversible attachment of RNA polymerase to DNA as a function of temperature. Biochem Biophys Res Commun. 1969 Jan 27;34(2):149–155. doi: 10.1016/0006-291x(69)90624-x. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- Modrich P., Lehman I. R., Wang J. C. Enzymatic joining of polynucleotides. XI. Reversal of Escherichia coli deoxyribonucleic acid ligase reaction. J Biol Chem. 1972 Oct 10;247(19):6370–6372. [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P. Some physical properties of RNA polymerase. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1616–1623. doi: 10.1073/pnas.55.6.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P. The binding of RNA polymerase to DNA. J Mol Biol. 1966 Oct 28;21(1):83–114. doi: 10.1016/0022-2836(66)90081-7. [DOI] [PubMed] [Google Scholar]
- Saucier J. M., Wang J. C. Angular alteration of the DNA helix by E. coli RNA polymerase. Nat New Biol. 1972 Oct 11;239(93):167–170. doi: 10.1038/newbio239167a0. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J Mol Biol. 1974 Aug 25;87(4):797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]


