Abstract
Inflammation is fundamental for protecting the organism against infection and injury. However, a failure to control immune response results in chronic inflammation and several associated disorders such as pain and loss of function. Initiation of inflammation is orchestrated by cytokines, among which IL-1β is particularly important. IL-1β is synthesized as an inactive protein that has to be processed by the inflammasome to generate the mature bioactive form. Conventional techniques cannot monitor IL-1β activation with high spatial and temporal resolution. In this study, we present a ratiometric biosensor that allows monitoring IL-1β processing in real time, with a temporal resolution of seconds and with a single-cell spatial resolution. Using this sensor, to our knowledge, we describe for the first time the kinetic of the inflammasome activity in living macrophages. With this new probe, we also demonstrated that the pro–IL-1β processing occurs all over the cytoplasm.
Introduction
Inflammation is an innate immune response aimed at recovering tissue homeostasis following injury or infection. The proinflammatory cytokine IL-1β is a key mediator of inflammation (1). It is produced and secreted by different cell types, with monocytes and macrophages being the main source. The IL-1β is synthesized as an inactive precursor, pro–IL-1β, which is accumulated in the cytosol. After cleavage by the cysteine protease caspase-1 into the biologically active form, mature IL-1β is secreted to the extracellular space (2). Caspase-1 activation is regulated by a supramolecular protein complex generally defined as the inflammasome (3). The best-characterized inflammasome contains the cytosolic innate receptor nucleotide-binding, leucine-rich repeat, receptor pyrin 3 (NLRP3) and the adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC). Various pathogens (e.g., the bacterial ionophore nigericin) and endogenous danger signals such as, for example, extracellular ATP, trigger the NLRP3 inflammasome-associated activation of caspase-1, resulting in pro–IL-1β processing and release. Subsequently, mature IL-1β binds to the IL-1 type I receptor on neighboring cells and initiates the inflammatory response. Aberrant IL-1β processing has been reported in different pathologies, including gout, rheumatoid arthritis, or type 2 diabetes (4).
Despite the importance of IL-1β in the host defense and in chronic inflammation, contemporary techniques for monitoring caspase-1 activation and IL-1β processing are limited in their spatiotemporal resolution. For example, Western blot (WB) and ELISA, which are widely used to detect and quantify IL-1β processing, do not allow monitoring IL-1β activation in real time and in situ. Alternatively, activation of caspase-1 can be detected using peptide substrates conjugated to fluorescent dyes; these, however, are not commonly used due to their lack of sensitivity and low specificity. Furthermore, these probes are not adaptable to follow the dynamics of caspase-1 activation in real time. Finally, all approaches outlined above are time and/or cell consuming and require cell lysis or fixation.
For this study, we engineered a genetically encoded biosensor based on bioluminescence resonance energy transfer (BRET) in which the precursor pro–IL-1β is fused at its terminals to RLuc8 (a variant of Renilla luciferase) and Venus (a variant of yellow fluorescent protein). The functional features of the sensor are similar to the endogenous IL-1β, which makes this probe an ideal tool for the characterization of pro–IL-1β processing and for the high-throughput screening of compounds interacting with the initiation of inflammation. Using macrophages as a model, we report that this probe allows detection of IL-1β processing in real time at the single-cell level and with a time resolution in the range of seconds, showing that full activation of NLRP3 inflammasome requires at least 5 min following nigericin stimulation.
Materials and Methods
Plasmids, cloning, and site-directed mutagenesis
NLRP3-flag construct was a gift of J. Tschopp (University of Lausanne, Epalinges, Switzerland). Caspase-1 and ASC-V5 constructs were a gift of G. Dubyak (Case Western Reserve University School of Medicine, Cleveland, OH). Pro–IL-1β sequence construct was cloned from LPS-treated THP-1 cells. The IL-1β sensor was constructed by amplifying out each of the components (RLuc8, pro–IL-1β, and Venus) by PCR using Platinum Taq polymerase (Invitrogen, Paisley, U.K.). RLuc8 and Venus were amplified from pcDNA3.1-RLuc8 and pcDNA3.1-Venus, respectively. Care was taken to ensure that there was only one starting methionine (Met) at the 5′ end of Rluc8 and one stop codon (TAG) at the 3′ end of Venus. Primers were designed to add: 1) NheI restriction site and retain the Met at the 5′ Rluc8, but replace the stop codon with a EcoRV site at its 3′ end (RLuc8 forward, 5′-ATATCAGCTAGCGCCACCATGGCTTCCAAGGTGTACGACCCCGAG-3′; R-Luc8 reverse, 5′-TGCTGAAGAAC GAGCAGGAAAGCGGCCGCATAGTG-3′); 2) to replace pro–IL-1β Met with a EcoRV site at the 5′ end and replace the stop codon with a XhoI site at the 3′ end (IL-1β forward, 5′-AGTTGTGCGGCCGCTTGGCAGAAGTACCTGAG-3′; IL-1β reverse, 5′-ACCATGCAATTTGTGTCTTCTCTCGAGTGTACGTCTAGAACCTC-3′); and 3) to add a XhoI site and replace the Met at the 5′ of Venus and to add XbaI and a stop codon, TAG, to the 3′ end (Venus forward, 5′-TCAGTACTCGAGTTGGTGAGCAAGGGCG-3′; Venus reverse, 5′-TCCGCATCTAGATTACTTGTACAGCTCG-3′). These were then sequentially ligated into pcDNA3.1+ vector (Invitrogen), using T4 DNA ligase (New England Biolabs). Between each stage, the intermediate products were transformed into chemically competent bacteria, grown overnight in culture. Plasmid DNA preparations were performed and digested with the relevant enzymes to ensure that the correct size insert was present. Single D27A and D116A and double D27A/D116A point mutations were generated from the above construct using the PCR overlap extension method and Phusion High-Fidelity DNA Polymerase (New England Biolabs). Final constructs were fully sequenced to discard random mutations during the amplification and cloning procedure.
Cell culture and transfection
Immortalized mouse bone marrow-derived macrophages (BMDM) from C57BL/6 (wild type [WT]), ASC−/−, or NLRP3−/− were a gift of E. Latz (5) and were cultured in DMEM; J774A.1 mouse macrophage cell line was cultured in RPMI 1640 and HEK293 in F-12/DMEM, all supplemented with 10% FCS and 1% Glutamax. All cell culture media components were from Invitrogen. For mouse primary BMDM, mice were killed by excess isoflurane inhalation (Baxter Healthcare). Experiments were carried out under the Animals (Scientific Procedures) Act 1986. Femurs and tibia were removed and dissociated from muscles, and DMEM was used to wash out the marrow cavity plugs. The bone marrow cells were resuspended in DMEM (Invitrogen) supplemented with 20% L cell-conditioned medium, 10% calf serum (Invitrogen), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen), plated onto T75 flasks. After 7 d, the resulting BMDM were detached by cells scraping and plated in the appropriate plate dishes.
HEK293 cells were transfected with Lipofectamine 2000 (Invitrogen), according to manufacturer’s instructions. Twenty-four hours after plating, primary BMDM and macrophage cell lines were transfected with Trans-IT-Neural (Mirus), according to manufacturer’s instructions. Twenty-four hours later, macrophages were primed with 1 μg/ml LPS (Sigma-Aldrich) for 4 h before any experiments.
BRET recordings from cell populations
Transfected HEK293 cells were plated in poly-l-lysine–coated white 96-well plates (Greiner). Macrophages were directly transfected in 96-well plates. Twenty-four hours later, cells were washed with E-total physiological solution (in mM: 147 NaCl, 2 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, and 13 d-glucose [pH 7.3]), and readings were then immediately performed after addition of 5 mM coelenterazine h substrate (Invitrogen). Signals were detected with two filter settings (R-Luc filter, 480 ± 20 nm; and YFP filter, 535 ± 25 nm) at 37°C using the Mithras LB940 plate reader (Berthold Biotechnologies). The BRET ratio was defined as the difference of the emission at 535/480 nm transfected R-Luc8 and Venus fusion proteins and the emission at 535/480 nm R-Luc8 protein alone. Results were expressed in milli BRET units (mBU). BRET recordings were performed using the ARPEGE Pharmacology Screening-Interactome platform facility at the Institute of Functional Genomics (Montpellier, France).
BRET recording on single cells
Single-cell BRET recording was performed, as described previously (6). Briefly, BMDM were transfected in 35-mm glass-bottom dishes. Twenty-four hours later, cells were washed and maintained in E-total solution. BRET imaging studies were performed at room temperature using an Axiovert 200M inverted fluorescence microscope (Zeiss, Jena, Germany) equipped with a Plan-Apochromat ×63/1.40 Oil M27 objective. Transfected cells were identified using filters to excite Venus (exciter HQ480/40 No. 44001, emitter HQ525/50 No. 42017; Chroma, Rockingam, VT). Coelenterazine h (20 μM) was applied 5 min before acquisition. Images were collected using an Evolve camera (equipped with an EMCCD detector, back-illuminated, On-chip Multiplication Gain) from Photometrics (Tucson, AZ). Sequential acquisitions of 15 or 30 s (depending on sensor expression) were performed with emission filters D480/60 nm (No. 61274; Chroma) and HQ535/50 nm (No. 63944; Chroma) to select em480 and em535 wavelengths, respectively. Acquisition and analysis were controlled and analyzed by Metamorph (Molecular Devices, Sunnyvale, CA) and ImageJ (NIH) software, respectively, as described previously (6). BRET signals were represented using a continuous 16-pseudocolor look-up table, as displayed in the figures.
Detection of emission spectra
Twenty-four hours after transfection, HEK293 cells were lysed with a hypotonic cell lysis buffer (25 mM HEPES [pH 7.5], 5 mM MgCl2, 5 mM EDTA, 5 mM DTT) for 10 min at 4°C. After spinning (13,200 rpm, 10 min, 4°C), supernatants were mixed with a 2× concentrated caspase-1 buffer (0.2% CHAPS, 0.2 M HEPES, 20% sucrose, 29 mM DTT [pH 7.5]). After addition of human recombinant caspase-1 (Calbiochem), samples were incubated at 37°C for 1 h. Emission spectra were recorded in 96-white–well plates in presence of 5 μM coelenterazine h using a Flexstation plate reader (Molecular Devices) and a 1-nm step increment from 400 to 600 nm.
Western blots
After washing with PBS, cells were scraped in lysis buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 5 mM EDTA, 1% Nonidet P-40), supplemented with the Halt protease and phosphatase inhibitor mixture (Pierce). After 30 min at 4°C, lysates were centrifuged (16,000 × g, 10 min, 4°C), and the supernatant was collected and mixed to NuPAGE LDS Sample Buffer (Invitrogen). Proteins were separated on 4–12% Bis-Tris gels (NuPage Novex, Invitrogen) or homemade 15% acrylamide gels and transferred to nitrocellulose membranes. Proteins were detected using HRP-coupled secondary Abs, appropriate primary Abs (GFP, Roche, 1/2000; RLuc, MBL International, 1/1000; ASC, Santa Cruz Biotechnology, 1/2500; NLRP3, Axxora, 1/1000; caspase-1 p10, Santa Cruz Biotechnology, 1/150), and SuperSignal West Pico/Femto substrates (Pierce).
Cell supernatants were clarified by brief centrifugation, concentrated using 10-kDa nominal molecular mass cutoff filters (Millipore), and then mixed to sample buffer, as described above.
Subcellular localization of R-proIL1β-V
Images of HEK293 and macrophage cell lines transfected with the BRET sensor were acquired using a Zeiss Axiovert 25 microscope equipped with a Zeiss Axiocam HRc camera. Venus fluorescence was detected at 515–565 nm following excitation at 450–490 nm (Filter set 10; Zeiss).
Statistical analysis
Statistical analysis was performed using an ANOVA, followed by Tukey-Kramer multiple comparison test using GraphPad Instat (GraphPad Software). Significance thresholds were always set at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). All data in graphs are shown as mean ± error bars representing SE (SEM) from the number of assays indicated in the figure legend.
Results
A BRET sensor to detect IL-1β processing
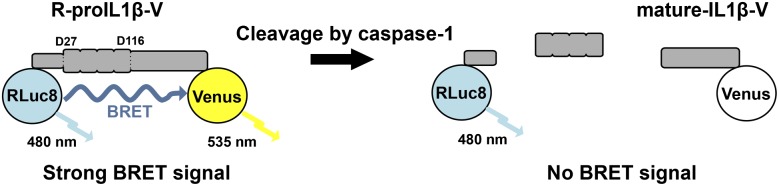
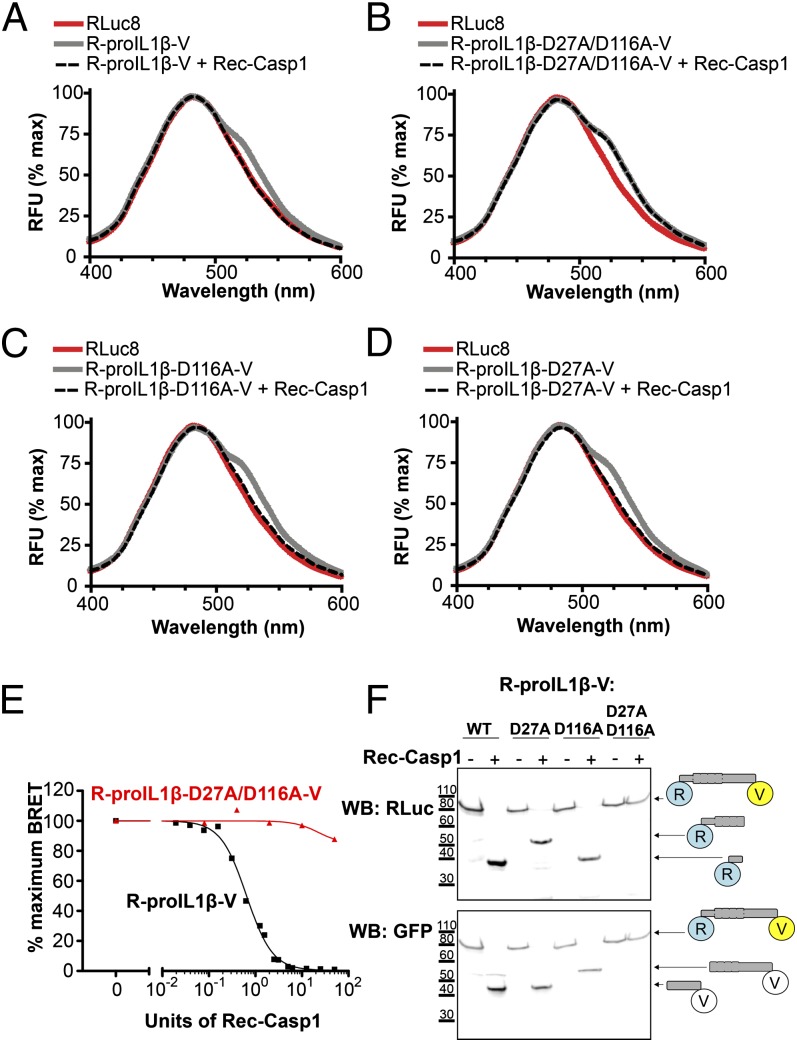
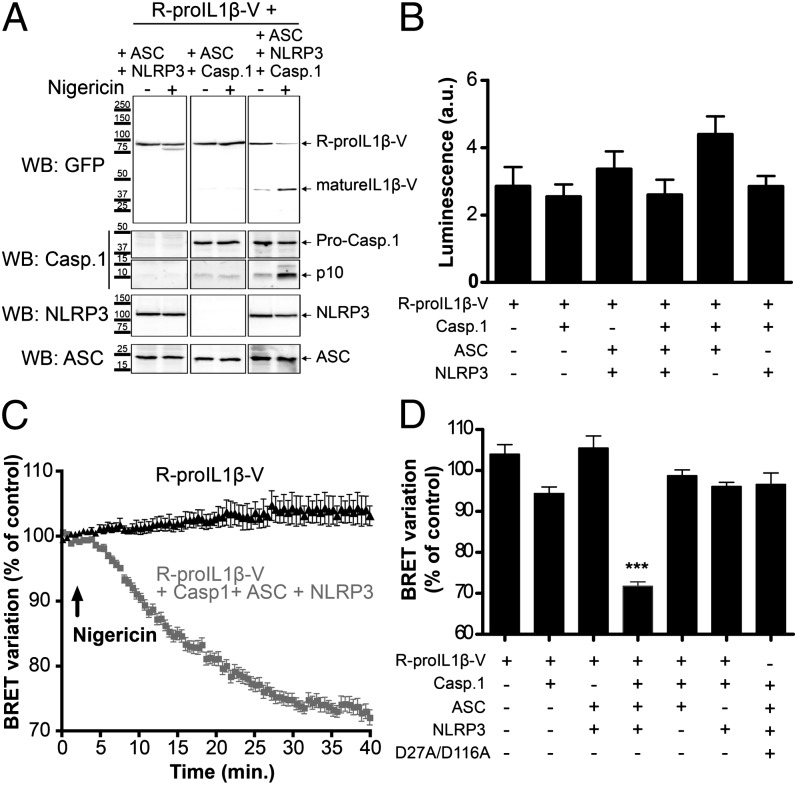
Human pro–IL-1β is a protein encoded by 269 aa that is cleaved by caspase-1 at two positions (Asp27 and Asp116) (7), resulting in the formation of three independent proteins. It is assumed that pro–IL-1β processing by caspase-1 forces the N and C terminus extremities of the protein to move apart. We therefore constructed a BRET-based sensor (termed R-pro–IL1β-V) in which the pro–IL-1β sequence has been fused to RLuc8 (the donor) on the N terminus and to Venus (the acceptor) on the C terminus (Fig. 1). When measured in vitro in the presence of the luciferase substrate coelenterazine h, the spectrum of the IL-1β sensor displayed an increase of fluorescence at 535 nm compared with the spectrum of the RLuc8 protein (Fig. 2A), indicating that the conformation of pro–IL-1β allows energy transfer between the donor and the acceptor within the sensor. Incubation of the sensor with recombinant caspase-1 results in a full cleavage of the protein, as confirmed by WB (Fig. 2F), which is accompanied with a complete loss of the energy transfer, as shown by the overlapping spectra of RLuc8 and R-proIL1β-V (Fig. 2A). The IL-1β BRET signal decreased with increasing concentrations of recombinant caspase-1 in a dose-dependent manner (Fig. 2E), with a EC50 of 0.644 U recombinant human caspase-1. To validate the specificity of caspase-1 cleavage, we constructed a pro–IL-1β biosensor in which both caspase-1 processing sites are absent. Double mutation of residues Asp27 and Asp116 to alanine completely abolished the cleavage of the biosensor by caspase-1, as corroborated by WB (Fig. 2F). These mutations also eliminated the BRET signal decrease observed after caspase-1 incubation, without changing the resting BRET signal (Fig. 2B). The corresponding single mutations on the pro–IL-1β BRET sensor did not impair the decrease of energy transfer after incubation with caspase-1 (Fig. 2C, 2D), and resulted in a partial cleavage of the sensor, as shown in WB (Fig. 2F). Altogether, these results demonstrate that R-proIL1β-V biosensor is efficiently cleaved by caspase-1, which results in decrease in BRET signals and allows direct monitoring of IL-1β processing.
FIGURE 1.
Design of a BRET sensor to detect IL-1β processing. Schematic representation of the RLuc8-IL-1β-Venus sensor (R-proIL1β-V). The pro–IL-1β protein is fused to RLuc8 and Venus at N and C terminus, respectively. Upon coelenterazine h degradation, the donor (RLuc8) emits with an emission peak at 480 nm. Close proximity (<100 Å) between the N and C terminus of pro–IL-1β allows energy transfer between the donor and the acceptor (Venus) that emits with a maximum peak at 535 nm. Cleavage by caspase-1 results in a decrease of BRET signal.
FIGURE 2.
Response of a BRET sensor to detect IL-1β processing in vitro. (A) Emission spectra of the R-proIL1β-V sensor. HEK293 cells expressing R-proIL1β-V or RLuc8 were solubilized and incubated for 1 h at 37°C with or without 50 U recombinant caspase-1 (Rec-Casp1). After adding coelenterazine h, emission spectra were measured. Note that the shoulder observed at 535 nm for the BRET sensor completely disappeared after addition of caspase-1. (B) Emission spectra of the R-proIL1β-V sensor mutated on the two caspase-1 cleavage sites on residues D27 and D166. Experiments were performed, as described in (A), on HEK293 cells expressing mutated R-proIL1β-V (R-proIL1β-D27A/D116A-V). Note that emission spectra of the double mutant superposed before and after incubation with caspase-1. (C and D) Emission spectra of the R-proIL1β-V sensor mutated on one of the two caspase-1 cleavage sites. Experiments were performed as described in (A). Note that shoulder observed at 535 nm for the BRET sensor completely disappeared after cleavage by recombinant caspase-1 for the two single mutants. Emission spectra from (A–D) are representative of three independent experiments. (E) Percentage of BRET signal measured for R-proIL1β-V and R-proIL1β-D27A/D116A-V constructs after incubation with different units of recombinant caspase-1. Dose-response curve is representative of three independent experiments. (F) WB analysis of R-proIL1β-V cleavage by Rec-Casp1. HEK293 cells expressing R-proIL1β-V, R-proIL1β-D27A-V, R-proIL1β-D116A-V, or R-proIL1β-D27A/D116A-V were treated as described in (A). R-proIL1β-V byproducts were analyzed by WB and compared with their expected size, as schematized on the right of the blot. The blot shown is representative of four independent experiments.
Real-time detection of IL-1β processing
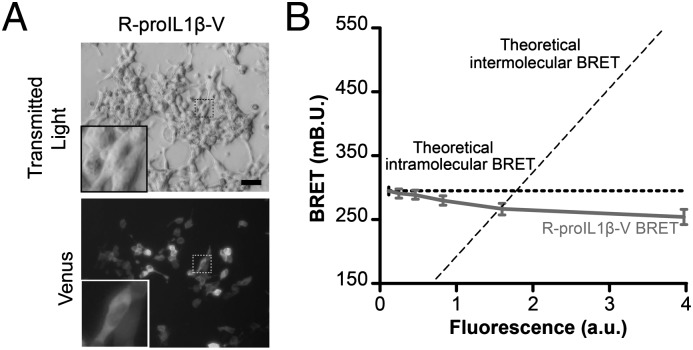
Next, we studied the pro–IL-1β BRET sensor in living cells. Transfection of HEK293 cells with R-proIL1β-V resulted in a widespread subcellular localization of the protein, as has been already described for endogenous IL-1β in macrophages (8) (Fig. 3A) and a strong BRET signal of 262.38 ± 25.68 mBU (n = 33 wells) (Fig. 4D). This BRET signal was stable regardless the amount of R-proIL1β-V expressed, indicating that the BRET signal generated by the sensor did not come from intermolecular energy transfer, but exclusively from intramolecular BRET (Fig. 3B). To reproduce a functional NLRP3 inflammasome necessary for caspase-1 activation, we coexpressed ASC, NLRP3, and caspase-1 in HEK293 cells. As shown in Fig. 5A, stimulation of transfected cells with nigericin induced activation of caspase-1, as indicated by the detection of the small subunit of active caspase-1 (p10), and resulted in the cleavage of R-proIL1β-V. When recording the kinetic of the BRET signal, we found that in the absence of caspase-1 activation, the signal was stable for up to 40 min (Fig. 5C). In contrast, in cells expressing functional NLRP3 inflammasome, the R-IL1β-V BRET signal progressively decreased after 5 min of nigericin stimulation (Fig. 5C). After 25 min, the energy transfer was reduced by ∼25% compared with the basal signal and reached a steady-state level (Fig. 5C). Neither changes in BRET signal nor R-proIL1β-V processing following nigericin stimulation were detectable in cells that lacked any single component of NLRP3 inflammasome (Fig. 5A, 5D, Supplemental Fig. 1A). Similarly, BRET signal was not sensitive to nigericin in cells transfected with construct in which caspase-1 cleavage sites were mutated from Asp to Ala (Fig. 5D, Supplemental Fig. 1B). The BRET changes observed in different transfection experiments did not correlate with different level of expression of R-proIL1β-V, because equal luminescence signal from the sensor was found in all conditions (Fig. 5B). Altogether, these results show that the R-proIL1β-V BRET sensor can be used to detect real-time caspase-1–dependent IL-1β processing in living cells.
FIGURE 3.
In living HEK293 cells, BRET from R-proIL1β-V sensor is due to intramolecular energy transfer. (A) Representative picture of HEK293 cells transfected with R-proIL1β-V. The sensor was detected after excitation of the Venus protein. Note the widespread localization of the protein in the higher magnification image, as already described for native pro–IL-1β (8). Scale bar, 30 μM. (B) R-proIL1β-V BRET signal is due to intramolecular energy transfer. BRET signals were measured on HEK293 cells expressing increasing amounts of the R-proIL1β-V sensor and plotted as a function of the amount of protein. As expected for intramolecular energy transfer, BRET does not increase in cells expressing higher amounts of sensor. The curves shown represent the mean ± SEM of n ≥ 6 wells.
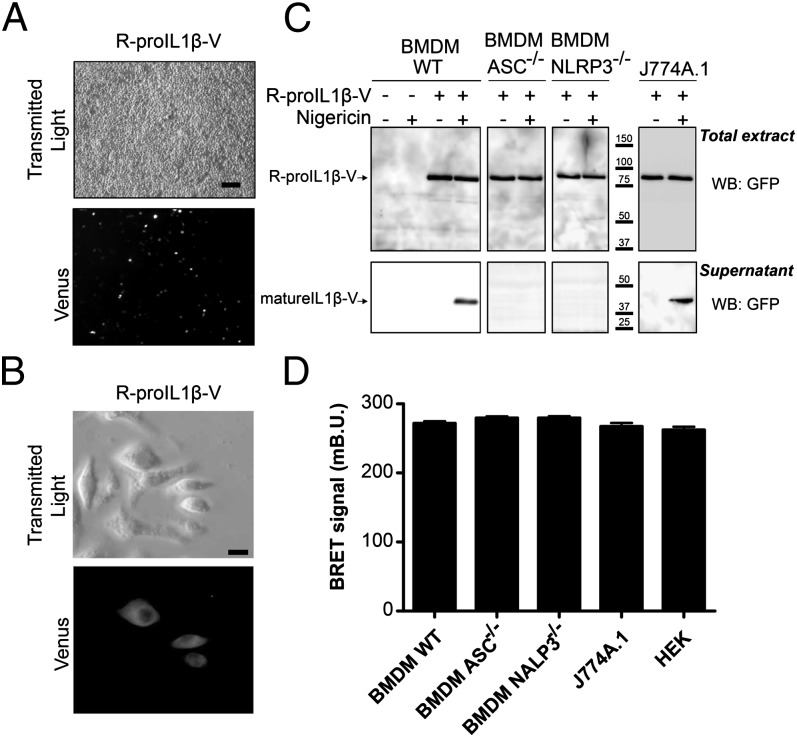
FIGURE 4.
R-proIL1β-V sensor has similar functional properties as endogenous IL-1β processing when transfected in macrophages. (A) Representative images of transfection efficiency of immortalized BMDM transfected with R-proIL1β-V. Twenty-four hours after transfection, R-proIL1β-V sensor was observed by microscopy after Venus excitation. Scale bar, 60 μM. (B) Representative image of R-proIL1β-V expression and its subcellular localization in J774A.1 macrophages. Experiments were realized as described in (A). Scale bar, 10 μM. (C) Macrophage activation induces R-proIL1β-V cleavage. Twenty-four hours after transfection with the R-proIL1β-V BRET sensor, immortalized WT BMDM or J774A.1 macrophages were primed with LPS (1 μg/ml for 4 h) and subsequently treated (or for controls not treated) with 5 μM nigericin for 30 min. Supernatant and lysates were analyzed by WB. In BMDM immortalized from ASC- or NLRP3-deficient mice, R-proIL1β-V was not cleaved after stimulation. The blot shown is representative of three independent experiments. (D) R-proIL1β-V displays similar BRET in resting macrophages. BRET signals were measured after R-proIL1β-V overexpression in HEK293, J774A.1, and immortalized BMDM from WT or ASC or NLRP3 knockout mice. Results are mean ± SEM of n ≥ 3 experiments and n ≥ 33 wells.
FIGURE 5.
R-proIL1β-V sensor allows real-time monitoring of IL-1β processing by BRET in living HEK293 cells. (A) R-proIL1β-V is efficiently cleaved by active caspase-1 in a mature form. HEK293 cells overexpressing NLRP3 inflammasome components and R-proIL1β-V were stimulated with the K+ ionophore nigericin (5 μM for 40 min). Cleavage of the R-proIL1β-V sensor to a mature form (mIL1β-V) was detected by WB using a GFP Ab. Note that IL-1β processing is only observed in cells overexpressing all the NLRP3 inflammasome. The blot shown is representative of three independent experiments. (B) R-proIL1β-V displays similar expression level after coexpression with NLRP3 inflammasome components. HEK293 cells were transfected with the BRET sensor and different combination of ASC, NLRP3, and caspase-1. Luminescence signals were measured after addition of coelenterazine h. Results are mean ± SEM of n ≥ 3 experiments and n ≥ 33 wells. (C) NLRP3 inflammasome activation results in a progressive decrease in BRET signal of the sensor. BRET signals were measured after stimulation with nigericin (5 μM, arrow) in HEK293 cells overexpressing R-proIL1β-V alone or in combination with NLRP3, ASC, and caspase-1. Results are mean ± SEM of n ≥ 3 experiments and n = 3 wells per experiment. (D) Histogram showing the BRET variation measured 30 min after nigericin stimulation in cells expressing R-proIL1β-V or R-proIL1β-D27A/D116A-V (D27A/D116A). Experiments were performed, as described above, with combination of transfection vectors for different inflammasome components. Note that decrease of the BRET signal is only observed in experimental conditions allowing activation of caspase-1 and IL-1β cleavage. Results are mean ± SEM of n ≥ 3 experiments and n = 3 wells per experiment. ***p < 0.001 compared with the other conditions.
Real-time monitoring of IL-1β processing in macrophages
We first examined the subcellular localization of the sensor in transfected macrophages. Transfection of J774A.1 or immortalized mouse bone marrow-derived macrophages (BMDM) resulted in a good transfection efficiency (Fig. 4A, Supplemental Fig. 1C) and a widespread localization of R-proIL1β-V (Fig. 4B), indicating appropriate intracellular trafficking and distribution of the protein.
The functional properties of the biosensor were subsequently analyzed in the two different macrophage cell lines treated with LPS to promote inflammasome priming (9, 10). In J774A.1 and in BMDM, nigericin stimulation induced cleavage of R-proIL1β-V sensor and release of a mature IL1β-V form into the supernatant (Fig. 4C). This processing of R-proIL1β-V was abolished in immortalized BMDM derived from ASC- or NLRP3-deficient mice (Fig. 4C). These results show that the IL-1β biosensor displays features similar to the endogenous pro–IL-1β cytokine.
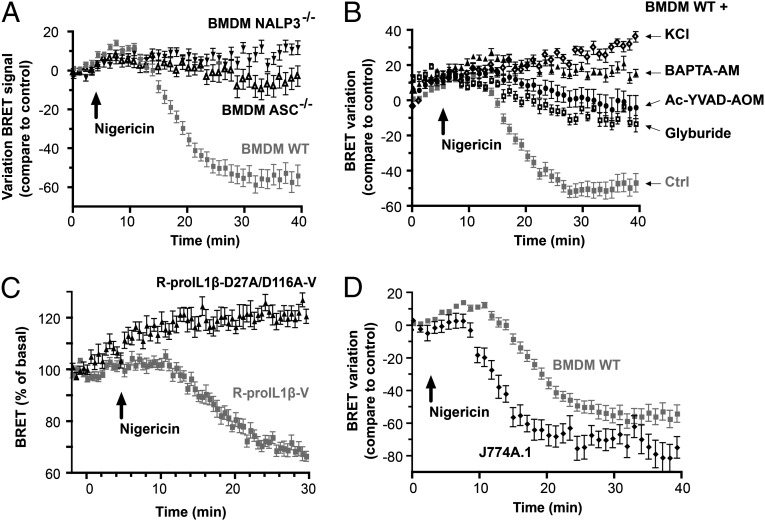
In resting conditions, the energy transfer for the R-proIL1β-V sensor measured in the different macrophage cell lines was similar to that observed in transfected HEK293 cells (Fig. 4D). After stimulation of BMDM with nigericin, the R-proIL1β-V BRET signal was stable for 10 min before a rapid decrease that reached a steady state 20 min after stimulation (Fig. 6A). In J774A.1, decrease in BRET signal occurred faster (in <6 min) before stabilizing 10 min after the stimulation (Fig. 6D). Despite the difference in the onset, the half life of the BRET between the two macrophages’ cell line was in the same range (3.88 and 3.34 min for BMDM and J774A.1, respectively). The reduction in energy transfer observed after nigericin stimulation was absent in macrophages deficient in either NLRP3 or ASC (Fig. 6A), and was abolished when NLRP3 inflammasome or caspase-1 was pharmacologically inhibited (Fig. 6B). Mutation of both caspase-1 cleavage sites on the R-proIL1β-V abolished the BRET signal decrease after nigericin treatment (Fig. 6C), suggesting that caspase-1 is the main protease processing IL-1β in macrophages. Altogether, these results show that the R-proIL1β-V sensor can be used to study caspase-1 activation and IL-1β processing kinetics with a time resolution of seconds in macrophages.
FIGURE 6.
Real-time monitoring of IL-1β processing in macrophages. (A and B) R-proIL1β-V BRET kinetic in macrophages. Macrophages were transfected and treated with LPS (1 μg/ml for 4 h). BRET was measured every 1 min in living macrophages after stimulation with 5 μM nigericin. BRET decrease was not observed in immortalized macrophages from ASC or NLRP3 knockout mice or in macrophages incubated with a caspase-1 inhibitor (Ac-YVAD-AOM, 100 μM), glyburide (150 μM), BAPTA-AM (100 μM), or an extracellular solution with high concentration of potassium (130 mM KCl). (C) BRET kinetic in BMDM. Immortalized BMDM from WT mice were transfected with R-proIL1β-V or R-proIL1β-D27A/D116A-V constructs and treated with LPS (1 μg/ml for 4 h). BRET was measured every 30 s before and after stimulation with 5 μM nigericin. (D) BRET kinetic in J774A.1 and BMDM macrophage cell lines. Macrophages were transfected with R-proIL1β-V and treated as described above. BRET was measured every 1 min. Results in (A–D) are mean ± SEM of n ≥ 3 experiments and n = 3 wells per experiment.
Monitoring real-time IL-1β processing in individual macrophages
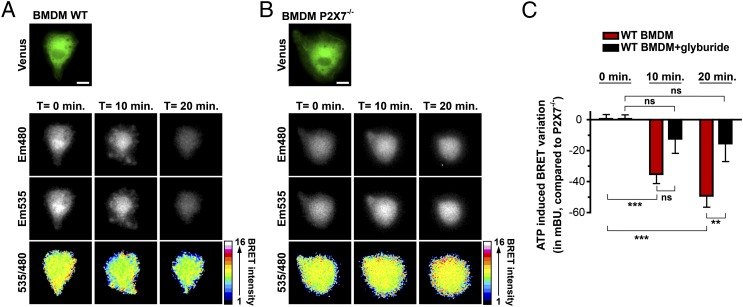
We used LPS-primed primary BMDM from WT and P2X7-deficient mice as positive and negative models of IL-1β processing induced by extracellular ATP (11, 12). After coelenterazine h addition, the emission of light at 480 nm (corresponding to the Rluc8 emission peak) and 535 nm (corresponding to the Venus emission peak) was acquired every minute from individual macrophages, before and after stimulation with 2 mM ATP (Fig. 7A, 7B). BRET signals were then represented as a 480/535 ratio image using a 16 pseudo-color scale (Fig. 7A, 7B). In WT macrophages, ATP stimulation decreased the R-proIL1β-V BRET signal, as illustrated by the color-coded changes at 10 and 20 min after stimulation (Fig. 7A). Changes were observed all over the cytoplasm and were more pronounced near the plasma membrane. In contrast, ATP failed to reduce BRET signals in macrophages deficient for the P2X7 receptor (Fig. 7B). Consistent with these observations, analysis of the 480/535 ratio after 10 and 20 min of ATP stimulation shows a reduction of 35.2 ± 6.0 and 49.2 ± 7.2 mBU, respectively, in BRET values (n ≥ 7 individual cells) in WT compared with P2X7-deficient macrophages (Fig. 7C). The BRET signal decrease for the R-proIL1β-V was inhibited by incubating WT macrophages with the NRLP3 inflammasome inhibitor glyburide (13) (Fig. 7C).
FIGURE 7.
Real-time monitoring of IL-1β processing in single primary macrophages. (A and B) BRET imaging of R-proIL1β-V transfected in primary BMDM derived from WT (A) or P2X7-deficient mice (P2X7−/−; B). Twenty-four hours after transfection with R-IL1β-V, cells were primed for 4 h with LPS (1 μg/ml). Transfected cells were identified after Venus excitation, and, subsequently, coelenterazine h (20 μM) was added. Images were acquired at 480 and 535 nm under basal condition or after ATP stimulation (2 mM) every 1 min for 20 min. A 480/535 ratio picture was obtained and presented in pseudo-colors. Scale bar, 10 μM. Note that for the WT BMDM, membrane blebbing observed after ATP stimulation as a hallmark of P2X7 activation. (C) BRET variation measured on single WT or P2X7−/−-deficient macrophages, as described above, after being incubated or not with glyburide (100 μM) and further stimulation with ATP. Results represent the BRET variation in WT macrophages compared with P2X7−/−-deficient macrophages. Results are mean ± SEM of n ≥ 3 experiments and n ≥ 7 cells for each experimental condition. **p < 0.01, ***p < 0.001. ns, Nonsignificant.
Discussion
In this work, we describe a new biosensor based on BRET to monitor, in real time, processing of IL-1β that reflects caspase-1 activity. Using macrophages as a model, we found that this new probe can specifically detect IL-1β cleavage by caspase-1 after activation of NLRP3 inflammasome. This new approach, when compared with WB or ELISA analysis, presents several advantages, as follows: 1) to our knowledge, for the first time, it allows to follow IL-1β processing and caspase-1 activity in real time; 2) it could be readily adapted to 96- or 384-well plate format suitable for automatized drug screening; and 3) by combining the BRET technique with live microscopy, IL-1β processing can be visualized in single cells.
Inflammasome and caspase-1 activation are associated with changes in the intracellular environment, such as production of reactive oxygen species (ROS) [e.g., after ATP stimulation (14)] and pH decrease following lysosome disruption [e.g., after silica crystals and aluminum salts activation (5)]. For all these reasons, we engineered a biosensor for which the properties of the donor and the acceptor are insensitive to changes in ROS and pH. RLuc8 (the donor) is a mutant of Renilla luciferase with enhanced stability (15) and with a low sensitivity to ROS (16). Venus (the acceptor) is a variant of enhanced YFP with a very low sensitivity to pH, which has been already used to detect caspase-3 and -9 activities by fluorescent resonance energy transfer (17). Therefore, RLuc8 and Venus provide the most appropriate donor–acceptor combination to monitor IL-1β processing in the specific cellular environment accompanying the activation of the inflammasome. Biophysical stability of R-proIL1β-V probe is demonstrated by experiments using the caspase-1–insensitive biosensor R-IL1β-D27A/D116A-V, which show stable BRET signals in conditions of inflammasome activation. Pro–IL-1β can be processed by different proteases (18–20), and the double-mutant biosensor demonstrates that, when stimulated with nigericin, caspase-1 is the main protein cleaving this cytokine.
After intracellular processing, mature IL-1β is rapidly released into the extracellular space. We rule out that the decrease in BRET signal observed after inflammasome stimulation could be due to secretion of the sensor because of the following: 1) the BRET signal is independent of sensor expression, because R-proIL1β-V is a ratiometric probe and BRET signal results solely from intramolecular energy transfer; 2) BRET decrease was observed in HEK293 cells that lack the machinery to release IL-1β, as confirmed by WB experiments showing that all processed IL-1β remain inside of the cell; and 3) the plate reader can record BRET signals from both intra- and extracellular compartments.
The kinetic of posttranslational processing of IL-1β in macrophages is usually analyzed using WB (21–23). However, these biochemical experiments present several limitations, including poor temporal resolution and a limited accuracy due to the analysis of different cell populations for each time point. To address these limitations, different biosensors detecting caspase-1 activity have been recently developed (24, 25). These sensors are based on the use of specific peptides or inhibitors for the cysteine proteases and can be used to detect caspase-1 activity in situ. However, these probes are still burdened with limitations, as follows: 1) they are not ratiometric; 2) due to the small peptide sequence used in these probes, they are often processed by other caspases; and 3) these probes have not been proven to detect capsase-1 activity in real time, specifically because the majority of them inhibit caspase-1 activity. The R-proIL1β-V probe reported in this work presents several improvements. First, being based on BRET detection, the R-proIL1β-V sensor is ratiometric; hence, the BRET signal provides a direct measure of actual processing of pro–IL-1β, this being the readout for the proteolytic activity of caspase-1. Second, the R-proIL1β-V probe is able to measure caspase-1 activity at the level of single cell. Finally, the R-proIL1β-V sensor has a temporal resolution in the range of the tens of second, which, due to the high signal given by RLuc8 and Venus, could potentially be increased to one frame every second using a plate reader. By using the R-proIL1β-V sensor, we found, for example, that the assembly of the NLRP3 inflammasome and activation of caspase-1 in nigericin-treated macrophages require ∼11 and 6 min in BMDM and J774A.1, respectively. These results suggest some differences in the formation of the inflammasome or in sensitivity to nigericin between the two cell lines that could probably also involve differences in signal transduction. Thus, this new approach might be helpful to characterize intracellular pathways linked to IL-1β processing and provides a better understanding of the molecular mechanisms leading to NLRP3 inflammasome activation.
Combining BRET with microscopy, we were able to monitor intracellular IL-1β processing in single macrophages. The decrease in the BRET signal observed in single cells has features similar to those described for endogenous IL-1β processing. The BRET decrease is induced by ATP stimulation in macrophages (26); it is absent in cells lacking P2X7 receptors (27); and it is blocked by glyburide, a NLRP3 inflammasome inhibitor (13). Hence, this new sensor allows monitoring IL-1β processing in transfection-resistance cells such as primary macrophages, where classical plate readers are not sensitive enough to detect low luminescence and fluorescence signals. The imaging system used in the current study has a spatial resolution (1 pixel/μm) insufficient to examine the precise subcellular compartmentalization of the sensor. However, it indicates that the processing of IL-1β occurs all over the cytoplasm and might be more pronounced in the vicinity of the plasma membrane. Monitoring BRET dynamics at single-cell level suggests that mature IL-1β is mainly generated immediately before or during the secretion of the cytokine, as already suggested by the membrane localization of the mature cytokine and caspase-1 observed by immunoelectron microscopy (28). However, we did not detect any particular decrease on pro–IL-1β BRET signal in specific speck, as ASC relocalizes in a small defined structure called pyroptosome to activate caspase-1 (29). Our results by BRET show that the pyroptosome is not the main site where caspase-1 is processing pro–IL-1β, and therefore could be the initial place where inflammasome activation occurs. Therefore, improving image acquisition parameters can be instrumental for using this biosensor to characterize controversial mechanisms leading to IL-1β release (30). This technical approach could also allow monitoring IL-1β processing in a more physiological context in which several cell types are interacting. For example, there is evidence indicating that different cell types in the brain, including neurons and neuroglia, can express IL-1β (31). However, in neuronal cultures, glial cells are necessary for neuronal survival, development, and function, and the current techniques to study IL-1β processing do not discriminate between the cell types responsible for maturation of this cytokine. Thus, monitoring biosensor cleavage by BRET using real-time microscopy can allow identification of types of the cells involved in IL-1β processing.
In conclusion, we developed a genetically encoded BRET-based sensor that can be used to detect IL-1β processing in single living cells. One of the advantages of the present approach is that the sensor shares features with the endogenous caspase-1 substrate, IL-1β, including subcellular localization and release to the supernatant, and, to our knowledge, for the first time, it allows to characterize in detail IL-1β processing kinetics in living macrophages. Using this sensor in transgenic animals may help in detection of sites where pro–IL-1β is produced and cleaved in different in vivo inflammatory models.
Supplementary Material
Acknowledgments
We thank F.A. Rassendren and R.A. North for helpful discussions and critical revisions of the manuscript. We thank Abhijit De and Andreas Loening for providing us the pcDNA3.1-Rluc8 plasmid. We thank Atsushi Miyawaki for providing the pcDNA3.1-Venus plasmid. We thank A.I. Gomez and R. Gaskell for both molecular and cellular technical assistance and L. Ulmann for animal handling.
This work was supported by grants from the Wellcome Trust and from Plan Nacional de Investigación Científica, Desarrollo e Innovación 2008-2011-Instituto Salud Carlos III-Fondo Europeo de Desarrollo Regional (EMER07/049 and PI09/0120) and Fundación Séneca (11922/PI/09).
The online version of this article contains supplemental material.
- ASC
- apoptosis-associated speck-like protein containing a caspase recruitment domain
- BMDM
- bone marrow-derived macrophage
- BRET
- bioluminescence resonance energy transfer
- mBU
- milli BRET unit
- NLRP3
- nucleotide-binding leucine-rich repeat receptor pyrin 3
- ROS
- reactive oxygen species
- WB
- Western blot
- WT
- wild type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Dinarello C. A. 1996. Biologic basis for interleukin-1 in disease. Blood 87: 2095–2147 [PubMed] [Google Scholar]
- 2.Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J., et al. 1992. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356: 768–774 [DOI] [PubMed] [Google Scholar]
- 3.Martinon F., Burns K., Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10: 417–426 [DOI] [PubMed] [Google Scholar]
- 4.Church L. D., Cook G. P., McDermott M. F. 2008. Primer: inflammasomes and interleukin 1beta in inflammatory disorders. Nat. Clin. Pract. Rheumatol. 4: 34–42 [DOI] [PubMed] [Google Scholar]
- 5.Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulon V., Audet M., Homburger V., Bockaert J., Fagni L., Bouvier M., Perroy J. 2008. Subcellular imaging of dynamic protein interactions by bioluminescence resonance energy transfer. Biophys. J. 94: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaan P. W., Knoell D. L., Helsper F., Wewers M. D. 2001. Sequential processing of human ProIL-1beta by caspase-1 and subsequent folding determined by a combined in vitro and in silico approach. Pharm. Res. 18: 1083–1090 [DOI] [PubMed] [Google Scholar]
- 8.Brough D., Rothwell N. J. 2007. Caspase-1-dependent processing of pro-interleukin-1beta is cytosolic and precedes cell death. J. Cell Sci. 120: 772–781 [DOI] [PubMed] [Google Scholar]
- 9.Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430: 213–218 [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto M., Yaginuma K., Tsutsui H., Sagara J., Guan X., Seki E., Yasuda K., Yamamoto M., Akira S., Nakanishi K., et al. 2004. ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells 9: 1055–1067 [DOI] [PubMed] [Google Scholar]
- 11.Solle M., Labasi J., Perregaux D. G., Stam E., Petrushova N., Koller B. H., Griffiths R. J., Gabel C. A. 2001. Altered cytokine production in mice lacking P2X(7) receptors. J. Biol. Chem. 276: 125–132 [DOI] [PubMed] [Google Scholar]
- 12.Labasi J. M., Petrushova N., Donovan C., McCurdy S., Lira P., Payette M. M., Brissette W., Wicks J. R., Audoly L., Gabel C. A. 2002. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J. Immunol. 168: 6436–6445 [DOI] [PubMed] [Google Scholar]
- 13.Lamkanfi M., Mueller J. L., Vitari A. C., Misaghi S., Fedorova A., Deshayes K., Lee W. P., Hoffman H. M., Dixit V. M. 2009. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J. Cell Biol. 187: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz C. M., Rinna A., Forman H. J., Ventura A. L., Persechini P. M., Ojcius D. M. 2007. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 282: 2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loening A. M., Fenn T. D., Wu A. M., Gambhir S. S. 2006. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19: 391–400 [DOI] [PubMed] [Google Scholar]
- 16.Czupryna J., Tsourkas A. 2011. Firefly luciferase and RLuc8 exhibit differential sensitivity to oxidative stress in apoptotic cells. PLoS One 6: e20073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takemoto K., Nagai T., Miyawaki A., Miura M. 2003. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schönbeck U., Mach F., Libby P. 1998. Generation of biologically active IL-1 beta by matrix metalloproteinases: a novel caspase-1-independent pathway of IL-1 beta processing. J. Immunol. 161: 3340–3346 [PubMed] [Google Scholar]
- 19.Mizutani H., Schechter N., Lazarus G., Black R. A., Kupper T. S. 1991. Rapid and specific conversion of precursor interleukin 1 beta (IL-1 beta) to an active IL-1 species by human mast cell chymase. J. Exp. Med. 174: 821–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmler M., Hertig S., MacDonald H. R., Sadoul R., Becherer J. D., Proudfoot A., Solari R., Tschopp J. 1995. Granzyme A is an interleukin 1 beta-converting enzyme. J. Exp. Med. 181: 1917–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perregaux D., Barberia J., Lanzetti A. J., Geoghegan K. F., Carty T. J., Gabel C. A. 1992. IL-1 beta maturation: evidence that mature cytokine formation can be induced specifically by nigericin. J. Immunol. 149: 1294–1303 [PubMed] [Google Scholar]
- 22.Perregaux D., Gabel C. A. 1994. Interleukin-1 beta maturation and release in response to ATP and nigericin: evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem. 269: 15195–15203 [PubMed] [Google Scholar]
- 23.Verhoef P. A., Kertesy S. B., Estacion M., Schilling W. P., Dubyak G. R. 2004. Maitotoxin induces biphasic interleukin-1beta secretion and membrane blebbing in murine macrophages. Mol. Pharmacol. 66: 909–920 [DOI] [PubMed] [Google Scholar]
- 24.Nishii W., Shoda T., Matsumoto N., Nakamura T., Kudo Y., Takahashi K. 2002. In situ visualization of caspase-1-like activity associated with promotion of hippocampal cell death. FEBS Lett. 518: 149–153 [DOI] [PubMed] [Google Scholar]
- 25.Kindermann M., Roschitzki-Voser H., Caglic D., Repnik U., Miniejew C., Mittl P. R., Kosec G., Grütter M. G., Turk B., Wendt K. U. 2010. Selective and sensitive monitoring of caspase-1 activity by a novel bioluminescent activity-based probe. Chem. Biol. 17: 999–1007 [DOI] [PubMed] [Google Scholar]
- 26.Ferrari D., Chiozzi P., Falzoni S., Dal Susino M., Melchiorri L., Baricordi O. R., Di Virgilio F. 1997. Extracellular ATP triggers IL-1 beta release by activating the purinergic P2Z receptor of human macrophages. J. Immunol. 159: 1451–1458 [PubMed] [Google Scholar]
- 27.Chessell I. P., Hatcher J. P., Bountra C., Michel A. D., Hughes J. P., Green P., Egerton J., Murfin M., Richardson J., Peck W. L., et al. 2005. Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain 114: 386–396 [DOI] [PubMed] [Google Scholar]
- 28.Singer I. I., Scott S., Chin J., Bayne E. K., Limjuco G., Weidner J., Miller D. K., Chapman K., Kostura M. J. 1995. The interleukin-1 beta-converting enzyme (ICE) is localized on the external cell surface membranes and in the cytoplasmic ground substance of human monocytes by immuno-electron microscopy. J. Exp. Med. 182: 1447–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes-Alnemri T., Wu J., Yu J. W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E. S. 2007. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 14: 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Castejon G., Brough D. 2011. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 22: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allan S. M., Tyrrell P. J., Rothwell N. J. 2005. Interleukin-1 and neuronal injury. Nat. Rev. Immunol. 5: 629–640 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.