Abstract
Virus-like particles (VLPs) are promising vaccine candidates because they represent viral antigens in the authentic conformation of the virion and are therefore readily recognized by the immune system. As VLPs do not contain genetic material they are safer than attenuated virus vaccines. In this study, herpes simplex virus type 1 (HSV-1) amplicon vectors were constructed to coexpress the rotavirus (RV) structural genes VP2, VP6, and VP7 and were used as platforms to launch the production of RV-like particles (RVLPs) in vector-infected mammalian cells. Despite the observed splicing of VP6 RNA, full-length VP6 protein and RVLPs were efficiently produced. Intramuscular injection of mice with the amplicon vectors as a two-dose regimen without adjuvants resulted in RV-specific humoral immune responses and, most importantly, immunized mice were partially protected at the mucosal level from challenge with live wild-type (wt) RV. This work provides proof of principle for the application of HSV-1 amplicon vectors that mediate the efficient production of heterologous VLPs as genetic vaccines.
Introduction
Rotaviruses (RVs) are segmented, double-stranded RNA viruses of the Reoviridae family and are the most common cause of acute viral gastroenteritis in infants around the world. Almost all children both in developing and developed countries are infected with RVs during their first years of life and even advanced levels of sanitation and hygiene appear unable to control the spread of RV infections. Death from RV infection is most prevalent in developing countries where timely health care is not always available, causing more than 600,000 deaths per year.1 Although the recently licensed human RV vaccines, which are based on orally administrated live attenuated strains, are very successful, data from clinical trials and post-licensure studies indicate that both vaccines are significantly less effective in low-income countries of Africa, Asia and Latin America.2 Additionally, potential safety issues like the risk of intussusception, inadvertent immunization of immunosuppressed individuals and generation of new pathogenic strains by reassortment of vaccine strains with wild-type (wt) human and animal RV, suggest that development of new RV vaccines is still needed. Due to the history of lower efficacy of all live oral vaccines in low-income countries, alternative approaches like parenteral vaccines should be pursued. Among these, inactivated RV particles, virus-like particles (VLPs), subunit and vector based vaccines have been tested in animal models.3,4
Mature infectious RV particles are nonenveloped, triple-layered icosahedral capsids. The innermost layer, composed of VP2 protein, encloses the 11 genomic segments of double-stranded RNA. The middle layer is composed of the major capsid protein VP6, and the outermost layer is made of the glycoprotein VP7 and spikes of VP4. During the replication cycle of RVs, discrete electron-dense structures, called viroplasms, appear in the cytoplasm of infected cells, where synthesis of double-stranded RNA segments and the initial steps of assembly of the new particles are taking place.5
The structural proteins of many viruses have the ability to assemble spontaneously into particles that are similar to the authentic viruses. Importantly, VLPs are replication-defective because they assemble without incorporating genetic material. Moreover, VLPs offer a promising approach to the production of vaccines against many diseases, because their repetitive and high-density native display of epitopes is often effective in eliciting strong immune responses.6 In addition, VLPs are generally more immunogenic than subunit or recombinant protein immunogens and are able to stimulate both the humoral and cellular arms of the immune system.7 VLPs provide the spatial structure for display of conformational epitopes and, in doing so, are most likely to mimic the native virus structure, thereby enhancing the production of neutralizing antibodies. A wide variety of VLPs have shown promising results when applied in small animal models and may offer great potential for the development of vaccines.8,9 To date, two VLP-based vaccines are licensed for application in humans, the papilloma virus vaccine and the hepatitis B virus vaccine.10
The synthesis of RV proteins using the well established baculovirus system facilitated the analysis of virus structure and, to some extent, of virus assembly. Core, double- and even triple-layered RV-like particles (RVLP) have been produced in insect cells infected with baculovirus vectors.11,12 However, the limitations of the baculovirus system include the inefficient infection of mammalian cells which prevents the direct use of baculovirus vectors for immunization; consequently, vaccination with baculovirus-derived RVLPs requires their prior purification from infected insect cells.
Herpes simplex virus type 1 (HSV-1) amplicons are versatile gene transfer vectors as they have a very large transgene capacity of up to 150 kbp and are capable of efficiently transducing a wide range of different cells, including professional antigen-presenting cells.13,14 Amplicon vectors have shown promising results in many preclinical gene- and cancer therapy applications, as well as in vaccination studies.15,16 HSV-1 amplicons have also been used for the synthesis of proteins from other viruses, e.g., amplicon vector mediated synthesis of the full set of structural proteins allowed the assembly of retrovirus VLPs17,18 and foot-and-mouth disease virus.19 In particular, the possibility of inducing local assembly of inert VLPs in the context of a quasi-infectious process holds great promises as new vaccine formulation.
The goal of this study was to construct and evaluate HSV-1 amplicon vectors encoding individual or multiple structural RV proteins from a polycistronic transgene cassette in mammalian cells. The expression of the RV genes was confirmed by western blot and immunofluorescence analysis, and the generation of RVLPs in vector-infected cells was demonstrated by electron microscopy. Mice vaccinated with these vectors were partially protected from challenge with wt RV. These results demonstrate that polycistronic HSV-1 based amplicon vectors encoding structural RV proteins are promising tools to study RV assembly in mammalian cells and may be useful as safe genetic vaccines against RV infections.
Results
Polycistronic HSV-1 amplicon vectors encode structural RV proteins
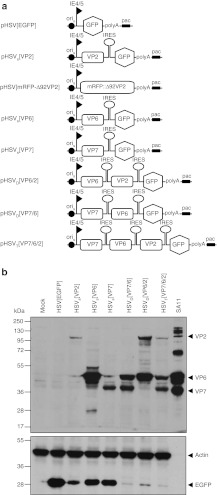
The HSV-1 amplicon vectors constructed for this study are shown in Figure 1a. The order of the individual RV genes in the polycistronic vectors was based on the composition of the mature RV particle: the major constituent of the outer layer is glycoprotein VP7, of which 780 copies are grouped as 260 trimers; the intermediate layer is formed by 780 copies of VP6 arranged as 260 trimers; the innermost layer is composed of 120 copies of VP2. We hypothesized therefore that an equimolar ratio of the structural proteins is not strictly required and placed the VP2 gene, which encodes the least abundant structural protein after the second or third internal ribosome entry site (IRES). Vector-mediated expression of RV genes VP2, VP6, and VP7 was confirmed by western analysis of total cell lysates harvested at 24 hours after infection (Figure 1b). As expected, in pHSVD and pHSVT-infected cells, expression of downstream cistrons was markedly reduced. Accordingly, the intensity of the enhanced green fluorescent protein (EGFP) band also decreased with increasing position number in the polycistron (Figure 1b, lower panel).
Figure 1.
Rotavirus (RV) genes expressed from herpes simplex virus type 1 (HSV-1) amplicon vectors. (a) Schematic representation of the HSV-1 amplicon vectors expressing or co-expressing RV genes VP2, VP6, and VP7. Polycistronic expression is facilitated by two picornavirus internal ribosome entry site (IRES) and an encephalomyocarditis virus (EMCV)-derived IRES, and controlled by the HSV-1 IE4/5 promoter. All vectors contain the enhanced green fluorescent protein (EGFP) reporter gene to support titration of vector stocks. The HSV-1 origin of DNA replication (oriS) and packaging/cleavage signal (pac) are indicated. The polyadenylation signal was from SV40. (b) Vero 2-2 cells were infected with the indicated HSV-1 amplicon vectors (MOI 1), and total cell lysates were harvested at 24 hours postinfection (hpi). Transgene expression was analyzed by western blot using a polyclonal rabbit anti-RV serum for detection of RV proteins or a monoclonal anti-GFP antibody to stain EGFP. Detection of actin was used as loading control. Purified and inactivated wt RV strain SA11 served as positive control. The positions of molecular weight markers (kDa) are indicated.
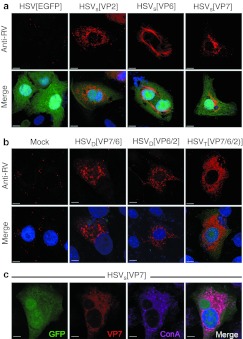
Next, we examined the synthesis and subcellular localization of the structural RV proteins in HSV-1 amplicon vector-infected cells by indirect immunofluorescence using a polyclonal rabbit anti-RV serum (Figure 2). EGFP fluorescence was used to identify vector-infected cells. VP2 and VP7 were observed as small foci with some aggregation around the nucleus (Figure 2a). As previously described,12 the major capsid protein VP6 formed fiber-like structures in the cytoplasm (Figure 2a). In cells infected with pHSVD[VP6/2], pHSVD[VP7/6], or pHSVT[VP7/6/2], the RV proteins were distributed in a punctuate pattern throughout the cytoplasm and no fiber-like structures were detected (Figure 2b). As observed by western blot (Figure 1b), the intensity of EGFP fluorescence decreased with increasing position number in the polycistron (Figure 2a,b). In wt RV-infected cells, the outer capsid glycoprotein VP7 is a membrane protein located at the endoplasmic reticulum.12 To determine whether VP7 localizes to the endoplasmic reticulum also when encoded by HSV-1 amplicon vectors, cells were infected with pHSVS[VP7] and, 24 hours later, stained with ConA (Figure 2c) and a polyclonal rabbit anti-RV serum. Confocal laser scanning microscopy revealed that VP7 expressed from HSV-1 amplicon vectors was indeed located at the endoplasmic reticulum as it colocalized with ConA staining.
Figure 2.
Intracellular distribution of herpes simplex virus type 1 (HSV-1) amplicon vector encoded rotavirus (RV) proteins. Vero 2-2 cells were infected with HSV-1 amplicon vectors (MOI 0.5). The cells were fixed 24 hpi, permeabilized, stained with a polyclonal anti-RV serum (anti-RV) and an Alexa Fluor594-conjugated secondary antibody (red), and analyzed using a confocal laser-scanning microscope. Enhanced green fluorescent protein (EGFP) fluorescence served to identify vector-infected cells (green); nuclei were stained with DAPI (blue). Bars = 5 µm. (a) Cells infected with HSVS encoding VP2, VP6, or VP7. (b) Cells infected with HSVD and HSVT encoding different combinations of VP2, VP6, and VP7. (c) HSV-1 amplicon vector encoded VP7 is localized at the endoplasmic reticulum (ER), as determined by staining with Alexa Fluor594-conjugated lectin ConA (purple).
Taken together, these results demonstrate that the three RV structural genes VP2, VP6, and VP7 are expressed from the different polycistronic vectors, however, with reduced levels when placed after IRES. The subcellular localization of the three vector-encoded proteins was comparable to that described for wt RV-encoded VP2, VP6, and VP7 proteins.20,21
HSV-1 amplicon vector-encoded VP2 induces the formation of viroplasm-like structures
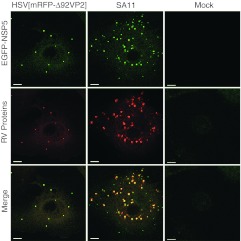
The nonstructural RV proteins NSP2 and NSP5 have been shown to be essential for viroplasm formation, and when they are co-expressed in uninfected cells, they form viroplasm-like structures (VLS).22 Formation of VLS has been demonstrated using the MA104 cell line stably expressing NSP5 fused to EGFP. Using this cell line, fluorescent NSP5-EGFP is concentrated in viroplasms following RV infection23 or upon transfection with plasmids expressing NSP5 and VP2, indicating that VP2 is also a VLS inducer.20 To investigate if VLS are formed also upon delivery of VP2 by HSV-1 amplicon vectors, MA104/NSP5-EGFP cells were infected with HSV[mRFP-Δ92VP2] which encodes a truncated VP2 fused with mRFP. After 24 hours, discrete green fluorescent dots, representing VLS, were observed in the cytoplasm of cells infected with either HSV[mRFP-Δ92VP2] or wt RV strain SA11 (Figure 3). HSV-1 amplicon vector-encoded RV VP2-mRFP (red) was recruited to VLS. Mock-infected cells showed no aggregation of NSP5-EGFP in the cytoplasm. Comparable results were obtained when cells were infected with HSVD[VP6/2] and HSVT[VP7/6/2] (data not shown). Taken together, the observed formation of discrete green dots in the cytoplasm indicated that HSV-1 amplicon vectors encoding the core protein VP2 are able to induce VLS in cells that express RV NSP5.
Figure 3.
Herpes simplex virus type 1 (HSV-1) amplicon vector encoded rotavirus (RV) proteins induce the formation of viroplasm-like structures. MA104/NSP5-EGFP cells, which stably express RV NSP5 fused to enhanced green fluorescent protein (EGFP), were mock-infected or infected with either HSV[mRFPΔ92VP2] (MOI 0.5) or RV SA11. Cells infected with RV SA11 were fixed 12 hpi, permeabilized, and stained with a polyclonal anti-RV serum (anti-RV) and an Alexa Fluor594-conjugated secondary antibody (red). Cells infected with HSV[mRFPΔ92VP2] were fixed 24 hpi, the red fluorescence corresponds to mRFP fused to VP2. EGFP fluorescence indicates VLS containing NSP5-EGFP (green). Samples were analyzed using a confocal laser-scanning microscope. Bars = 10 µm.
HSV-1 amplicon vectors support the generation of RVLPs in vector-infected cells
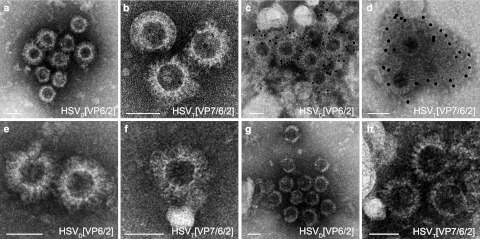
In order to examine the assembly of the vector encoded structural proteins into RVLPs, total cell lysates were harvested at 48 hours after infection and the purified and concentrated RVLPs were prepared for electron microscopy. Assembled empty RV particles were observed in lysates of HSVD[VP6/2] or HSVT[VP7/6/2] infected Vero 2-2 cells (Figure 4a,b) and confirmed by immunogold staining using a polyclonal RV-specific antiserum and a secondary antibody coupled to 12-nm gold particles (Figure 4c,d). To assess whether vector encoded structural RV proteins can assemble also in human cells, HeLa cells (Figure 4e,f) and HEK293 cells (Figure 4g,h) were infected with HSVD[VP6/2] or HSVT[VP7/6/2]. In both human cell lines, RVLPs were readily detected. Western analysis confirmed that the observed RVLPs consisted of structural RV proteins (data not shown). Taken together, these data demonstrate that the HSV-1 amplicon vectored delivery of structural RV genes supports the assembly of RVLPs in mammalian cells.
Figure 4.
Electron micrographs of herpes simplex virus type 1 (HSV-1) amplicon vector encoded rotavirus-like particles (RVLPs). Purified RVLPs from (a–d) Vero 2-2 cells, (e,f) HeLa cells, and (g,h) Hek293 cells infected with HSV-1 amplicon vectors (MOI 1). Two days postinfection, RVLPs were purified over a sucrose cushion and the concentrated particles were analyzed by electron microscopy. (a,b) Negative staining of RVLPs from Vero 2-2 cells infected with (a) HSVD[VP6/2] and (b) HSVT[VP76/2]. (c,d) Immunogold staining using a polyclonal anti-RV serum and a secondary antibody coupled to 12-nm gold particles. (c) Double-layered RVLPs from cells infected with HSVD[VP6/2]. (d) RVLPs from cells infected with HSVT[VP7/6/2]. (e,f) Negative staining of RVLPs from HeLa cells infected with (e) HSVD[VP6/2] and (f) HSVT[VP76/2]. (g,h) Negative staining of RVLPs from Hek293 cells infected with (g) HSVD[VP6/2] and (h) HSVT[VP76/2]. Bars = 50 nm.
Splicing of RV genes expressed from HSV-1 amplicon vectors
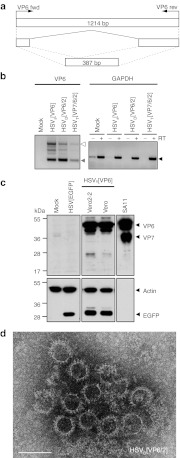
As HSV-1-derived vectors deliver transgenes into the host cell nucleus whereas RV replicates in the cytoplasm, and because several splicing donor and acceptor sites are predicted on the VP6 sequence (Figure 5a), we examined the VP6 RNA from vector-infected cells for possible splicing events. For this, total RNA was extracted from vector-infected or mock-infected cells at 24 hours postinfection and reverse transcribed. Subsequent PCR was performed to amplify the complete open-reading frame of VP6 with a size of ~1,214 bp (Figure 5b). RT-PCR of GAPDH in presence or absence of reverse transcriptase served as control (Figure 5b, right panel). Interestingly, RT-PCR with RNA isolated from cDNA prepared from pHSVS[VP6], pHSVD[VP6/2], or pHSVT[VP7/6/2]-infected cells yielded a prominent band of ~400 bp in addition to the full-length band (Figure 5b, left panel). Sequence analysis of the 400 bp band revealed a truncated VP6 gene, in which the middle part of the VP6 ORF was deleted, as predicted from the splice donor and acceptor sites. The start and stop codons of the VP6 ORF were still in frame and can potentially give rise to a truncated protein with a calculated molecular weight of ~14 kDa. Besides VP6, there are splicing sites predicted also for VP2, but no splicing was detected for either VP2 or VP7 by RT-PCR of RNA isolated from vector-infected cells (data not shown).
Figure 5.
The herpes simplex virus type 1 (HSV-1) amplicon vector encoded VP6 RNA is spliced. (a) Schematic representation of the splicing of the VP6 RNA. Primers used for detection of the VP6 open-reading frame (ORF) are indicated as VP6 fwd and VP6 rev (see Table 1). The amplified full-length VP6 ORF (1,214 bp) and the truncated form of VP6 (387 bp) are shown. (b) Reverse transcription PCR of total RNA isolated from infected cells (see Table 1 for primer sequences). Left panel: Amplification of VP6 sequences using the primers VP6 fwd and VP6 rev. The detected bands correspond to the sizes predicted for full length (white arrowhead) and truncated (gray arrowhead) VP6 sequences. Right panel: Amplification of GAPDH sequences was used as control (black arrowhead). Reactions were performed in presence (+) or absence (–) of reverse transcriptase (RT). Negative control: mock-infected cells. (c) Immunoblot of HSVS[VP6] infected Vero and Vero 2-2 cells. The polyclonal anti-RV serum was used for detection of rotavirus (RV) proteins, a monoclonal anti-GFP antibody to stain enhanced green fluorescent protein (EGFP), and actin staining as loading control. Purified and inactivated wt RV strain SA11 served as positive control. (d) Electron micrograph (negative staining) of double-layered RVLPs produced in Vero cells infected with HSVD[VP6/2]. RVLPs purified from total cell lysates were concentrated over a sucrose cushion. Bar = 100 nm.
The Vero 2-2 cells used for the experiments described above express the HSV-1 ICP27 gene. ICP27 is an essential immediate-early (IE) protein, which, besides other functions, can inhibit splicing of both viral and cellular RNA.24 To find out whether HSV-1 amplicon vector-mediated production of RV proteins and RVLPs is possible only in cells in which splicing is inhibited, we next analyzed protein synthesis and RVLP production in vector infected parental Vero cells. No differences concerning RV protein synthesis (Figure 5c) or RVLP structure (Figure 5d) were observed between Vero and Vero 2-2 cells infected with HSVS[VP6] (Figure 5c) or HSVD[VP6/2] (Figure 5d). The intensity of the band corresponding to full-length VP6 protein was comparable between Vero and Vero 2-2 cells, indicating that even if splicing of VP6 RNA occurred, there was no major decrease in the production of the full-length protein. Moreover, the use of splicing inhibitors did not result in increased accumulation of vector encoded VP6 protein (data not shown).
Immunization of mice with HSVT[VP7/6/2] induced partial protection from virus challenge
After demonstrating the assembly of the vector-encoded RV proteins into RVLPs, the potential usefulness of these HSV-1 amplicon vectors for vaccination against RV infection was evaluated. For this, BALB/c mice were inoculated twice intramuscularly (i.m.) at days 0 and 21 (Figure 6a) with 5 × 105 or 1 × 106 transducing units (TU) of amplicon vectors encoding (i) three RV proteins, HSVT[VP7/6/2], (ii) a single RV protein, HSVS[VP6], (iii) a mixture of HSVS[VP7], HSVS[VP6], and HSVS[VP2] (HSVSComposite, 1:1:1), (iv) the empty vector HSV[EGFP], or (v) phosphate-buffered saline (PBS). Another group of mice was vaccinated with 1.5 × 105 fluorescent focus units of cell culture-adapted live RV strain EC. As RV antigen and RVLPs are synthesized during the production of HSV-1 amplicon vector stocks in Vero 2-2 cells and are expected to be copurified with the vector particles, we analyzed the presence of preformed antigen and RVLPs in the vector stocks by western blot and electron microscopy. Both RV antigen and RVLPs (106 RVLPs/ml of vector stock) were indeed observed (data not shown). Therefore, in order to assess the contribution of the preformed antigen to the immune response, mice were inoculated twice with 1 × 106 TU of HSVT[VP7/6/2] inactivated with binary ethyleneimine (BEI). We confirmed that BEI-treatment eliminated transgene expression but did not affect the levels of preformed RV antigen present in the vector stock (data not shown). RV-specific antibodies in serum and feces collected from mice at days 0 (preimmune), 21 (before second dose), and 42 (before challenge) were determined by enzyme-linked immunosorbent assay (ELISA) and western analysis. Amplicon vector-induced RV-specific fecal immunoglobulin A (IgA) or serum IgG antibody levels were low or undetectable when tested by ELISA after the first dose (data not shown). However, after the second dose, VP6-specific serum IgG antibodies were detected by ELISA (data not shown) and western blot (Figure 6b) in all serum samples, except for serum of mice inoculated with either empty vector or BEI-inactivated HSVT[VP7/6/2]. As BEI-inactivated HSVT[VP7/6/2] did not express any transgenes (data not shown), we concluded that the observed antibody responses were due to de novo synthesized antigen encoded by the vectors while preformed RV antigen present in the vector stock did not induce a detectable antibody response. VP2-specific IgG was also detected in sera of some animals inoculated with RV strain EC or HSVT[VP7/6/2].
Figure 6.
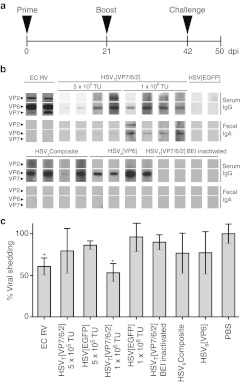
Immunization of mice with HSVT[VP7/6/2] confers partial protection from challenge with wt rotavirus (RV). (a) Mice (4–6 per group) were immunized (i.m.) at day 0 and boosted at day 21. Stool and serum samples were collected on days 0, 21, and 42 after the first immunization. Three weeks after the second immunization, mice were orally challenged with 104 SD50 of wt RV strain EC, and stool samples were collected every day for eight days post challenge. (b) Western analysis of RV specific antibodies in sera and feces of immunized mice. RV (strain RRV) proteins were separated by SDS-PAGE and blotted onto nitrocellulose membranes. Membranes individually probed with sera diluted 1/100 or fecal samples 1/10 from different groups of mice immunized with amplicon vectors are shown. The figure shows RV-specific IgG in sera (upper panel) and RV-specific IgA in feces (lower panel). The results shown represent one of two independent experiments. Mouse serum against RV strain EC was used as positive control to identify VP2, VP6, and VP7. (c) Fecal samples were collected for 8 days after the challenge and the presence of RV antigen was measured by ELISA. Viral antigen shedding curves of each animal were plotted, and the area under the curve (AUC) for each animal was calculated and compared to that of the control group (PBS, 100%). Mean AUC per group is shown. Error bars indicate the standard deviation of the mean AUC per group. *P < 0.05, Student's t-test.
RV-specific fecal or serum IgA antibody levels were not detectable even after the second dose when tested by ELISA (data not shown). However, western analysis of feces revealed RV VP6-specific IgA antibodies in all mice and RV VP2-specific IgA antibodies in one of four mice inoculated twice with 1 × 106 TU of HSVT[VP7/6/2] (Figure 6b). Importantly, no IgA antibody was detected in mice inoculated with all other vectors or with cell culture-adapted live RV strain EC. This result shows that de novo synthesized antigen is required but not sufficient for the observed antibody response, because no fecal IgA was detected in mice immunized with HSVSComposite which is a 1:1:1 mixture of vectors expressing VP7, VP6, or VP2 genes individually (Figure 6b).
Next, we addressed the question whether vector immunized mice were protected from oral challenge with wt RV. In the adult mouse model, infection with RV does not induce disease. Protection is therefore defined as the absence of detectable fecal viral antigen following challenge, and partial protection is defined as reduced quantities of fecal viral antigen compared to that shed by PBS-inoculated control mice.25,26 Accordingly, protection from RV infection upon challenge was evaluated by comparing antigen shedding in mice vaccinated i.m. with two doses of amplicon vectors encoding individual or combined RV proteins or HSV[EGFP] and mock vaccinated mice (PBS animals were orally challenged three weeks after the second immunization with live wt RV strain EC (104 SD50) (see Figure 6a). Of the mice immunized with HSV-1 amplicon vectors, only the HSVT[VP7/6/2] group inoculated with 1 × 106 TU were partially protected from RV infection. This group showed a significant decrease (P < 0.05) in the shedding of RV (46.89% average reduction, range 40.09–63.21%) in feces after challenge compared to control mice (Figure 6c). Partial protection in the HSVT[VP7/6/2] group was comparable to that induced in mice immunized with cell culture-adapted live RV strain EC (39.39% average reduction, range 29.41–53.07%). Mice immunized with the lower dose, 5 × 105 TU of HSVT[VP7/6/2] showed a nonsignificant level of reduction of RV antigen shedding in feces (Figure 6c; 18.7% average reduction, range 0–55.84%). Taken together, immunization of mice with HSVT[VP7/6/2] resulted in a dose-dependent partial protection of vaccinated mice that correlated with RV specific IgA detection and was as potent as that achieved with cell culture-adapted live RV strain EC. Moreover, the data shows that de novo synthesized antigen is required but not sufficient also for conferring partial protection, as no decrease in RV antigen shedding was observed in animals immunized with either BEI-inactivated HSVT[VP7/6/2] or HSVSComposite.
Discussion
Here we demonstrate the efficient expression of up to four different transgenes from a single polycistronic HSV-1 amplicon vector. More precisely, amplicon vectors that encode multiple structural RV genes supported the production of RVLPs in vector-infected cells and, when injected into mice, induced RV-specific immune responses and partial protection from challenge with wt RV. Interestingly, despite the facts that (i) RV replication normally takes place in so-called viroplasms in the cytoplasm whereas HSV-1 amplicon vectors deliver the RV genes into the nucleus and (ii) several splicing sites are present in the RV genes, full-length RV proteins were nevertheless efficiently synthesized and splicing inhibitors did not result in increased production of vector-encoded RV proteins or RVLPs. Although other vector systems, including recombinant vaccinia virus vectors27 and Semliki Forest virus vectors,28 have previously been used to (co-)express the RV VP2 and VP6 genes, the vaccine potential of those vectors has not been investigated.
The use of VLPs as vaccines is very promising because of their high safety profile compared to live attenuated virus vaccines. Despite the lack of genetic material, VLPs retain the high immunogenicity of the parental virus particle. The use of a mammalian delivery system, such as HSV-1 amplicon vectors, to launch the production of heterologous VLPs, provides the additional advantages that time consuming and expensive purification of VLPs is not required. Moreover, as opposed to injection of purified VLPs, the delivery of VLP encoding genes into host cells results in the intracellular production of antigens. This may induce more potent immune responses, as it mimics the situation in which cells support replication of the live pathogen. Although the HSV-1 amplicon vector does not express any HSV-1 genes, it can further boost the immune responses by providing adjuvants function via the structural HSV-1 proteins that make up the vector particle.8
Indeed, we show that when delivered i.m. to adult mice, the HSV-1 amplicon vaccine vectors expressing the structural RV genes VP7, VP6, and VP2 from a single transcription unit, a RV-specific antibody response was induced; both systemic IgG and fecal IgA were detected in all immunized animals. Most importantly, antigen shedding after live RV challenge was significantly reduced when compared to control mice. VP7-specific antibodies were not detected in any of the vaccinated mice and VP2-specific antibodies only in some of the animals. This may reflect a lower immunogenicity of VP2 and VP7 compared to VP6, which has indeed been shown to be the most immunogenic RV protein.29
Importantly, mice vaccinated with HSVT[7/6/2] were protected on a level comparable to that achieved with a vaccine dose of cell culture-adapted live RV strain EC, although HSVT[7/6/2] delivers an incomplete set of antigens and indeed induced lower serum levels of RV specific antibodies than cell culture-adapted live RV strain EC, or HSVS[VP6], or a mixture of vectors expressing individual RV genes (HSVSComposite). Immune responses were not due to preformed antigen present in vector stocks because immunization with BEI-inactivated HSVT[7/6/2] stocks, which did not express any transgenes but contained the same levels of preformed RV antigen as noninactivated vector stocks, did not induce detectable antibody responses nor partial protection (Figure 6c). Antigen de novo synthesis appears to be important but not sufficient for conferring partial protection from challenge with live RV strain EC, because vectors expressing an individual RV antigen (VP6) induced VP6-specific antibody responses but not protection (Figure b,c). Moreover, HSVSComposite, which is a 1:1:1 mixture of vectors expressing VP7, VP6, and VP2 genes individually also induced RV-specific antibody responses but no protection. These data indicate that partial protection induced by HSVT[7/6/2] was due to de novo cosynthesis and interaction of all three RV antigens in the same cell, a prerequisite that is much less likely when the transgenes are expressed from three different vectors, in particular because expression of the different transgenes in different cells would likely not support the assembly of VP7/6/2 RVLPs in vivo.
Examination of injected muscle tissue by electron microscopy and western analysis did not reveal any RVLPs or RV antigen (data not shown). However, we neither detected any virus particles at 24 hours after i.m. injection of as much as 109 inactivated RV particles, suggesting that virus particles are rapidly degraded or transported out of the muscle tissue. Previous work performed with amplicon vectors expressing HIV Gag protein reported that preformed antigen indeed rapidly decayed in vivo.30 Another possibility is that the vectors can efficiently infect antigen-presenting cells, including dendritic cells, which are migrating out of the muscle tissue, thereby making detection of de novo synthesized RVLPs and RV antigen in muscle tissue difficult. Support for this hypothesis comes from the fact that HSV-1 amplicons have indeed been shown to efficiently infect professional antigen-presenting cells,13 whereas they do not efficiently transduce muscle cells of adult and even young adult mice.31 The inefficiency of transducing mouse muscle has been attributed to the basal lamina, which wraps the surface of muscle fibers, and is believed to act as a physical barrier for large viruses, such as HSV-1 and adenoviruses, and prevents them from contacting the cell membrane32. The choice of the i.m. route of injection was based on a pilot experiment in which i.m. injection resulted in the highest RV-specific antibody response (data not shown) and not because muscle cells are efficiently transduced by the vectors. Although, we did not detect RVLPs in vivo, there is strong evidence that the immune responses observed in this work were due to the de novo synthesis of multiple RV transgenes in a single cell, a situation that at least in cell culture was demonstrated to support the assembly of RVLPs.
As discussed above, HSV-1 amplicons are versatile gene transfer vectors which have a very large transgene capacity of up to 150 kbp and are capable of efficiently transducing a wide range of different cells, including antigen-presenting cells. It has been suggested that antigen-presenting B cells for example can migrate from the periphery to gut-associated lymphoid tissue.33 This might contribute to the generation of a mucosal IgA response in feces after i.m. administration of antigen.
High levels of fecal VP6 specific IgA have been the main correlate for protection in the mouse model of RV infection, and it has also been suggested that VP6 specific IgA antibodies can neutralize RV intracellularly.34 Although RV-specific IgA were detected and did correlate with partial protection in our study, other mechanisms may have contributed as well. At least in the mouse, different immune effectors appear to be responsible for protection, depending on whether immunity is elicited by (i) natural infection, (ii) vaccination with attenuated virus strains, or (iii) immunization with inactivated vaccines. The protective mechanisms are also different depending on the inoculation route, the type of adjuvant used and even on the source and purity of the preparation.35,36 While protection induced by subunit vaccines is dependent exclusively on CD4+ T cells,37 the same does not apply when attenuated RV is used for vaccination. In this case, protection does not solely rely on either IgA38 or T cells.39
In previously tested models using RV particles,40 VLPs41 or DNA42 for immunization, the parenteral route proved to be effective in protecting against RV infection in mice. Of note, in all of the preceding examples, an adjuvant was absolutely required to induce protection, while in the present study, amplicon vectors were inoculated without additives. In future work, it will be important to characterize the inductive site and immune effectors of the observed protective response upon different routes of immunization, including parenteral and mucosal. Specific local humoral and T cell responses through cytokines, cytotoxicity and the duration of protection (memory) need to be investigated. Moreover, structural proteins from different RV serotypes may be combined in the polycistronic transgene cassette. For example, RVLPs produced with VP2 and VP6 from strain RRV may serve as a scaffold for chimeric RVLPs containing the outer structural protein VP7 and VP4 from other RV strains. Moreover, HSV-1 amplicon vector induced RVLPs may serve also as carriers of foreign epitopes from bacterial or viral pathogens, to enhance the RV specific immune response or induce immune responses against multiple pathogens. In addition to structural proteins, HSV-1 amplicon vectors may also encode nonstructural RV proteins, such as the viral enterotoxin NSP4, which has been shown to provide adjuvant activity in mice.43
In conclusion, the present work suggests that HSV-1 amplicon vectors are highly suitable to launch the production of heterologous VLPs and can be used as platforms for the generation of genetic vaccines against both DNA and RNA viruses.
Materials and Methods
Cells and viruses. MA104 (embryonic African green monkey kidney), HEK293 (human embryonic kidney), HeLa, Vero (African green monkey kidney epithelium, ATCC) and Vero 2-244 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml of penicillin G, 100 µg/ml of streptomycin, 0.25 µg/ml of amphotericin B, and, for VERO 2-2 cells, 500 µg/ml of G418 (Invitrogen, Carlsbad, CA). The MA104/NSP5-EGFP cell line (embryonic African green monkey kidney cells stably expressing the fusion protein NSP5-EGFP) was obtained from Dr Catherine Eichwald and cultured in Dulbecco's modified Eagle's medium supplemented with 800 µg/ml of G418.23
The murine wt RV strain EC was obtained from Dr Harry Greenberg (Department of Medicine and Microbiology and Immunology, Stanford University School of Medicine, Stanford, CA). The production and titration of wt RV EC used to challenge mice after vaccination was described previously.34 The simian RV strain SA11 was obtained from Dr Catherine Eichwald (University of Zurich, Zurich, Switzerland); and strain RRV was obtained from Dr Viviana Parreño (Instituto de Virología, CICVyA, INTA Castelar, Argentina); they were propagated in MA104 cells and inactivated as previously described.45
Construction of HSV-1 amplicon vectors. All HSV-1 amplicon vectors used in this study encode EGFP to facilitate titration of the vector stocks and identification of vector-infected cells. In addition, the vectors express individual or multiple RV genes and were constructed as follows: (i) pHSVD: The poliovirus IRES element was amplified with primers containing ClaI and XbaI restriction sites at the 5′ ends, respectively (for primer sequences, see Table 1), using plasmid pQuattro (kindly provided by M. Fussenegger, Institute of Biotechnology, Swiss Federal Institute of Technology, ETH Zurich, Switzerland) as template. The resulting PCR product (IRES2) was inserted between the ClaI and XbaI sites of pHSVS, which contains a transcription unit consisting of the HSV-1 IE 4/5 promoter, an IRES, and the SV40 polyadenylation signal19 (ii) pHSVT: A third IRES sequence, derived from encephalomyocarditis virus was amplified with primers containing SpeI and Ecl136II restriction sites at the 5′ ends, respectively, using plasmid pQuattro as the template. The resulting PCR product was inserted between the SpeI and Ecl136II restriction sites of pHSVD. The resulting HSV-1 amplicon plasmid pHSVT contains three IRES signals between the HSV-1 IE 4/5 promoter and the SV40 polyadenylation signal.
Table 1. Vector construction details.

The HSV-1 amplicon vectors encoding the structural RV proteins of strain RRV have been generated as follows (see also Table 1 and Figure 1a): (i) pHSVS[VP2]: The VP2 gene was amplified with primers containing SalI restriction sites at the 5′ ends using plasmid pVP2_RRV_Lopez (kindly provided by S. Lopez, Universidad Nacional Autonoma de Mexico (UNAM), Morelos, Mexico) as the template. The resulting PCR product was inserted into the SalI restriction site of pHSVS. (ii) pHSVS[VP6]: The VP6 gene was amplified with primers containing SalI restriction sites at the 5′ ends, using plasmid pENTR-VP6_RRV as the template. The resulting PCR product was inserted into the SalI restriction site of pHSVS. (iii) pHSVS[VP7]: The VP7 gene was amplified with primers containing AsuII restriction sites at the 5′ ends, using plasmid pVP7_RRV_Lopez (S. Lopez, Morelos, Mexico) as template. The resulting PCR product was inserted into the AsuII restriction site of pHSVS. (iv) pHSVD[VP6/2]: The VP6 PCR product described under (ii) was also subcloned into the SalI site of pHSVD resulting in pHSVD[VP6]. VP2 was amplified from plasmid pVP2_RRV_Lopez with primers containing AscI and ClaI restriction sites at the 5′ ends, respectively, and inserted between the AscI and ClaI sites of pHSVD[VP6]. (v) pHSVD[VP7/6]: The VP7 PCR product described under (iii) was inserted also into the AsuII site of pHSVD resulting in pHSVD[VP7]. VP6 was amplified from plasmid pENTR-VP6_RRV with primers containing AscI and ClaI restriction sites at the 5′ ends, respectively, and inserted between the AscI and ClaI sites of pHSVD[VP7]. (vi) pHSVT[VP7/6/2]: The VP7 PCR product described under (iii) was inserted also into the AsuII site of pHSVT resulting in pHSVT[VP7]. The VP6 PCR product described under (v) was also inserted between the AscI and ClaI sites of pHSVT[VP7] resulting in pHSVT[VP7/6]. VP2 was amplified from plasmid pVP2_RRV_Lopez with primers containing XhoI restriction sites at the 5′ ends, and inserted into the XhoI site of pHSVT[VP7/6]. (vii) pHSV[mRFP-Δ92VP2]: Δ92VP246 was amplified with primers containing SalI restriction sites (fwd primer starting at aa 93 of VP2 and containing an additional NheI site plus a linker sequence for the fusion with mRFP, see Table 1) at the 5′ ends using pHSVS[VP2] as template and the PCR product was inserted into the SalI restriction site of pHSVS resulting in pHSVS[Δ92VP2]. The mRFP was amplified with primers containing NheI restriction sites at the 5′ ends using pcDNAmRFPN1 (U.F. Greber, University of Zurich, Zurich, Switzerland) as template and the PCR product was inserted into the NheI restriction site of pHSVS[Δ92VP2] resulting in pHSVS[mRFP-Δ92VP2]. In the final step, EGFP and the IRES were excised from the plasmid with BbrPI and EcoRI, blunt-ended with T4 DNA polymerase (New England Biolabs, Ipswich, MA) and religated resulting in pHSV[mRFP-Δ92VP2].
Production of HSV-1 amplicon vector stocks. Helper virus-free HSV-1 amplicon vector stocks were prepared as previously described.47 The HSV-1 genome was provided in trans by a bacterial artificial chromosome containing the HSV-1 genome with deletions in the DNA cleavage/packaging signals and the essential ICP27 gene (fHSVΔpacΔICP27). Briefly, Vero 2-2 cells were cotransfected with amplicon plasmid DNA, the fHSVΔpacΔICP27 bacterial artificial chromosome DNA, and plasmid pEBHICP27 (which provides the HSV-1 ICP27 gene in trans), using Lipofectamine and Plus Reagent (Invitrogen). After 72 hours, cells were scraped into the medium, sonicated, and the cell debris was removed by centrifugation. For titration, Vero 2-2 cells were infected with the amplicon vectors and, after 24 hours, green fluorescent cells were counted using an inverted fluorescence microscope (Axio Observer inverted microscope; Zeiss AG, Feldbach, Switzerland). The titers were determined as TU/ml and ranged between 2–8 × 106 TU/ml.
For virus vector inactivation, aliquots of HSVT[VP7/6/2] amplicon vectors were treated with BEI at a final concentration of 1.4 mmol/l during 24 hours at room temperature. BEI was prepared as described.48 Inactivation was confirmed by the lack of detectable gene expression after transduction of Vero 2-2 cells and subsequent western analysis.
Western analysis of vector encoded structural RV proteins. Cells were seeded at a concentration of 1 × 105 cells per well in 24-well tissue culture plates, and amplicon vectors were added at a multiplicity of infection (MOI) of 0.5 or 1 TU per cell. Total cell lysates were harvested at different time points after infection and separated on 10% SDS-polyacrylamide gels (PAGE). The fractionated proteins were transferred to nitrocellulose membranes, probed with the primary antibodies, and stained using anti-mouse (Sigma-Aldrich, St Louis, MO) or anti-rabbit (Southern Biotech, Birmingham, IL) IgG antibodies conjugated with horseradish peroxidase, followed by detection with Amersham ECL western Blotting Analysis System (GE Healthcare, Zurich, Switzerland). Rabbit anti-RV polyclonal serum raised against whole virus (strain RF, provided by D. Poncet, CNRS/INRA, Gif-sur-Yvette, France), mouse anti-GFP monoclonal antibody (Jl-8, Santa Cruz Biotechnology, Santa Cruz, CA) and mouse anti-actin monoclonal antibody (Sigma-Aldrich, St Louis, MO) were used as primary antibodies. For stripping, membranes were incubated for 15 minutes with Stripping Buffer (Thermo Scientific, Rockford, IL) and washed three times with PBS.
Immunofluorescence assays. Vero 2-2 cells or MA104/NSP5-EGFP cells were grown on 12-mm coverslips (0.17-mm thick) in 24-well plates and either mock infected or infected with amplicon vectors at a MOI of 0.5 TU/cell. The cells were fixed with 3.7% formaldehyde in PBS and treated with 0.1 mol/l glycine in PBS. After permeabilization with PBS containing 0.2% Triton X-100 (PBS-T), the cells were treated with PBS supplemented with 3% bovine serum albumin (PBS-BSA). Cells were incubated with rabbit anti-RV serum (strain RF, provided by D. Poncet, Gif-sur-Yvette, France) diluted in PBS-BSA (1:400) and then washed three times with PBS. As secondary antibodies, goat anti-rabbit IgG(H+L)-Alexa Fluor 594 or 633 (Molecular Probes, Invitrogen) were used at a dilution of 1:400. Cells were incubated with DAPI (1 µg/ml in PBS; Roche, Rotkrenz, Switzerland) to visualize nuclei. The endoplasmic reticulum was stained using lectin ConcanavalinA (ConA) conjugated with Alexa Fluor 594 (20 µg/µl in PBS; Molecular Probes, Invitrogen). After washing the cells with PBS and water, the coverslips were mounted in Glycergel (Dako Cytomation, Denmark) containing 25 mg/ml DABCO (Sigma-Aldrich) to retard discoloration. Samples were analyzed using a confocal laser-scanning microscope SP2 (Leica Microsystems, Wetzlar, Germany, 63× oil objective).
Electron microscopy. Cells were seeded at a density of 1.2 × 106 cells per plate into 6 cm2 tissue culture plates. The following day, cells were mock-infected or infected with HSV-1 amplicon vectors at a MOI of 1 or 2 TU per cell. After 48 hours, the cells were scraped into the medium and prepared for purification of RVLPs by repeated cycles of freezing/thawing to disrupt the cell membranes. The cell debris was removed by centrifugation and filtration through a 0.45-µm filter. The cleared supernatant was loaded onto a 10% sucrose cushion and concentrated for 2 hours at 100,000g and 20 °C. For protection, protease inhibitor (Protease inhibitor cocktail tablets complete, Mini, EDTA-free; Roche Diagnostics, Mannheim, Germany) was added to the supernatant. For immune electron microscopy, samples were adsorbed to carbon-coated parlodion films mounted on 300 mesh/inch copper grids (EMS, Fort Washington, PA) for 10 minutes, blocked with PBS-containing 0.1% BSA (PBS-BSA/0.1%) for 10 minutes, incubated with the polyclonal anti-RV serum at a dilution of 1:1 000 PBS-BSA/0.1% for 1 hour, washed several times with PBS-BSA/0.1%, incubated with goat anti-rabbit IgG coupled to 12-nm colloidal gold particles (Jackson ImmunoResearch, West Grove, PA), washed several times with PBS and water, and stained with 2% phosphotungstic acid, pH 7.0 (Aldrich, Steinheim, Germany) for 1 minute. Specimens were analyzed in a transmission electron microscope (CM12; Philips, Eindhoven, the Netherlands) equipped with a CCD camera (Ultrascan 1000; Gatan, Pleasanton, CA) at an acceleration voltage of 100 kV.
RNA isolation and reverse transcription PCR. Cells were seeded at a density of 5x105 cells per well in a six-well plate and, one day later, mock infected or infected with HSV-1 amplicon vectors at a MOI of 1. Total RNA from infected cells was harvested 24 hours later using the total RNA purification kit NucleoSpin RNA II (Macherey-Nagel, Oensingen, Switzerland). Then, the RNA was treated with 1 µl of DNaseI (Roche) per 50 µl of RNA. The samples were incubated for 15 minutes at 37 °C, inactivated for 10 minutes at 75 °C, and stored at –20 °C. Reverse transcription of total RNA was performed using the Reverse Transcription System (Promega, Madison, WI) with random primers provided in the kit. As control, the reaction was performed without the enzyme. Per reaction, 1 µg of total RNA was used and incubated for 10 minutes at room temperature and for 30 minutes at 42 °C; then the enzyme was inactivated for 5 minutes at 95 °C. The cDNA was used immediately for PCR or stored at –20 °C. PCR of the cDNA was performed using the REDTaq ReadyMix PCR Reaction Mix (Sigma-Aldrich) and the primers shown in Table 1.
Immunization of mice and sample collection. Five-weeks-old BALB/c mice were previously confirmed to be negative for anti-RV antibodies by ELISA. Mice were i.m. inoculated at days 0 and 21 with: (i) 5 × 105 TU or 1 × 106 TU of HSVT[VP7/6/2], (ii) 1 × 106 TU of BEI inactivated HSVT[VP7/6/2], (iii) 1 × 106 TU of HSVS[VP6], (iv) 1 × 106 TU of a vector composite containing equal amounts of HSVS[VP2] HSVS[VP6], and HSVS[VP7] (HSVSComposite), (v) 5 × 105 TU or 1 × 106 TU of HSV[EGFP], or (vi) PBS buffer. Another group of mice was immunized once at day 0 with cell culture-adapted live RV strain EC as a positive control of partial protection.25 Individual serum and fecal samples were collected from all mice at 0, 21 and 42 days after the first immunization. All animal procedures were conducted in accordance with the regulations of the Quilmes University Ethic Committee.
Analysis of antibody responses. Purified and concentrated RV strain RRV proteins were separated by SDS-PAGE (10%) and transferred to nitrocellulose membranes. After blocking with PBS containing 1% casein, the membranes were incubated with 1/100 dilutions of serum samples or 1/10 dilutions of fecal samples obtained at day 42 after immunization. Mouse hyperimmune serum against RV strain RRV was used as positive control. After washing, membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce Biotechnology, Rockford, IL) for IgG detection. To detect IgA, membranes were incubated with rabbit anti-mouse IgA (USBiological, Swampscott, MA) and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (DakoCytomation, Glostrup, Denmark) followed by detection with a chemiluminescent substrate (PBL, Bernal, Argentina).
Virus challenge and detection of virus shedding. Three weeks after the second immunization, mice were orally challenged with 104 shedding doses (SD50) of wt RV strain EC.34 To measure RV shedding, stool pellets were collected from each mouse every day for 8 days after challenge and stored at –80 °C. The collected samples were thawed and 10% dilutions were made in TNC buffer containing 0.05% Tween-20 and protease inhibitor cocktail (Sigma-Aldrich), and mixed well before debris was removed by centrifugation (2,500g, 10 minutes). The presence of RV antigen in fecal samples was determined by ELISA as described previously.49 Measurement of protective efficacy of amplicon vectors was based on both the duration and amplitude of virus antigen shedding. Therefore, virus antigen shedding curves (absorbance versus days postchallenge) of each animal were plotted and the area under the shedding curve for each animal was calculated and compared to that of the control group.50
Statistical analysis. Statistical analysis was performed using the program GraphPad Prism (La Jolla, CA). Comparison of the viral shedding and differences between animal groups were compared by Student's t-test. P values were considered to be significant if <0.05 (P < 0.05).
Acknowledgments
We thank Catherine Eichwald, Martin Fussenegger, Urs Greber, Harry Greenberg, Viviana Parreño, Susana Lopez, and Didier Poncet for providing reagents. We acknowledge Bernd Vogt for technical assistance and Peter Wild for critically reading the manuscript. This work was supported by the European Research Commission 6th framework program HEVAR (A.L.E., G.G., J.A., M.A., C.F.) and the Swiss National Science Foundation No. 31003A_124938 (C.F.). L.E. is the recipient of a scholarship from CONICET (Comisión Nacional de Investigaciones Científicas y Técnicas), Argentina. The authors declare no conflict of interest.
REFERENCES
- Ward RL, McNeal MM., and, Steele AD. Why does the world need another rotavirus vaccine. Ther Clin Risk Manag. 2008;4:49–63. doi: 10.2147/tcrm.s821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Shane AL, Parashar UD, Jiang B, Gentsch JR., and, Glass RI. Oral rotavirus vaccines: how well will they work where they are needed most. J Infect Dis. 2009;200 Suppl 1:S39–S48. doi: 10.1086/605035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AH, McNeal MM, Flint JA, Basu M, Lycke NY, Clements JD.et al. (2002The level of protection against rotavirus shedding in mice following immunization with a chimeric VP6 protein is dependent on the route and the coadministered adjuvant Vaccine 201733–1740. [DOI] [PubMed] [Google Scholar]
- Johansen K, Schröder U., and, Svensson L. Immunogenicity and protective efficacy of a formalin-inactivated rotavirus vaccine combined with lipid adjuvants. Vaccine. 2003;21:368–375. doi: 10.1016/s0264-410x(02)00617-5. [DOI] [PubMed] [Google Scholar]
- Silvestri LS, Taraporewala ZF., and, Patton JT. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow Y., and, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Mol Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P., and, Noad R. Virus-like particles as a vaccine delivery system: myths and facts. Hum Vaccin. 2008;4:5–12. doi: 10.4161/hv.4.1.5559. [DOI] [PubMed] [Google Scholar]
- Brun A, Bárcena J, Blanco E, Borrego B, Dory D, Escribano JM.et al. (2011Current strategies for subunit and genetic viral veterinary vaccine development Virus Res 1571–12. [DOI] [PubMed] [Google Scholar]
- Grgacic EV., and, Anderson DA. Virus-like particles: passport to immune recognition. Methods. 2006;40:60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings GT., and, Bachmann MF. The coming of age of virus-like particle vaccines. Biol Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- Crawford SE, Labbé M, Cohen J, Burroughs MH, Zhou YJ., and, Estes MK. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes MK., and, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos K, Simon DA, Conway E, Bowers WJ, Mitra S, Foster TH.et al. (2007Spatial and temporal expression of herpes simplex virus type 1 amplicon-encoded genes: implications for their use as immunization vectors Hum Gene Ther 1893–105. [DOI] [PubMed] [Google Scholar]
- Willis RA, Bowers WJ, Turner MJ, Fisher TL, Abdul-Alim CS, Howard DF.et al. (2001Dendritic cells transduced with HSV-1 amplicons expressing prostate-specific antigen generate antitumor immunity in mice Hum Gene Ther 121867–1879. [DOI] [PubMed] [Google Scholar]
- Frazer ME, Hughes JE, Mastrangelo MA, Tibbens JL, Federoff HJ., and, Bowers WJ. Reduced pathology and improved behavioral performance in Alzheimer's disease mice vaccinated with HSV amplicons expressing amyloid-beta and interleukin-4. Mol Ther. 2008;16:845–853. doi: 10.1038/mt.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorantla S, Santos K, Meyer V, Dewhurst S, Bowers WJ, Federoff HJ.et al. (2005Human dendritic cells transduced with herpes simplex virus amplicons encoding human immunodeficiency virus type 1 (HIV-1) gp120 elicit adaptive immune responses from human cells engrafted into NOD/SCID mice and confer partial protection against HIV-1 challenge J Virol 792124–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish E, Peltékian E, Dickson G, Epstein AL., and, Garcia L. Cell engineering for muscle gene therapy: Extemporaneous production of retroviral vector packaging macrophages using defective herpes simplex virus type 1 vectors harbouring gag, pol, env genes. Cytotechnology. 1999;30:173–180. doi: 10.1023/A:1008022713466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Saeki Y, Camp SM, Chiocca EA., and, Breakefield XO. Single-step conversion of cells to retrovirus vector producers with herpes simplex virus-Epstein-Barr virus hybrid amplicons. J Virol. 1999;73:10426–10439. doi: 10.1128/jvi.73.12.10426-10439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antuono A, Laimbacher AS, La Torre J, Tribulatti V, Romanutti C, Zamorano P.et al. (2010HSV-1 amplicon vectors that direct the in situ production of foot-and-mouth disease virus antigens in mammalian cells can be used for genetic immunization Vaccine 287363–7372. [DOI] [PubMed] [Google Scholar]
- Contin R, Arnoldi F, Campagna M., and, Burrone OR. Rotavirus NSP5 orchestrates recruitment of viroplasmic proteins. J Gen Virol. 2010;91 Pt 7:1782–1793. doi: 10.1099/vir.0.019133-0. [DOI] [PubMed] [Google Scholar]
- López T, Camacho M, Zayas M, Nájera R, Sánchez R, Arias CF.et al. (2005Silencing the morphogenesis of rotavirus J Virol 79184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbretti E, Afrikanova I, Vascotto F., and, Burrone OR. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol. 1999;80 (Pt 2):333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- Eichwald C, Rodriguez JF., and, Burrone OR. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J Gen Virol. 2004;85 Pt 3:625–634. doi: 10.1099/vir.0.19611-0. [DOI] [PubMed] [Google Scholar]
- Hardy WR., and, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin SE, Moser CA, Cohen S, Clark HF., and, Offit PA. Immunologic correlates of protection against rotavirus challenge after intramuscular immunization of mice. J Virol. 1997;71:7851–7856. doi: 10.1128/jvi.71.10.7851-7856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, McNeal MM., and, Sheridan JF. Development of an adult mouse model for studies on protection against rotavirus. J Virol. 1990;64:5070–5075. doi: 10.1128/jvi.64.10.5070-5075.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González SA., and, Affranchino JL. Assembly of double-layered virus-like particles in mammalian cells by coexpression of human rotavirus VP2 and VP6. J Gen Virol. 1995;76 (Pt 9):2357–2360. doi: 10.1099/0022-1317-76-9-2357. [DOI] [PubMed] [Google Scholar]
- Nilsson M, von Bonsdorff CH, Weclewicz K, Cohen J., and, Svensson L. Assembly of viroplasm and virus-like particles of rotavirus by a Semliki Forest virus replicon. Virology. 1998;242:255–265. doi: 10.1006/viro.1997.8987. [DOI] [PubMed] [Google Scholar]
- Franco MA, Angel J., and, Greenberg HB. Immunity and correlates of protection for rotavirus vaccines. Vaccine. 2006;24:2718–2731. doi: 10.1016/j.vaccine.2005.12.048. [DOI] [PubMed] [Google Scholar]
- Santos K, Duke CM, Rodriguez-Colon SM, Dakwar A, Fan S, Keefer MC.et al. (2007Effect of promoter strength on protein expression and immunogenicity of an HSV-1 amplicon vector encoding HIV-1 Gag Vaccine 251634–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mukherjee S, Fraefel C, Breakefield XO., and, Allen PD. Herpes simplex virus type 1 amplicon vector-mediated gene transfer to muscle. Hum Gene Ther. 2002;13:261–273. doi: 10.1089/10430340252769789. [DOI] [PubMed] [Google Scholar]
- Huard J, Feero WG, Watkins SC, Hoffman EP, Rosenblatt DJ., and, Glorioso JC. The basal lamina is a physical barrier to herpes simplex virus-mediated gene delivery to mature muscle fibers. J Virol. 1996;70:8117–8123. doi: 10.1128/jvi.70.11.8117-8123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin SE, Clark SL, Bos NA, Brubaker JO., and, Offit PA. Migration of antigen-presenting B cells from peripheral to mucosal lymphoid tissues may induce intestinal antigen-specific IgA following parenteral immunization. J Immunol. 1999;163:3064–3070. [PubMed] [Google Scholar]
- Burns JW, Krishnaney AA, Vo PT, Rouse RV, Anderson LJ., and, Greenberg HB. Analyses of homologous rotavirus infection in the mouse model. Virology. 1995;207:143–153. doi: 10.1006/viro.1995.1060. [DOI] [PubMed] [Google Scholar]
- Jiang JQ, He XS, Feng N., and, Greenberg HB. Qualitative and quantitative characteristics of rotavirus-specific CD8 T cells vary depending on the route of infection. J Virol. 2008;82:6812–6819. doi: 10.1128/JVI.00450-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimerink JH, Boshuizen JA, Einerhand AW, Duizer E, van Amerongen G, Schmidt N.et al. (2007Systemic immune response after rotavirus inoculation of neonatal mice depends on source and level of purification of the virus: implications for the use of heterologous vaccine candidates J Gen Virol 88Pt 2604–612. [DOI] [PubMed] [Google Scholar]
- McNeal MM, VanCott JL, Choi AH, Basu M, Flint JA, Stone SC.et al. (2002CD4 T cells are the only lymphocytes needed to protect mice against rotavirus shedding after intranasal immunization with a chimeric VP6 protein and the adjuvant LT(R192G) J Virol 76560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal CM, Harriman GR., and, Conner ME. Protection of the villus epithelial cells of the small intestine from rotavirus infection does not require immunoglobulin A. J Virol. 2000;74:4102–4109. doi: 10.1128/jvi.74.9.4102-4109.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco MA., and, Greenberg HB. Immunity to rotavirus in T cell deficient mice. Virology. 1997;238:169–179. doi: 10.1006/viro.1997.8843. [DOI] [PubMed] [Google Scholar]
- McNeal MM, Rae MN, Conner ME., and, Ward RL. Stimulation of local immunity and protection in mice by intramuscular immunization with triple- or double-layered rotavirus particles and QS-21. Virology. 1998;243:158–166. doi: 10.1006/viro.1998.9060. [DOI] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A, Ciarlet M, Crawford SE, Conner ME., and, Estes MK. Immunogenicity and protective efficacy of rotavirus 2/6-virus-like particles produced by a dual baculovirus expression vector and administered intramuscularly, intranasally, or orally to mice. Vaccine. 2003;21:3885–3900. doi: 10.1016/s0264-410x(03)00308-6. [DOI] [PubMed] [Google Scholar]
- Yang K, Wang S, Chang KO, Lu S, Saif LJ, Greenberg HB.et al. (2001Immune responses and protection obtained with rotavirus VP6 DNA vaccines given by intramuscular injection Vaccine 193285–3291. [DOI] [PubMed] [Google Scholar]
- Kavanagh OV, Ajami NJ, Cheng E, Ciarlet M, Guerrero RA, Zeng CQ.et al. (2010Rotavirus enterotoxin NSP4 has mucosal adjuvant properties Vaccine 283106–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IL, Hardwicke MA., and, Sandri-Goldin RM. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- Estes MK, Graham DY, Gerba CP., and, Smith EM. Simian rotavirus SA11 replication in cell cultures. J Virol. 1979;31:810–815. doi: 10.1128/jvi.31.3.810-815.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpilienne A, Nejmeddine M, Berois M, Parez N, Neumann E, Hewat E.et al. (2001Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells J Biol Chem 27629361–29367. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Fraefel C, Ichikawa T, Breakefield XO., and, Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol Ther. 2001;3:591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- Aarthi D, Ananda Rao K, Robinson R., and, Srinivasan VA. Validation of binary ethyleneimine (BEI) used as an inactivant for foot and mouth disease tissue culture vaccine. Biologicals. 2004;32:153–156. doi: 10.1016/j.biologicals.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Argüelles MH, Villegas GA, Castello A, Abrami A, Ghiringhelli PD, Semorile L.et al. (2000VP7 and VP4 genotyping of human group A rotavirus in Buenos Aires, Argentina J Clin Microbiol 38252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J., and, Desselberger U. Humana Press; 2000. Rotaviruses: Methods and Protocols. [Google Scholar]