Abstract
Adaptive immune responses may be vital in the overall efficacy of oncolytic viruses in human malignancies. However, immune responses to oncolytic adenoviruses are poorly understood because these viruses lack activity in murine cells, which precludes evaluation in immunocompetent murine cancer models. We have evaluated human adenovirus activity in murine cells. We show that a panel of murine carcinoma cells, including CMT64, MOVCAR7, and MOSEC/ID8, can readily be infected with human adenovirus. These cells also support viral gene transcription, messenger RNA (mRNA) processing, and genome replication. However, there is a profound failure of adenovirus protein synthesis, especially late structural proteins, both in vitro and in vivo, with reduced loading of late mRNA onto ribosomes. Our data also show that in trans expression of the nonstructural late protein L4-100K increases both the amount of viral mRNA on ribosomes and the synthesis of late proteins, accompanied by reduced phosphorylation of eIF2α and improved anticancer efficacy. These results suggest that murine models that support human adenovirus replication could be generated, thus allowing evaluation of human adenoviruses in immunocompetent mice.
Introduction
Oncolytic viruses multiply selectively within infected cancer cells and cause death, with release of mature viruses that infect neighboring cells. The second generation adenovirus mutants dl922-947 and Δ24 contain a deletion in the E1A CR2 region, which binds to E2F–pRb complexes, thereby dissociating E2F to drive cells into an S phase-like state allowing transactivation of genes necessary for viral DNA replication.1 We have previously shown that dl922-947 has considerable activity in ovarian cancer and is more potent than E1A wild-type adenoviruses and the E1B-55K mutant dl1520 (Onyx-015, H101).2,3 dl922-947 replicates selectively in cells with abnormalities of the Rb pathway and consequent G1-S checkpoint, defects seen in many human malignancies,4 including ovarian cancer.5,6 Phase I trials of Δ24 derivatives have been completed with encouraging results.7
Ovarian cancer is an immunogenic disease.8 The extent and nature of immune responses are prognostic: for example, high intraepithelial CD8+:Treg (CD4+CD25+FoxP3+) ratios are associated with favorable outcome in high-grade serous disease.9,10 However, the nature of the tumor antigens remains unclear and attempts at clinical immunotherapy in ovarian cancer have been largely unsuccessful.
In preclinical models, immune responses contribute to the efficacy of oncolytic reovirus, herpes simplex virus, vesicular stomatitis virus, vaccinia and Newcastle disease virus.11 Adenoviruses can stimulate robust innate and adaptive (both cellular and humoral) immune responses. Multiple pathways contribute to the innate response12 and the major capsid protein Hexon contains immunodominant epitopes for both CD4+ and CD8+ T cells that are common across multiple adenovirus subtypes.13,14,15 However, nearly all work on adenovirus immunology has utilized replication-defective vaccine vectors.16 Knowledge on the role of immune responses to replicating adenoviruses is very limited, as murine cells do not support productive replication of human adenoviruses (hAd). Syrian hamsters are partially permissive to hAd and may be a useful model,17 but reagents and tools are few, and specifically there is no transplantable Syrian hamster ovarian cancer cell line. Another potential option is to use mouse adenovirus 1 (MAV1). However, this virus causes fatal hemorrhagic encephalitis in C57Bl/6 mice, rendering it less attractive.18
Productive infection of human adenoviruses in murine cells is significantly lower than in human cells,19 although infection and some replication can be seen.20 However, the mechanisms for lack of infectious virion formation remain unclear. In monkey cells, it was postulated that aberrant messenger RNA (mRNA) splicing or compartmentalization are responsible for abortive infection with hAd.21,22
We have investigated hAd type 5 (hAd5) species specificity in murine cells as a prelude to studying immune responses to oncolytic adenovirus in intraperitoneal ovarian cancer models. We find that murine malignant cells can be infected readily with hAd5. Both early and late viral genes are transcribed efficiently and there is evidence of viral genome replication. However, a profound failure of productive virion production is observed with late protein expression particularly poor. Ribosome fractionation assays show reduced viral mRNA loading in murine cells, resulting in failure of translation, especially of late transcripts. Our results indicate that this translation failure can be partially overcome by ectopic expression of the nonstructural L4 protein 100K.
Results
Activity of human oncolytic adenoviruses in murine carcinoma models
We have previously shown that E1A CR2-deleted human oncolytic adenovirus vectors have significant activity in human ovarian cancer cells and ovarian cancer xenografts in nude mice.3,23 However, immunocompromised animals do not permit evaluation of the role of immune responses to virus efficacy. As a first step towards developing immunocompetent murine models, we first examined the efficacy of dl922-947 in murine cancers. In vitro, the E1A-CR2 deleted vector dl922-947 was between 15- and 1,800-fold more potent in human ovarian cancer cells and HeLa cells than in a panel of five murine carcinoma lines (Figure 1a). Similar data were obtained with Ad5 wild-type and dl309 (Supplementary Figure S1). Moreover, delivery of dl922-947 to C57Bl/6 mice bearing intraperitoneal CMT64 (Figure 1b) and ID8 tumors (data not shown) produced no antitumor activity at all, in contrast to our previous data with human ovarian intraperitoneal xenografts.3
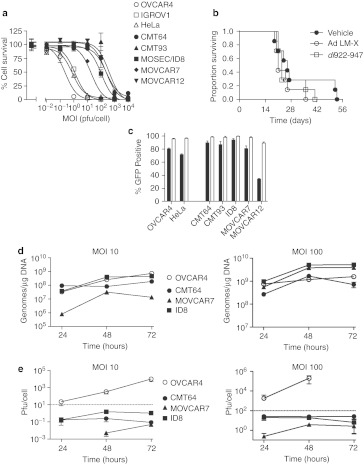
Figure 1.
Activity of human adenoviruses in murine cells. (a) 104 cells were infected with dl922-947 in triplicate at MOI (multiplicity of infection) 0.01–10,000 pfu/cell. Cell survival was assessed 120 hours postinfection by MTT assay. (b) 5 × 106 CMT64 cells were injected intraperitoneally into female C57Bl/6 mice. dl922-947 or control virus Ad LM-X (both 5 × 109 particles/day) was injected on days 4–8 inclusive. Mice were killed when ascites was clinically evident as per UK Home Office guidelines. (c) Human and murine cells were infected with Ad CMV-GFP (MOI 5 (black) and 50 (white) pfu/cell). Green fluorescence was assessed by flow cytometry 24 hours postinfection. (d) 5 × 105 cells were infected with dl922-947 at either MOI 10 (left) or MOI 100 (right). DNA was extracted from infected cells up to 72 hours postinfection and subjected to quantitative PCR using Hexon region primers and probes. Purified dl922-947 genomic DNA was used to generate standard curves. (e) 105 cells were infected with dl922-947 at either MOI 10 (left) or MOI 100 (right). Intracellular virion production was titered by TCID50 assay up to 72 hours postinfection. Dotted line represents input dose. Cell death precluded assessment of virion titer in OVCAR4 cells infected at MOI 100 for 72 hours. CMV, cytomegalovirus; GFP, green fluorescent protein; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; pfu, plaque-forming unit.
We investigated why human adenoviruses lack activity in murine cells, by evaluating individual steps of the adenovirus life cycle in murine cells. Four of the five murine carcinoma cells were at least as infectable as human OVCAR4 and HeLa cells in a fluorescence green fluorescent protein (GFP) assay, and the fifth (MOVCAR12) could still be infected at higher multiplicities of infection (MOI) (Figure 1c). Therefore, failure of infection did not explain lack of human virus activity in these murine cells.
We next quantified genome replication in three murine lines using quantitative PCR (qPCR) using Hexon region primers. There was demonstrable replication of the dl922-947 genome in all three murine lines at MOI 10 plaque-forming unit (pfu)/cell (Figure 1d, left), which, in the case of ID8, matched replication in human OVCAR4 cells. Identical patterns of genome replication were seen with E1A region primers, indicating that replication occurred throughout the genome (data not shown). When the input MOI was increased to 100, replication in the murine cells matched or even exceeded that in human cells (Figure 1d, right). However, despite this genome replication, there was profound failure of production of infectious virions in murine cells. At neither MOI 10 nor MOI 100 (Figure 1e) did the number of virions produced in any of the murine cells exceed the input dose, in sharp contrast to human OVCAR4 cells and other human ovarian cancer cells as we have previously demonstrated.3,23
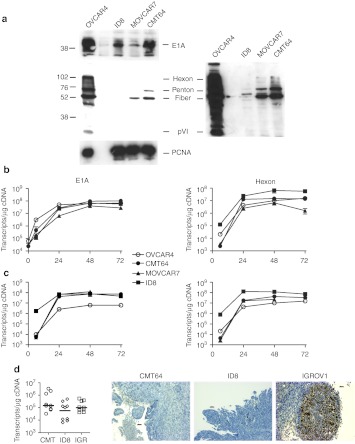
We then examined viral protein expression. E1A was expressed at demonstrable levels in ID8 and CMT64 cells, albeit at levels lower than in human OVCAR4 cells, but there was almost total absence of late structural protein expression in all three murine cell lines tested (Figure 2a). The same patterns of protein expression were seen following infection with the wild-type control virus dl309 (data not shown), indicating that these results were not specific to dl922-947. There was definite evidence that early and late viral genes were being transcribed in murine cells—at MOI 10, both E1A and Hexon were transcribed at levels comparable to OVCAR4 cells (Figure 2b), while at MOI 100, transcript number in murine cells exceeded that in human cells (Figure 2c). We also analyzed gene expression in vivo. A single dose of dl922-947 was administered intraperitoneally to immunodeficient mice bearing murine CMT64 and ID8 tumors as well as human IGROV1 tumors. Again, there was evidence of adenovirus gene transcription, with no statistical difference in Hexon transcripts between human and murine tumours (Figure 2d, left). However, there was almost no detectable structural adenovirus protein expression by immunohistochemistry in the murine tumors, in sharp contrast to the human xenograft (Figure 2d, right).
Figure 2.
Failure of adenovirus protein expression in murine cells. (a) OVCAR4, ID8, MOVCAR7, and CMT64 cells were infected with dl922-947 (MOI 10); protein was harvested 48 hours postinfection and expression of E1A and adenovirus structural proteins assessed by immunoblot. A long exposure image of the structural protein blot is also presented. (b,c) 5 × 105 cells were infected with dl922-947 at either (b) MOI 10 or (c) MOI 100. Total cellular RNA was extracted from infected cells up to 72 hours postinfection and DNAse I treated. One microgram was reverse transcribed using random hexanucleotide primers; 50 ng cDNA was then subjected to quantitative PCR using both E1A (left) and Hexon (right) primers and probes. Purified dl922-947 genomic DNA was used to generate standard curves. (d) 5 × 106 cells were injected intraperitoneally into ICRF nude female mice in 200 µl PBS. When ascites was clinically evident, mice received a single intraperitoneal injection of dl922-947 (1010 particles in 400 µl 20% icodextrin). Mice were killed 48 hours later and all visible tumor dissected out. Half was snap-frozen in dry ice, while half was fixed in 5% formaldehyde. RNA was extracted from the snap frozen specimens, DNAse I treated and reverse transcribed using random hexanucleotide primers; 50 ng cDNA was then subjected to quantitative PCR using Hexon region primers and probes as before (left). Expression of adenovirus structural proteins was assessed in formaldehyde-fixed tissue by immunohistochemistry (right). Bar represents 50 µm. cDNA, complementary DNA; MOI, multiplicity of infection; PBS, phosphate-buffered saline.
These results suggested that adenovirus genes are transcribed into mRNA in murine cells, but not translated into protein before assembly into whole virions. We therefore examined potential causes of failed translation.
Viral mRNA is correctly processed in murine cells but not loaded onto ribosomes
We firstly addressed whether hAd5 protein was simply being targeted for rapid proteasomal degradation. However, MG132 treatment was unable to increase late structural protein expression in murine cells in CMT64 (Supplementary Figure S2) and MOVCAR7 cells (data not shown). After transcription, viral mRNA leaves the nucleus for translation in the cytoplasm. Nuclear and cytoplasmic fractions were generated from CMT64, MOVCAR7, and OVCAR4 infected with dl922-947 (MOI 10) and subjected to quantitative reverse transcription-PCR (qRT-PCR) analysis for E1A and Hexon transcripts. The cytoplasmic:nuclear transcript ratios were similar in human and murine cells; indeed, E1A transcripts appeared more abundant in the cytoplasm of CMT64 and MOVCAR7 cells than in OVCAR4 (Figure 3a). There was also evidence that both E1A and Hexon transcripts exited the nucleus in ID8 cells (Supplementary Figure S3). Overall, these results indicate that failure of mRNA export from the nucleus was not the cause of absence viral protein expression in murine cells.
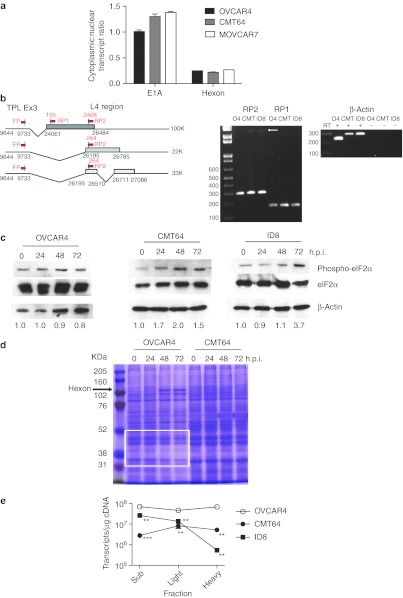
Figure 3.
Translation of adenovirus late mRNA in murine cells. (a) OVCAR4, CMT64, and MOVCAR7 cells were infected with dl922-947. Forty-eight hours postinfection, cells were fractionated into nuclear and cytoplasmic compartments. RNA from each compartment was reverse transcribed using random hexanucleotide primers; 50 ng cDNA was subjected to quantitative PCR using both Hexon and E1A region primers and probes with purified dl922-947 genomic DNA used to generate standard curves. (b) RNA was extracted from OVCAR4, CMT64, and ID8 cells 48 hours after infection with dl922-947 (MOI 10). After DNAse I treatment and reverse transcription, 50 ng cDNA underwent PCR using TPL exon3 sense primer and either L4 reverse primer 1 or 2 as depicted. Expected band sizes were 155 bp for RP1, and both 264 and 2,408 bp (arrowed) for RP2. (c) Cells were infected with dl922-947 (MOI 10). Protein was extracted up to 72 hours postinfection. Expression of eIF2α and Ser-51 phospho eIF2α was assessed by immunoblot. Extent of phosphorylation was assessed using ImageJ—numbers below each blot represent phospho-eIF2α:total eiF2α ratio normalized to uninfected cells. (d) Twenty microgram total cellular protein from OVCAR4 and CMT64 cells infected with dl922-947 (MOI 10) for up to 72 hours was electrophoresed on and 8% SDS-PAGE gel and stained with Coomassie blue. Expression of cellular proteins diminishes over time in human cells (white box) as Hexon expression becomes more evident (arrow) in human cells only. (e) OVCAR4, CMT64, and ID8 cells were infected with dl922-947 (MOI 10) or mock-infected. Forty-eight hours later, cells were treated with cycloheximide and lysed. After sucrose gradient ultracentrifugation, 11 ribosome fractions were generated—fractions 1–5 inc represent subpolysomes, 6–8 inc are light polysomes, while fractions 9–11 inc are heavy polysomes. RNA was precipitated from each fraction, pooled, and reverse transcribed. Hexon transcript number was quantified in 10 ng cDNA using qPCR. h.p.i., hours postinfection; inc, inclusive; MOI, multiplicity of infection; qPCR, quantitative PCR; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TPL, tripartite leader sequence. **P < 0.01; ***P < 0.001.
All late genes in adenovirus are transcribed from the Major Late Promoter as a single 28,000 nucleotide pre-mRNA, which is polyadenylated at separate sites to generate five separate mRNA species. The tripartite leader sequence (TPL) is then spliced to different 5′ sites within each mRNA species to generate at least 15 different mRNA transcripts. It was previously suggested that there was aberrant splicing of hAd transcripts in non-human cells.21 To investigate whether murine cells correctly spliced late hAd5 mRNA, RT-PCR was used, with a sense primer in the TPL and two separate reverse primers in L4 as depicted in Figure 3b. Results indicated that the same processed species are seen in both murine and human cells following infection with dl922-947 (MOI 10), indicating that RNA processing and splicing occur efficiently in the murine cells (Figure 3b). Indeed, the larger L4 100K transcript is more prominent in murine cells than human 48 hours postinfection.
Adenovirus infection can induce robust type I interferon responses, which lead to a global shutdown of host protein translation as a result of activation of protein kinase R (PKR), which in turn phosphorylates translation initiating factor eIF2α. Ad5 encodes two highly structured RNA molecules, VAI and VAII, whose role is to inhibit PKR.24 There was no increase in either interferon-α or -β released from OVCAR4 cells over a 48 hours time course after dl922-947 infection (data not shown). In keeping with this, phospho-eIF2α immunoblotting indicated no significant increase in eIF2α phosphorylation in OVCAR4 cells over 72 hours (Figure 3c). In CMT64 and ID8 cells, we were unable to detect any type I interferon release over 48 hours (data not shown), although there was some increase in eIF2α phosphorylation. However, Coomassie staining suggested that this phosphorylation in CMT64 cells did not result in a global shutdown of host protein synthesis, nor did viral protein bands emerge, in keeping with the immunoblot data (Figure 3d). Similar Coomassie staining was seen with other murine cells (data not shown). These data imply that the failure of adenovirus protein expression in murine cells did not result from a host cell interferon response despite some evidence of eIF2α phosphorylation. In human OVCAR4 cells by contrast, there was a reduction in host protein expression over time as prominent viral protein bands emerged, indicating a selective reduction of cellular protein synthesis with a selective production of viral proteins (Figure 3d).
We then investigated loading of viral mRNA onto ribosomes. OVCAR4, CMT64, and ID8 cells were infected with dl922-947 or mock-infected. Forty-eight hours later, they were lysed and subjected to sucrose gradient density ultracentrifugation to separate ribosomes into 11 fractions ranging from subpolysomal to heavy (polysomal) fractions (Supplementary Figure S4). In human cells, viral infection increased the amount of total RNA associated with the polysomal fractions (Supplementary Figure S4), whereas there was no change, or even a decrease, in total ribosomal RNA in murine cells. qRT-PCR indicated that there were significantly fewer Hexon transcripts across all fractions in murine cells compared to human especially in polysomal fractions (Figure 3e). Therefore, in murine cells, late viral mRNA is transcribed but does not associate with polysomes and thus cannot be translated.
Coinfection with MAV1 increases human adenovirus late protein expression in murine cells
Having determined that there was failure of translation of human adenoviral mRNA in murine cells, we then investigated whether this failure could be overcome.
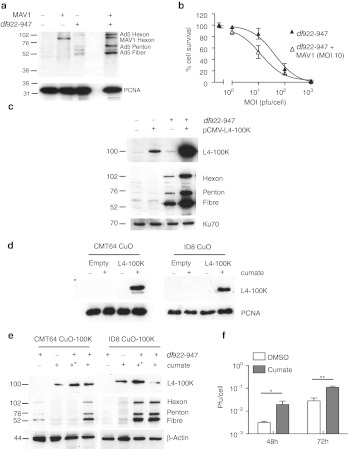
Murine cells are sensitive to infection with murine adenovirus 1 (MAV1) (Supplementary Figure S5). MAV1 is genetically similar to human Ad5 but infects via a non-CAR (coxsackie adenovirus receptor) receptor and causes fatal hemorrhagic encephalitis in C57Bl/6 mice.18 To investigate whether MAV1 proteins expressed in trans could promote expression of late hAd5 genes and act to sensitize murine cells to hAd5 cytotoxicity, murine cells were coinfected with dl922-947 and MAV1 (both MOI 10). There was some immunoblot cross-reactivity between MAV1 Hexon and hAd5 Hexon, but the blot showed a clear increase in human late protein expression in the presence of both viruses in MOVCAR7 cells (Figure 4a). There was also an increase in dl922-947-specific cytotoxicity when MOVCAR7 cells were coinfected with MAV1 at MOI 10 (Figure 4b)—similar sensitization was also seen with CMT64 cells (data not shown). This implied that one or more MAV1 proteins was able to augment the specific translation of hAd5 and demonstrated that murine cells could support production of human viral late proteins.
Figure 4.
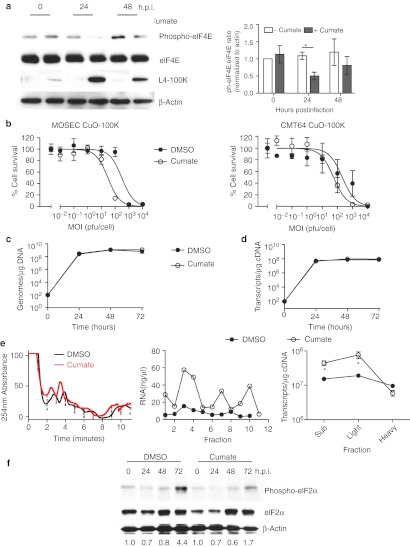
L4-100K expression increases adenovirus protein expression in murine cells. (a) MOVCAR7 cells were infected with dl922-947 (MOI 10) and mouse adenovirus 1 (MAV1–MOI 10) either separately or together. Forty-eight hours postinfection, expression of human adenovirus structural proteins was assessed by immunoblot. (b). MOVCAR7 cells were infected with dl922-947 (MOI 1–1,000) in triplicate in the presence and absence of MAV1 (MOI 10) coinfection. Cell survival was assessed 120 hours thereafter by MTT assay. Survival of cells infected with dl922-947 and MAV1 is normalized to that of cells infected with MAV1 alone, to control for the cytotoxic effects of MAV1 infection. (c) ID8 cells were infected with dl922-947 (MOI 10). Two hours later, they were also transfected with pCMV-L4-100K. Expression of L4 100K and late structural proteins was assessed 42 hours thereafter by immunoblot. (d) CMT64 and ID8 cells expressing L4 100K under the control of a cumate responsive promoter (CMT64 CuO-100K and ID8 CuO-100K respectively) were treated with 200 µg/ml cumate for 16 hours. Expression of L4 100K was then assessed by immunoblot. (e) CMT64 CuO-100K and ID8 CuO 100K were infected with dl922-947 (MOI 10). Cumate (200 µg/ml) was added 2 hours postinfection and either washed off 2 hours later (*) or left on cells. Forty-eight hours after virus infection, expression of L4-100K, and adenovirus structural proteins was assessed by immunoblot. (f) Intracellular production of infectious virions was assessed in ID8-CuO-100K cells 48 hours after infection with dl922-947 (MOI 10) in the presence and absence of cumate, using TCID50 assay. *P < 0.05; **P < 0.01. DMSO, dimethyl sulfoxide; MOI, multiplicity of infection; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; pfu, plaque-forming unit.
Expression of L4-100K alone increases human adenovirus late protein expression in murine cells
In the absence of specific tools and reagents for MAV1, we turned back to human adenovirus. Given its role in promoting the preferential translation of viral mRNA in infected cells, we hypothesized that expression of Ad5 L4-100K in trans in murine cells could increase the permissivity of the murine cells to human virus infection.
First, simple immunoblotting confirmed that L4-100K was expressed at very low levels in murine cells after hAd5 infection, as with other late gene products (Supplementary Figure S6). We then infected ID8 cells with dl922-947 4 hours after transfection with a plasmid encoding L4-100K. By immunoblot, there was a marked increase in expression of all late structural proteins (Figure 4c). There was also greater L4-100K expression in the presence of both virus and plasmid, implying positive feedback. Next, we used a novel inducible system to generate ID8 and CMT64 cells in which expression of L4-100K lay under the control of a cumate-inducible promoter.25 Exposure of CMT64 CuO-100K and ID8 CuO-100K cells to 200 µg/ml cumate induced demonstrable L4-100K expression (Figure 4d), which allowed greater expression of late structural proteins (Figure 4e) after hAd5 infection, without altering E1A expression (Supplementary Figure S7). There was also a significant increase in infectious virion production by TCID50 assay (Figure 4f). CMT64 cells required continuous exposure to cumate to increase late protein expression, whereas in ID8 cells, transient exposure was optimal (Figure 4e).
The mechanism by which L4-100K promotes preferential translation of viral mRNA is complex.26,27 However, it can displace the kinase Mnk1 from the cap-initiation complex eIF4F, thus blocking phosphorylation of eIF4E28 and preventing cap-dependent translation of cellular mRNA while promoting translation of viral mRNA via the process of ribosome shunting. Therefore, we investigated eIF4E phosphorylation in dl922-947–infected ID8 cells following cumate-induced L4-100K expression. Twenty-four hours after cumate addition, there was a significant reduction in eIF4E phosphorylation (Figure 5a). Addition of cumate also increased dl922-947 cytotoxicity (Figure 5b), which occurred despite no increase in viral genome replication (Figure 5c) or late gene transcription (Figure 5d).
Figure 5.
L4-100K promotes viral mRNA translation. (a) ID8 CuO-100K cells were infected with dl922-947 (MOI 10) in the presence or absence of cumate (200 µg/ml) for up to 48 hours. Expression of eIF4E, phospho-eIF4E, and L4-100K was assessed by immunoblot. Relative eIF4E phosphorylation in two separate blots was quantified using ImageJ (right). (b) ID8 CuO-100K and CMT64 CuO-100K were infected with dl922-947 (MOI 0.01–10,000) in the presence and absence of cumate. Cell survival was assessed 120 hours postinfection by MTT assay. (c,d) ID8 CuO 100K were infected with dl922-947 (MOI 10) in the presence and absence of cumate. (c) Viral genomes and (d) Hexon transcripts were quantified using quantitative PCR up to 72 hours postinfection. (e) ID8 CuO-100K cells were infected with dl922-947 (MOI 10) in the presence of DMSO or cumate. Forty-eight hours later, ribosomal fractions were prepared as before (left). RNA concentration was quantified in each fraction (center). Hexon transcript number in pooled fractions was quantified using qRT-PCR (right). *P < 0.05. (f) ID8 CuO-100K cells were infected with dl922-947 (MOI 10) in the presence of DMSO or cumate for up to 72 hours. Expression of eIF2α and phospho-eIF2α was assessed by immunoblot. Extent of phosphorylation was assessed using ImageJ—numbers below each lane represent phospho-eIF2α:total eiF2α ratio normalized to uninfected cells. DMSO, dimethyl sulfoxide; h.p.i., hours postinfection; MOI, multiplicity of infection; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; pfu, plaque-forming unit; qRT-PCR, quantitative reverse transcription-PCR.
We then repeated the ribosome fractionation assays using ID8 CuO-100K cells infected with dl922-947 for 48 hours in the presence and absence of cumate (Figure 5e). Cumate-induced 100K expression increased the amount of RNA associated with ribosomes and significantly increased the amount of Hexon mRNA associated with light ribosomal fractions, although not heavy polysome fractions. Cumate-induced 100K expression also reduced the extent of eIF2α phosphorylation in ID8 CuO-100K cells infected with dl922-947 (Figure 5f).
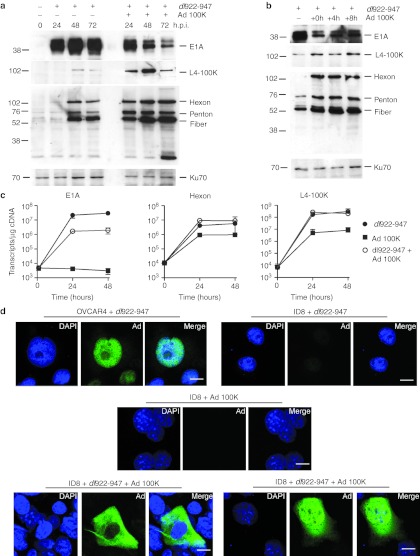
Because data on the use of cumate in vivo are limited, we next generated an E1-deleted, non-replicating adenovirus vector, in which L4-100K expression lies under the control of the cytomegalovirus (CMV) immediate early promoter in the E1 region. As with cumate, there was a marked increase in late protein expression after simultaneous coinfection with dl922-947 and Ad 100K (both MOI 10; Figure 6a). However, this was marked by a reduction in E1A expression, in contrast to the inducible cumate system. Addition of Ad 100K 4 or 8 hours after dl922-947 infection still allowed late protein expression but also reduced the inhibitory effects on E1A (Figure 6b). In keeping with these data, coinfection with dl922-947 and Ad 100K reduced E1A gene transcription (Figure 6c). However, as with the cumate system, addition of Ad 100K to dl922-947 did not significantly increase Hexon transcription, although, interestingly, infection with Ad 100K alone did support some Hexon transcription in ID8 cells, although this Hexon mRNA was not translated into protein to any significant extent (Supplementary Figure S8).
Figure 6.
Ad L4-100K increases late viral protein expression. (a) ID8 cells were infected with dl922-947 (MOI 10) in the presence or absence of simultaneous Ad 100K (MOI 10) coinfection. Expression of E1A, L4 100K, and adenovirus structural proteins was assessed up to 72 hours postinfection by immunoblot. (b) ID8 cells were infected with dl922-947 (MOI 10). Ad 100K (MOI 10) was added 0, 4 or 8 hours later. Expression of E1A, L4 100K, and adenovirus structural proteins was assessed 48 hours after initial dl922-947 infection. (c) The number of E1A, Hexon, and L4 100K transcripts in ID8 cells infected with dl922-947 (MOI 10), Ad 100K (MOI 10) either alone or together was measured using quantitative reverse transcription PCR up to 48 hours postinfection. (d) OVCAR4 and ID8 cells were infected with dl922-947 (MOI 10). In addition, ID8 cells were infected with Ad 100K (MOI 10), either alone or 16 hours after dl922-947 infection. Forty-eight hours after addition of dl922-947, cells were fixed in 3.7% paraformaldehyde, stained for adenovirus structural protein expression and counterstained with DAPI. Bars indicate 10 µm. DAPI, 4′,6-diamidino-2-phenylindole; h.p.i., hours postinfection; MOI, multiplicity of infection.
We also examined structural protein expression by immunofluorescence (Figure 6d). In OVCAR4 cells, late proteins are expressed almost exclusively in the nucleus, while there is no detectable late protein in ID8 cells infected with dl922-947 or Ad 100K alone. Dual infection with dl922-947 and Ad 100K does increase late protein expression markedly, in keeping with the immunoblots in Figure 6a. However, the pattern of expression is largely cytoplasmic.
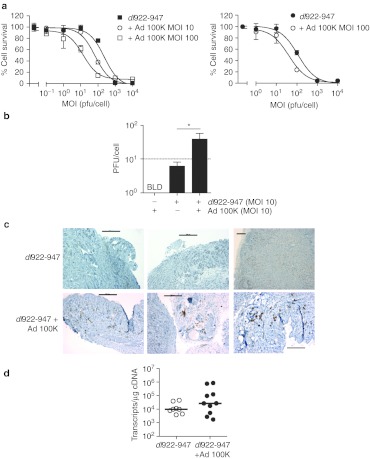
Finally, we showed that there was greater cell killing (Figure 7a) and increased intracellular infectious virion production (Figure 7b) in vitro following coinfection with dl922-947 and Ad 100K, but not with control hAd5 vector Ad LM-X (Supplementary Figure S9). In vivo, coinfection with dl922-947 and Ad 100K increased expression of late proteins in subcutaneous ID8 tumours (Figure 7c), despite no increase in late gene transcription (Figure 7d).
Figure 7.
Ad L4-100K increases dl922-947 activity in vitro and in vivo. (a) ID8 (left) and MOVCAR7 (right) cells were infected with dl922-947 in the presence or absence of Ad 100K. Cell survival was assessed 120 hours later by MTT assay. Data for dual-infected cells are normalized to survival following infection with Ad 100K alone. (b) Infectious intracellular virions generated 48 hours postinfection in ID8 cells were titered by TCID50 assay (as shown in b). *P < 0.05, dotted line indicates input MOI of dl922-947. BLD = below limit of detection. (c,d) Subcutaneous ID8 tumors were grown on the left flank of ICRF nude female mice. Once tumors reached ~7 mm in diameter, dl922-947 was injected intratumorally (5 × 109 particles in 50 µl PBS on days 1–3 inclusive). Mice also received a single intratumoral dose of Ad 100K (3 × 109 particles in 50 µl PBS) or PBS alone on day 4. Tumors were excised on day 5, dissected in two and either snap frozen in dry ice or fixed in 5% formaldehyde. Expression of adenovirus structural proteins was assessed by IHC (c; bars represent 200 µm). The number of Hexon transcripts was measured using quantitative reverse transcription PCR (as shown in d). IHC, immunohistochemistry; MOI, multiplicity of infection; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; pfu, plaque-forming unit; PBS, phosphate-buffered saline.
Discussion
The ability to assess novel biological therapies in immunocompetent models is very important. This is particularly true for oncolytic viruses, given the potential importance of immune response to overall efficacy.11
In this study, we attempted to dissect the cause of failed productivity of human adenoviruses in murine cells. We excluded primary infection, transcription of either early or late viral genes, mRNA splicing, and viral genome replication as potential causes. We did, however, identify that there were very low levels of viral protein expression, especially late proteins, with reduced loading of viral mRNA onto ribosomes. These combined to cause a profound failure of infectious virion production.
Complete understanding of the mechanisms controlling adenoviral protein production remains incomplete. However, two steps are required—prevention of translation of cellular mRNA with preferential translation of viral mRNA; the nonstructural late protein L4-100K appears pivotal in both.29
L4-100K prevents translation of capped cellular mRNA by uncoupling the cap-binding complex eIF4F via competitive antagonism of Mnk1, thus preventing Mnk1-dependent phosphorylation of the cap-binding protein eIF4E.28 Mnk1 displacement requires amino acids 280–345 within 100K, but does not require 100K to bind directly to mRNA.26 L4-100K also promotes selective translation of late viral mRNA by coordinating the recruitment of necessary components into a translation complex. It binds to eIF4G and Poly A binding protein (PABP) after displacing Mnk127 and also binds to the 5′ noncoding TPL of viral late mRNA species.30 In addition, phosphorylation of L4-100K tyrosine residues, specifically Y365 and Y682, promotes the process of ribosomal shunting, whereby ribosome subunits bound to the 5′ mRNA cap translocate directly to the downstream AUG start codon rather than via the usual process of ribosome scanning.30
Our data show that there are equal quantities of viral mRNA in human and murine cells following hAd5 infection, but significantly less is recruited to ribosomes in murine cells. Given that our data show adequate nuclear exit of viral mRNA in murine cells, the untranslated transcripts must sit within the murine cytoplasm, where they trigger PKR-mediated phosphorylation of eIF2α, which would tend to promote a global translation shutdown. Following in trans L4-100K expression, there is decreased eIF4E phosphorylation and an increased association of late mRNA with ribosomes. By inference, this lowers the cytosolic pool of viral mRNA, which reduces eIF2α phosphorylation and prevents global translation blockade. Thus L4-100K permits recruitment of viral mRNA to ribosomes, the specific translation of viral mRNA and appearance of structural viral proteins on immunoblot.
While L4-100K was able to increase production of late proteins, the rescue was not complete, and also the levels of late mRNA associated with heavy polysomal fractions remain low. We were unable to assess the phosphorylation status of L4-100K at Y365 and Y682 and the nature of the kinases that phosphorylate these residues is unclear.30 Multiple kinase pathways are activated in adenovirus-infected cells,31,32,33 but extracellular signal-related kinase (ERK) activity appears able to increase hAd5 viral protein expression in H1299 cells.34 Overall, it is possible that there is insufficient tyrosine phosphorylation in murine cells for maximum translation, potentially due to suboptimal ERK activity.
Inadequate phosphorylation may also explain another finding, namely that L4-100K did not greatly increase the number of infectious virion progeny: we were able to increase the number of infectious progeny above input MOI only following coinfection with dl922-947 and Ad 100K, but still not to the levels seen in human cells. Previously, inhibition of MEK (mitogen-activated protein kinase kinase)/ERK signaling in H1299 cells was shown to reduce virion production at least 100-fold.34 In addition, L4-100K undergoes arginine methylation under the influence of PRMT1,35 which promotes another important 100K function, namely interaction with Hexon monomers to assist their maturation to trimers and shuttling these trimers into the nucleus for incorporation into the capsid.36 It is noticeable that the expression of Hexon in tumors infected with both dl922-947 and Ad 100K did not show the same nuclear localization as seen in human xenografts infected with dl922-947 alone (see Figure 6d and compare Figure 7a with IGROV1 tumor in Figure 2d), suggesting that there may be insufficient arginine methylation in murine cells. Infectious virion production is a complex process, requiring proper capsid assembly as well as viral genome packaging, in which viral proteins L1 52/55K, IVa2 and IIIa play important roles.37,38 Further work will be required to dissect the roles of L4-100K phosphorylation and methylation as well as other viral proteins in human virion assembly in murine cells. Similarly, the mechanism of cell death following adenovirus infection remains unclear.39 We have shown previously that there is an incomplete relationship between cell death and infectious virion production and that host cell factors, including DNA damage responses may also contribute to cell sensitivity to virus-induced death.23,40 However, we show here that it is possible to increase viral cytotoxicity with only a modest increase in the number of infectious virions generated.
E1A, the first viral gene to be expressed, is an absolute prerequisite for productive infection. It drives infected cells into an S phase-like state required for viral genome replication. Although we found lower levels of E1A protein in murine cells compared to human, these levels were evidently adequate to permit viral DNA replication, as qPCR indicated equal quantities of viral DNA in murine and human cells. This lower level of E1A protein was also evident with wild-type Ad5 infection, so the 24 bp E1A CR2 deletion in dl922-947 was not responsible for any reduction in either E1A expression or productive infection. It was noticeable, however, that coinfection with dl922-947 and Ad 100K caused reduction in E1A protein expression compared to dl922-947 infection alone, but not gene transcription. This may reflect L4 100K-induced preferential translation of mRNA species that have 5′ TPL sequence (i.e., late viral genes) compared to early. This E1A inhibition was less evident with the cumate-inducible system, even though cumate generally generated more L4-100K than Ad 100K. Therefore, E1A suppression is unlikely to be a dose-effect of 100K, but may reflect inhibitory feedback from other viral genes expressed in Ad 100K.
In conclusion, we have identified that there is failure of translation of late human adenovirus mRNA in murine cells that can be partially overcome by in trans expression of L4-100K, which promotes loading of viral mRNA onto ribosomes. The possibility now opens that murine models, with coexpression of L4-100K, could permit evaluation of oncolytic adenovirus activity in purely murine systems.
Materials and Methods
Cell lines, viruses, and plasmids. IGROV1 and OVCAR4 are human ovarian carcinoma cell lines. HeLa cells were obtained from Cancer Research UK Cell Services (Herts, UK). All three lines were authenticated by 16 locus STR analysis (LGC Standards, Middlesex, UK) and discarded after every eighth passage. MOSEC/ID8 cells are murine ovarian surface epithelial cells that underwent transformation after in vitro culture in epidermal growth factor, and were obtained from Dr Katherine Roby, University of Kansas.41 MOVCAR7 and MOVCAR12 cells, kind gifts from Dr Denise Connolly (Fox Chase Cancer Center), were derived from tumors that arose in mice chimeric for the SV40 Tag under the control of the MISIIR promoter.42 CMT64 and CMT93 were both obtained from Cancer Research UK Cell Services. CMT64 was isolated from a primary alveogenic lung carcinoma tumor mass in C57Bl/lcrf mouse,43 while CMT93 is a line derived from a chemically induced rectal carcinoma also from C57Bl/lcrf.44 IGROV1, OVCAR4, HeLa, CMT64, and CMT93 were maintained in Dulbecco's modified Eagle's medium supplemented with penicillin and streptomycin and 10% FCS, (PAA Laboratories, Pasching, Austria). ID8, MOVCAR7, and MOVCAR12 were maintained in Dulbecco's modified Eagle's medium plus 4% FCS together with 1% transferrin and selenium (Invitrogen, Paisley, UK).
dl922-947 and dl309 are both hAd5 vectors deleted in E3B. In addition, dl922-947 is also deleted in amino acids 122–129 of E1A-CR2. Ad GFP is an E1-deleted hAd5 vector containing GFP in the E1 position under the control of the CMV immediate early promoter. Ad LM-X is an E1-deleted hAd5 vector with no transgene in the E1 region as described previously.45 MAV-1 was kindly provided by Dr Katherine Spindler (University of Michigan, Ann Arbor, MI). Viral particle counts were quantified using A260,46 while infectious titer was measured using TCID50 assay. The particle:infectivity ratio for the dl922-947 stock used in all experiments presented was 13.
Plasmid pCMV 100K Flag was kindly donated by Dr Keith Leppard (University of Warwick, Warwick, UK). The virus Ad 100K was generated by PCR amplification of the L4-100K open-reading frame from pCMV 100K Flag (forward 5′-gtcgacatggagtcagtcgagaagaaggacagcc, reverse 5′-gtcgacctacggtt gggtcggcgaacgggc, SalI recognition sequence underlined). After sequencing confirmation, L4 100K was excised from pCRII-Blunt-TOPO (Invitrogen) as a SalI fragment and ligated into pShuttle CMV, followed by recombination with pAdEasy in electrocompetent Escherichia coli as described previously.47 Following transfection into 293 cells, recombinant vector was amplified and purified by double density CsCl ultracentrifugation.
Inducible L4-100K gene expression was obtained using SparQ cumate switch (Systems Biosciences, Mountain View, CA). L4-100K was excised from pCMV 100K Flag and ligated into pCDH-CuO-IRES-copGFP. CMT64 and ID8 cells were first transduced with pCDH-EF1-CymR-T2A-Puro and cells selected with puromycin (500 ng/ml, 10 days). pCDH-CuO-100K-IRES-copGFP construct was packaged into lentiviral particles using ViraPower packaging mix (Invitrogen). Cells were then treated with cumate (50 µg/ml, 3 days) before cell sorting using ARIAII Cell Sorter (Beckton Dickinson, Franklin Lakes, NJ). When cells were infected with dl922-947, cumate (200 µg/ml) was added 4 hours before infection and again 2 hours postinfection.
Cell viability and infectivity assays. 2 × 104 cells were infected in serum-free medium at MOI 0.001–10,000 pfu/cell. After 2 hours, cells were re-fed with medium containing 5% FCS. Cell viability was assayed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay using a Victor3 plate reader (Perkin Elmer, Beaconsfield, UK). All viability assays were done in triplicate and experiments repeated at least twice. For infectivity, 105 cells were infected with Ad GFP (MOI 5 and 50 pfu/cell)—GFP fluorescence was assessed 24 hours postinfection on a FACS Caliber (Becton Dickinson, Oxford, UK) and analyzed using CellQuest Software (Becton Dickinson).
qPCR and TCID50 assays. Real-time PCR was performed on an ABI Prism 7700 (Applied Biosystems, Foster City, CA). Oligonucleotides and probes are as below:
E1A: sense primer: 5′-CCACCTACCCTTCACGAACTG; antisense primer: 5′-GCCTCCTCGTTGGGATCTTC; probe: ATGATTTAGACGT GACGGCC. Hexon: sense 5′-AGCGCGCGAATAAACTGCT; antisense 5′-AGGAGACCACTGCCATGTTGT; probe 5′-CCGCCGCTCCGTCC TGCA. L4-100K: sense 5′-TGGAGTGTGTCACTGTCGGCTGC; antisense 5′-CTGCGAATTGCAAACCAGG; probe: 5′-ACCTATGCACCCCGC ACCGCT.
PCR conditions were: 50 °C for 2 minutes, 95 °C for 10 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 60 seconds. Where stated, a standard curve using 103–109 dl922-947 genomes was used for quantification. Samples were also analyzed for 18S RNA as internal quality control. For TCID50 assays, 105 cells were infected. Cells were harvested into 0.5 ml 0.1 mol/l Tris pH 8.0 and subjected to three rounds of freeze/thawing (liquid N2/37 °C), after which they were centrifuged. The supernatant was titered on JH293 cells by serial dilution.
Nuclear and cytoplasmic subcellular fractionation. Cells were fractionated into nuclear and cytoplasmic compartments using a Protein And RNA Isolation System (PARIS; Ambion, Cambridgeshire, UK) 48 hours after infection. Nuclear and cytoplasmic RNA samples were then evaluated using qRT-PCR. Subcellular sample purity was confirmed by immunoblot (lamin for nuclear fraction, β-tubulin for cytoplasmic).
Polysome assays. Virus-infected cells were treated with cycloheximide (100 µg/ml; Sigma, Poole, UK) for 5 minutes, scraped into cold phosphate-buffered saline (PBS) supplemented with cycloheximide, pelleted and lysed in lysis buffer (300 mmol/l NaCl, 15 mmol/l MgCl2, 15 mmol/l Tris-HCl, 100 µg/ml cycloheximide, 1 mg/ml heparin, and 1% Triton-X). Samples were centrifuged and supernatants subjected to 10–60% sucrose gradient density ultracentrifugation in order to separate ribosomes into 11 fractions ranging from subpolysomal to heavy polysomal fractions with continuous monitoring at 254 nm. TRIzol (Invitrogen) was added to each fraction and RNA isolated according to manufacturer's instructions and quantified. RNA was cleaned up using RNeasy (Qiagen, Crawley, UK), and pools of subpolysomal, light, and heavy polysomal fractions made according to the 254 nm trace. Pooled RNA was precipitated using LiCl (5 mol/l, Sigma) and then with NaOAc (3 mol/l, Sigma) before treatment with AMV reverse transcriptase (Roche, Welwyn, UK). Ten nanogram cDNA was then used for qPCR as described above for hexon and β-actin.
Immunoblotting and Coomassie staining. Protein lysates were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels and transferred onto nitrocellulose membranes by semi-dry blotting. Antibody binding was visualized using enhanced chemiluminescence (GE Healthcare, Buckinghamshire, UK). Antibodies were obtained as follows: E1A (Santa Cruz Biotechnology, Heidelberg, Germany), anti-adenovirus (Abcam, Cambridge, UK), eIF2α and phospho-eIF2α, eIF4E and phospho-eIF4E (all Cell Signaling, Beverly, MA), Ku70, actin, and PCNA (all Santa Cruz Biotechnology, Santa Cruz, CA). The rabbit anti-Ad5 L4-100K Ab was kindly provided by Dr William Russell (University of St Andrews, St Andrews, UK). For Coomassie staining, cells were infected with dl922-947 (MOI 10) for up to 72 hours; 20 µg protein was run on an 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel, fixed in isopropanol/acetic acid at room temperature for 30–60 minutes and stained overnight with Rapid Coomassie blue (10% acetic acid, 0.006% Coomassie Brilliant Blue R-250; Bio-Rad, Hercules, CA) overnight at room temperature, followed by destaining in 10% acetic acid.
Immunofluorescence. 2 × 105 cells were grown overnight on coverslips, infected with dl922-947 (MOI 10), Ad 100K (MOI 10) alone or together. Forty-eight hours later, cells were fixed in 3.7% formaldehyde, permeabilized in 0.5% Triton in PBS for 10 minutes. Primary anti-adenovirus antibody (BD Biosciences, Oxford, UK) was added at 1:2,000 in 3% BSA, 0.2% Triton in PBS for 1 hour. After secondary Alexa488-conjugated secondary antibody, cells were stained with DAPI (4′,6-diamidino-2-phenylindole) (1:10,000) for 1 minute and viewed on a Zeiss LSM510 confocal microscope (×63 objective).
In vivo experiments. All experiments were performed under suitable UK Home Office personal and project licence authority. These studies were also reviewed and approved by the ethical review board of the Biological Services Unit, Queen Mary University of London, London, UK. For intraperitoneal experiments, 5 × 106 cells were injected intraperitoneally. When ascites was clinically evident, adenovirus was injected intraperitoneally in 400 µl 20% icodextrin. For subcutaneous experiments, 5 × 106 cells were injected subcutaneously in 100 µl PBS. Once tumors reached ~7 mm in maximum diameter, virus was injected intratumorally in 20 µl PBS. Mice were killed 24–96 hours after virus injection. Tumors were either fixed in 4% paraformaldehyde or snap-frozen in liquid nitrogen. Fixed tumors were transferred to ice cold 70% ethanol after 24 hours, 4 µm sections were cut and processed. Adenovirus structural protein expression was detected using a rabbit anti-Adenovirus Ab (AbCam, Cambridge, UK). Total RNA was extracted from snap-frozen tumors using TRizol. After DNAse I digestion, 1 µg total RNA was reverse transcribed using random hexonucleotide primers (First-strand synthesis kit; Roche, Welwyn, UK) and analyzed by qPCR.
SUPPLEMENTARY MATERIAL Figure S1. 104 human and murine ovarian cancer cells were infected with Ad5 WT in triplicate. Figure S2. CMT64 cells were infected with dl922-947 (MOI 10) for 48 hours. Figure S3. IGROV1 (IGR) and ID8 cells were infected with dl922-947 (MOI 10). Figure S4. OVCAR4, CMT64, and ID8 cells were infected with dl922-947 (MOI 10) or mock-infected. Figure S5. 2 × 104 cells were infected with mouse adenovirus 1 (MAV1) in triplicate. Figure S6. OVCAR4, ID8, MOVCAR7, and CMT64 cells were infected with dl922-947 (MOI 10); protein was harvested 48 hours postinfection and expression of L4-100K assessed by immunoblot. Figure S7. ID8 CuO-100K or control cells were infected with dl922-947 (MOI 10) in the presence and absence of cumate (200 µg/ml). Figure S8. CMT64 and ID8 cells were mock-infected, or infected with Ad 100K (MOI 1), dl922-947 (MOI 10) alone and together for 48 hours. Figure S9. 104 ID8 cells were infected with dl922-947 in triplicate.
Acknowledgments
This work is supported by the Medical Research Council (grant reference G0601891) and Cancer Research UK (Barts CRUK Centre, grant reference C236/A11795). We thank Mohammed Ikram for assistance with immunohistochemistry. The authors declared no conflict of interest.
Supplementary Material
104 human and murine ovarian cancer cells were infected with Ad5 WT in triplicate.
CMT64 cells were infected with dl922-947 (MOI 10) for 48 hours.
IGROV1 (IGR) and ID8 cells were infected with dl922-947 (MOI 10).
OVCAR4, CMT64, and ID8 cells were infected with dl922-947 (MOI 10) or mock-infected.
2 × 104 cells were infected with mouse adenovirus 1 (MAV1) in triplicate.
OVCAR4, ID8, MOVCAR7, and CMT64 cells were infected with dl922-947 (MOI 10); protein was harvested 48 hours postinfection and expression of L4-100K assessed by immunoblot.
ID8 CuO-100K or control cells were infected with dl922-947 (MOI 10) in the presence and absence of cumate (200 µg/ml).
CMT64 and ID8 cells were mock-infected, or infected with Ad 100K (MOI 1), dl922-947 (MOI 10) alone and together for 48 hours.
104 ID8 cells were infected with dl922-947 in triplicate.
REFERENCES
- Fattaey AR, Harlow E., and, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes A, Williams A.et al. (2000An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy Nat Med 61134–1139. [DOI] [PubMed] [Google Scholar]
- Lockley M, Fernandez M, Wang Y, Li NF, Conroy S, Lemoine N.et al. (2006Activity of the adenoviral E1A deletion mutant dl922-947 in ovarian cancer: comparison with E1A wild-type viruses, bioluminescence monitoring, and intraperitoneal delivery in icodextrin Cancer Res 66989–998. [DOI] [PubMed] [Google Scholar]
- Sherr CJ., and, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- D'Andrilli G, Kumar C, Scambia G., and, Giordano A. Cell cycle genes in ovarian cancer: steps toward earlier diagnosis and novel therapies. Clin Cancer Res. 2004;10:8132–8141. doi: 10.1158/1078-0432.CCR-04-0886. [DOI] [PubMed] [Google Scholar]
- TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball KJ, Preuss MA, Barnes MN, Wang M, Siegal GP, Wan W.et al. (2010A phase I study of a tropism-modified conditionally replicative adenovirus for recurrent malignant gynecologic diseases Clin Cancer Res 165277–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P.et al. (2004Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival Nat Med 10942–949. [DOI] [PubMed] [Google Scholar]
- Clarke B, Tinker AV, Lee CH, Subramanian S, van de Rijn M, Turbin D.et al. (2009Intraepithelial T cells and prognosis in ovarian carcinoma: novel associations with stage, tumor type, and BRCA1 loss Mod Pathol 22393–402. [DOI] [PubMed] [Google Scholar]
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F.et al. (2005Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer Proc Natl Acad Sci USA 10218538–18543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher A, Parato K, Rooney CM., and, Bell JC. Thunder and lightning: immunotherapy and oncolytic viruses collide. Mol Ther. 2011;19:1008–1016. doi: 10.1038/mt.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee EG, Blattman JN, Kasturi SP, Kelley RP, Kaufman DR, Lynch DM.et al. (2011Multiple innate immune pathways contribute to the immunogenicity of recombinant adenovirus vaccine vectors J Virol 85315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leen AM, Sili U, Vanin EF, Jewell AM, Xie W, Vignali D.et al. (2004Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells Blood 1042432–2440. [DOI] [PubMed] [Google Scholar]
- Olive M, Eisenlohr L, Flomenberg N, Hsu S., and, Flomenberg P. The adenovirus capsid protein hexon contains a highly conserved human CD4+ T-cell epitope. Hum Gene Ther. 2002;13:1167–1178. doi: 10.1089/104303402320138952. [DOI] [PubMed] [Google Scholar]
- Tang J, Olive M, Pulmanausahakul R, Schnell M, Flomenberg N, Eisenlohr L.et al. (2006Human CD8+ cytotoxic T cell responses to adenovirus capsid proteins Virology 350312–322. [DOI] [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H.et al. (2008HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis Lancet 3721894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MA, Spencer JF, La Regina MC, Dhar D, Tollefson AE, Toth K.et al. (2006Syrian hamster as a permissive immunocompetent animal model for the study of oncolytic adenovirus vectors Cancer Res 661270–1276. [DOI] [PubMed] [Google Scholar]
- Guida JD, Fejer G, Pirofski LA, Brosnan CF., and, Horwitz MS. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J Virol. 1995;69:7674–7681. doi: 10.1128/jvi.69.12.7674-7681.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair GE, Dixon SC, Griffiths SA., and, Zajdel ME. Restricted replication of human adenovirus type 5 in mouse cell lines. Virus Res. 1989;14:339–346. doi: 10.1016/0168-1702(89)90026-9. [DOI] [PubMed] [Google Scholar]
- Ganly I, Mautner V., and, Balmain A. Productive replication of human adenoviruses in mouse epidermal cells. J Virol. 2000;74:2895–2899. doi: 10.1128/jvi.74.6.2895-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KP., and, Klessig DF. Altered mRNA splicing in monkey cells abortively infected with human adenovirus may be responsible for inefficient synthesis of the virion fiber polypeptide. Proc Natl Acad Sci USA. 1984;81:4023–4027. doi: 10.1073/pnas.81.13.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KP, Wong EA., and, Klessig DF. Microinjection of mRNA enhances translational efficiency of human adenovirus fiber message in monkey cells. Mol Cell Biol. 1985;5:2870–2873. doi: 10.1128/mcb.5.10.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell CM, Shibata A, Tookman LA, Archibald KM, Flak MB, Pirlo KJ.et al. (2011Genomic DNA damage and ATR-Chk1 signaling determine oncolytic adenoviral efficacy in human ovarian cancer cells J Clin Invest 1211283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajewski J, Schneider RJ, Safer B, Munemitsu SM, Samuel CE, Thimmappaya B.et al. (1986Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase Cell 45195–200. [DOI] [PubMed] [Google Scholar]
- Mullick A, Xu Y, Warren R, Koutroumanis M, Guilbault C, Broussau S.et al. (2006The cumate gene-switch: a system for regulated expression in mammalian cells BMC Biotechnol 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q., and, Schneider RJ. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J Virol. 2004;78:7707–7716. doi: 10.1128/JVI.78.14.7707-7716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R., and, Schneider RJ. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R, Xi Q., and, Schneider RJ. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 2000;19:3465–3474. doi: 10.1093/emboj/19.13.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BW, Telling GC, Myat MM, Williams JF., and, Flint SJ. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R., and, Schneider RJ. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J Virol. 2005;79:5676–5683. doi: 10.1128/JVI.79.9.5676-5683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M, Nakano MY, Boucke K, Keller S., and, Greber UF. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001;20:1310–1319. doi: 10.1093/emboj/20.6.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, White LR, Clark SA, Heffner DJ, Winston BW, Tibbles LA.et al. (2005Akt/protein kinase B activation by adenovirus vectors contributes to NFkappaB-dependent CXCL10 expression J Virol 7914507–14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala MS, Rajala RV, Astley RA, Butt AL., and, Chodosh J. Corneal cell survival in adenovirus type 19 infection requires phosphoinositide 3-kinase/Akt activation. J Virol. 2005;79:12332–12341. doi: 10.1128/JVI.79.19.12332-12341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümann M., and, Dobbelstein M. Adenovirus-induced extracellular signal-regulated kinase phosphorylation during the late phase of infection enhances viral protein levels and virus progeny. Cancer Res. 2006;66:1282–1288. doi: 10.1158/0008-5472.CAN-05-1484. [DOI] [PubMed] [Google Scholar]
- Koyuncu OO., and, Dobner T. Arginine methylation of human adenovirus type 5 L4 100-kilodalton protein is required for efficient virus production. J Virol. 2009;83:4778–4790. doi: 10.1128/JVI.02493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SS, Szolajska E, Schoehn G, Franqueville L, Myhre S, Lindholm L.et al. (2005The 100K-chaperone protein from adenovirus serotype 2 (Subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses J Mol Biol 352125–138. [DOI] [PubMed] [Google Scholar]
- Hearing P., and, Shenk T. The adenovirus type 5 E1A enhancer contains two functionally distinct domains: one is specific for E1A and the other modulates all early units in cis. Cell. 1986;45:229–236. doi: 10.1016/0092-8674(86)90387-9. [DOI] [PubMed] [Google Scholar]
- Christensen JB, Byrd SA, Walker AK, Strahler JR, Andrews PC., and, Imperiale MJ. Presence of the adenovirus IVa2 protein at a single vertex of the mature virion. J Virol. 2008;82:9086–9093. doi: 10.1128/JVI.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SK, Aerts JL, Eddaoudi A, Lockley M, Lemoine NR., and, McNeish IA. Oncolytic adenoviral mutants induce a novel mode of programmed cell death in ovarian cancer. Oncogene. 2008;27:3081–3090. doi: 10.1038/sj.onc.1210977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flak MB, Connell CM, Chelala C, Archibald K, Salako MA, Pirlo KJ.et al. (2010p21 Promotes oncolytic adenoviral activity in ovarian cancer and is a potential biomarker Mol Cancer 9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby KF, Taylor CC, Sweetwood JP, Cheng Y, Pace JL, Tawfik O.et al. (2000Development of a syngeneic mouse model for events related to ovarian cancer Carcinogenesis 21585–591. [DOI] [PubMed] [Google Scholar]
- Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X.et al. (2003Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer Cancer Res 631389–1397. [PubMed] [Google Scholar]
- Franks LM, Carbonell AW, Hemmings VJ., and, Riddle PN. Metastasizing tumors from serum-supplemented and serum-free cell lines from a C57BL mouse lung tumor. Cancer Res. 1976;36:1049–1055. [PubMed] [Google Scholar]
- Franks LM., and, Hemmings VJ. A cell line from an induced carcinoma of mouse rectum. J Pathol. 1978;124:35–38. doi: 10.1002/path.1711240108. [DOI] [PubMed] [Google Scholar]
- McNeish IA, Tenev T, Bell S, Marani M, Vassaux G., and, Lemoine N. Herpes simplex virus thymidine kinase/ganciclovir-induced cell death is enhanced by co-expression of caspase-3 in ovarian carcinoma cells. Cancer Gene Ther. 2001;8:308–319. doi: 10.1038/sj.cgt.7700305. [DOI] [PubMed] [Google Scholar]
- Mittereder N, March KL., and, Trapnell BC. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW., and, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
104 human and murine ovarian cancer cells were infected with Ad5 WT in triplicate.
CMT64 cells were infected with dl922-947 (MOI 10) for 48 hours.
IGROV1 (IGR) and ID8 cells were infected with dl922-947 (MOI 10).
OVCAR4, CMT64, and ID8 cells were infected with dl922-947 (MOI 10) or mock-infected.
2 × 104 cells were infected with mouse adenovirus 1 (MAV1) in triplicate.
OVCAR4, ID8, MOVCAR7, and CMT64 cells were infected with dl922-947 (MOI 10); protein was harvested 48 hours postinfection and expression of L4-100K assessed by immunoblot.
ID8 CuO-100K or control cells were infected with dl922-947 (MOI 10) in the presence and absence of cumate (200 µg/ml).
CMT64 and ID8 cells were mock-infected, or infected with Ad 100K (MOI 1), dl922-947 (MOI 10) alone and together for 48 hours.
104 ID8 cells were infected with dl922-947 in triplicate.