Roughly two decades after its discovery and cloning, hepatitis C virus (HCV)—a pathogen infecting up to 180 million people worldwide and capable of causing severe liver disease, including cirrhosis and hepatocellular carcinoma—remains a frustrating target for development of antiviral therapies. Two articles in this issue of Molecular Therapy now report combinatorial RNA interference (coRNAi) strategies1 against multiple HCV genotypes that aim to balance in vivo safety, efficiency, and specificity, and that promise to restrict viral escape mutants. In the first study, Chandra et al.2 used lipid nanoparticles called nanosomes to encapsulate two preselected potent anti-HCV small interfering RNAs (siRNAs), and show that six daily combined systemic injections sufficed to significantly block HCV replication in a liver xenograft mouse model. In the second report, Suhy et al.3 optimized an adeno-associated viral (AAV) vector of serotype 8 for robust and safe expression of three short hairpin RNAs (shRNAs) targeting conserved regions of the HCV genome, and demonstrate its sustained performance in the livers of nonhuman primates (NHPs) following a single intravenous injection. By validating the concept of nonviral- or viral vector–mediated coRNAi against rapidly evolving targets in different animal models, the studies represent an important step forward in our efforts to translate newer vector and RNAi designs into clinical therapies for HCV.
HCV is a prototype RNA virus characterized by short generation times, inaccurate reproduction, and intrahost recombination, and it is therefore equipped with a strong advantage in the molecular genetic arms race with its human host. The virus comprises seven genotypes, more than 50 subtypes, and billions of quasi species, presenting tremendous difficulties for both the immune system and ectopic therapies. Accordingly, current treatments for HCV infection are ineffective (pegylated interferon-α and ribavirin fail in up to 60% of patients), creating an urgent need for newer approaches such as coRNAi. In 2002, Kay's lab demonstrated proof of concept for potent silencing of an HCV NS5B-luciferase fusion in livers of adult mice via hydrodynamically injected siRNAs or shRNA expression vectors.4 Concomitantly, others identified and validated additional siRNA or shRNA targets in the HCV genome, especially the highly conserved 5′ untranslated region (5′ UTR). However, it also soon became clear that a single RNAi target would ultimately fail to control the virus because of its propensity for rapid mutational escape. This was confirmed by Wilson and Richardson,5 who deliberately bred escape mutants via five successive electroporations of HCV replicon cells with a single anti-HCV siRNA. Notably, spiking in a second siRNA targeting a distinct HCV sequence dramatically diminished the emergence of resistant replicons, demonstrating the potential of coRNAi to counter rapidly mutating targets. Many other labs then also used multiple independent siRNAs or shRNAs to inhibit HCV transcripts, combined RNAi with other modes of gene silencing, or cotargeted cellular HCV cofactors assuming that these are less prone to mutation than the virus itself.6,7,8,9,10,11,12
Importantly, the two reports in this issue address several challenges and concerns that have persisted despite this promising previous work (Figure 1). These reflect the difficulties of in vivo delivery and the need to validate the efficacy, specificity, and safety of coRNAi strategies in small and large animal models of HCV infection. Chandra et al. made use of lipid nanoparticle carriers dubbed nanosomes based on cholesterol and DOTAP encapsulation of protamine sulfate–condensed siRNAs at an optimized lipid-to-siRNA ratio.2 The formulation was sonicated to reduce the particle size to 100 nm so as to prevent capillary clogging while permitting the nanosomes to cross the endothelial barrier to reach the HCV-infected hepatocytes. In human hepatoma cells, the siRNA–nanosomes achieved close to 100% transduction for over 7 days and induced only marginal cytotoxicity. Notably, although monotreatment of HCV replicon cells with a single type of siRNA–nanosome led to rapid emergence of escape mutants, combinatorial application of two types of siRNA–nanosomes cleared the culture of HCV for up to 60 days, and consistent data were reported in an infectious HCV cell culture system.2 siRNA–nanosome delivery and HCV inhibition were also very efficient in mice. In a subcutaneous tumor xenograft model of HCV infection, six peritumoral siRNA–nanosome injections every other day resulted in an approximately three-log drop in HCV RNA. Lower but still notable efficiency—a roughly two-log drop in HCV RNA—was also demonstrated in a second xenograft model in which replicon cells were injected intrasplenically to form liver tumors, followed a month later by six consecutive daily intravenous siRNA–nanosome infusions at 5 mg/kg body weight. The lack of complete HCV clearance in this model was shown not to be due to escape mutations; instead, the authors suspected suboptimal siRNA dosing. Finally, the authors assessed toxicity in 35 mice up to 1 week after the last of three siRNA–nanosome injections. They found no changes in body weight or histopathology and only minor elevations of serum transaminases, altogether implying that the formulation was well tolerated.
Figure 1.
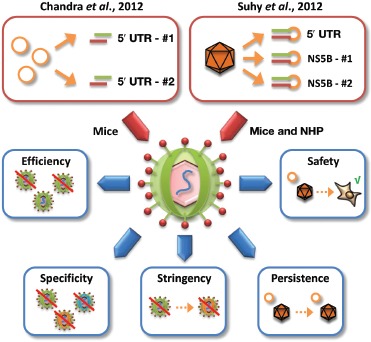
Two new coRNAi strategies against HCV. Represented at top are the two new coRNAi approaches reported in this issue, comprising either nanosome-mediated codelivery of two siRNAs against two regions within the HCV 5′ UTR (left box) or AAV8-mediated coexpression of three independent shRNAs against the HCV 5′ UTR or NS5B (two different target sites) (right box). The blue boxes show the critical parameters and aspects that must be considered in the context of coRNAi strategies and that were, accordingly, addressed in the two new HCV studies. They include (counterclockwise from left) the efficiency of target knockdown, the specificity against multiple naturally occurring viral variants, the stringency with which the coRNAi strategy can suppress resistant mutants, the persistence of the nonviral or viral vectors and hence the duration of target knockdown, and last, but not least, the safety of the vector–RNAi formulation in the liver and in potential off-target organs. 5′ UTR, 5′ untranslated region; coRNAi, combinatorial RNA interference; HCV, hepatitis C virus; NHP, nonhuman primate.
Suhy et al.3 engineered a viral vector based on AAV8, an AAV serotype with a high efficiency of hepatic transduction, to achieve potent in vivo codelivery of three shRNAs against conserved HCV regions. In a prior study,13 this vector potently inhibited HCV replicons of genotype 1b and, to a lesser extent due to reduced shRNA homology, also of genotype 2a. Congruent with the Chandra data and supporting the coRNAi concept, serial replicon treatment with a single shRNA yielded resistant HCV point mutants that could still be inhibited with the other two shRNAs. Moreover, the group tested a panel of 18 clinical isolates of common genotype 1 variants and found that more than 80% were inhibited by all three shRNAs, underscoring their vector's potential to widely suppress HCV. Most remarkable in the study by Suhy et al. are the data on vector performance in livers of cynomolgus monkeys, representing the first extensive analysis of a viral coRNAi vector in adult NHPs. The authors found nearly 100% hepatocyte transduction, equal vector distribution over all three liver lobes, and stable shRNA delivery for up to 180 days following one intravenous dose of 1.25 × 1012 vector genomes per kilogram of body weight. Also notably, a biodistribution analysis comprising more than 30 tissues showed that about 90% of the vector localized to the liver.
Finally, Suhy et al.3 assessed in vivo toxicity after a single AAV8 injection in 18 monkeys and found mild transaminase elevations only in the high-dose group (6.25 × 1012 genomes/kg body weight), the causes of which remained unclear. Histopathological samples showed no signs of hepatotoxicity, and vector safety was validated in a parallel mouse study. Curiously, the results strikingly differed for another vector encoding stronger promoters, which caused rapid and severe liver toxicity, culminating in morbidity in one high-dose monkey. Affected animals also showed continuous drops in shRNA and vector copy numbers that were probably due to cell death and liver repopulation. Lowering vector doses was not useful to avert toxicity, as it would have left a portion of the hepatocytes untreated and thus vulnerable to HCV re-infection. Instead, the authors engineered their promoters to mediate lower shRNA levels still able to inhibit HCV, resulting in the “safe” second-generation vector used in the other studies described above. These findings confirm and extend prior observations in murine livers and the central nervous system14,15,16,17 and are important not only because they represent the first report of dose-dependent RNAi toxicity in NHPs but also because they concurrently imply that this type of adverse event may be conserved in humans.
Bühler and Bartenschlager18 recently proposed a set of criteria that an ideal future therapy of chronic hepatitis C should (i) be free of interferon to reduce side effects, (ii) impose a high barrier of drug resistance, (iii) require only short treatment durations, and (iv) provide >90% sustained viral response. Clearly, both of the “all (siRNAs or shRNAs) for one (target), one (vector) for all (HCV isolates)” strategies in the two new Molecular Therapy articles have the potential to comply with at least three of these criteria. Notwithstanding these promises, a central aim for follow-up work must be to complete the characterization of the new vectors. Arguably, this should include a demonstration that the siRNA–nanosome is truly selective for hepatocytes, efficient and nontoxic in vivo also for prolonged periods of time, and under fewer, and thus more tolerable, infusion regimens. These vital aspects must be thoroughly analyzed—ideally, in chronically infected chimpanzees as the best available preclinical model of HCV infection—before one can rigorously assess the therapeutic potential of this approach in humans. Until then, the work by Chandra and colleagues provides a promising proof of concept that effective HCV inhibition in mammalian livers is feasible with systemically administered nonviral vectors. Likewise, it is necessary to confirm that the AAV–shRNA vector not only is safe in livers but also able to potently and stably suppress HCV gene expression and replication in vivo as well as, ideally, the evolution of resistance. Optimism is fueled by data from Yang et al.,19 who recently reported a similar vector expressing multiple anti-HCV RNAi triggers and demonstrated more than 90% inhibition of HCV reporters in murine livers, in the absence of hepatotoxicity. In this light, the two new studies are certainly welcome, compelling, and promising additions to our arsenal of therapeutic options that can be combined with the latest direct-acting antivirals and that may help to finally tip the balance of power between HCV and its human host in our favor.
REFERENCES
- Grimm D., and, Kay MA. Combinatorial RNAi: a winning strategy for the race against evolving targets. Mol Ther. 2007;15:878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra PK, Kundu AK, Hazari S, Chandra S, Bao L, Ooms T.et al. (2012Inhibition of hepatitis C virus replication by intracellular delivery of multiple siRNAs by nanosomes Mol Ther 201724–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy DA, Kao S-C, Mao T, Whiteley L, Denise H, Souberbielle B.et al. (2012Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model Mol Ther 201737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ., and, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Wilson JA., and, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronke J, Kittler R, Buchholz F, Windisch MP, Pietschmann T, Bartenschlager R.et al. (2004Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs J Virol 783436–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K.et al. (2006Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype Gene Ther 13883–892. [DOI] [PubMed] [Google Scholar]
- Shin D, Lee H, Kim SI, Yoon Y., and, Kim M. Optimization of linear double-stranded RNA for the production of multiple siRNAs targeting hepatitis C virus. RNA. 2009;15:898–910. doi: 10.1261/rna.1268209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczak D, Korf M, Beger C, Manns MP., and, Kruger M. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J. 2005;272:5910–5922. doi: 10.1111/j.1742-4658.2005.04986.x. [DOI] [PubMed] [Google Scholar]
- Korf M, Jarczak D, Beger C, Manns MP., and, Kruger M. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol. 2005;43:225–234. doi: 10.1016/j.jhep.2005.02.046. [DOI] [PubMed] [Google Scholar]
- Jahan S, Khaliq S, Samreen B, Ijaz B, Khan M, Ahmad W.et al. (2011Effect of combined siRNA of HCV E2 gene and HCV receptors against HCV Virol J 8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ., and, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Lavender H, Brady K, Burden F, Delpuech-Adams O, Denise H, Palmer A.et al. (2012In vitro characterization of the activity of PF-05095808, a novel biological agent for hepatitis C virus therapy Antimicrob Agents Chemother 561364–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR.et al. (2006Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways Nature 441537–541. [DOI] [PubMed] [Google Scholar]
- Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K.et al. (2010Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver J Clin Invest 1203106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I.et al. (2008Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi Proc Natl Acad Sci USA 1055868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler S., and, Bartenschlager R.2012New targets for antiviral therapy of chronic hepatitis C Liver Int 32Suppl 1: 9–16. [DOI] [PubMed] [Google Scholar]
- Yang X, Haurigot V, Zhou S, Luo G, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877–1887. doi: 10.1002/hep.23908. [DOI] [PubMed] [Google Scholar]


