Abstract
Squamous cell carcinoma of the head and neck (SCCHN) is a leading cause of cancer deaths worldwide. Epidermal growth factor receptor (EGFR), an upstream mediator of signal transducer and activator of transcription (STAT)-3 is overexpressed in a variety of cancers, including SCCHN. Therapies such as monoclonal antibodies and tyrosine kinase inhibitors targeting EGFR have demonstrated limited antitumor efficacy, which may be explained, in part, by persistent STAT3 activation despite EGFR inhibition. STAT3 activation induces expression of target genes in SCCHN, including Bcl-XL, a mediator of antiapoptotic activity. Bcl-XL is commonly overexpressed in SCCHN where it correlates with chemoresistance, making it a potential therapeutic target. Targeting the EGFR-STAT3-Bcl-XL pathway at several levels, including the upstream receptor, the intracellular transcription factor, and the downstream target gene, has not been investigated previously. Using erlotinib, an EGFR-specific reversible tyrosine kinase inhibitor in combination with a STAT3 transcription factor decoy, we found enhanced antitumor effects in vitro and in vivo. The combination of the STAT3 decoy and gossypol, a small molecule targeting Bcl-XL, also yielded enhanced inhibition of cell proliferation. The triple combination of erlotinib, STAT3 decoy, and gossypol further enhanced cell growth inhibition and apoptosis in vitro, and it down-regulated signaling molecules further downstream of the EGFR-STAT3 signaling pathway, such as cyclin D1. These results suggest that combined targeting of several components of an oncogenic signaling pathway may be an effective therapeutic strategy for SCCHN.
Approximately 500,000 cases of squamous cell carcinoma of the head and neck (SCCHN) are diagnosed annually worldwide, and they account for approximately 3% of all cancers in the United States. SCCHN, an epithelial malignancy that affects the upper aerodigestive tract mucosa, has been linked to chronic tobacco and alcohol use. Conventional therapeutic strategies including surgery, chemotherapy, and radiation are effective in only 50% of cases, underscoring the need for new approaches to treat this malignancy.
Recent studies have focused on combining inhibitors that target several molecules in a single signaling pathway known to contribute to cancer progression to enhance antitumor efficacy. Epidermal growth factor receptor (EGFR) overexpression has been detected in a variety of human malignancies, including SCCHN in which expression levels in the tumor are correlated with decreased patient survival (Rubin Grandis et al., 1998; Ang et al., 2002). Signal transducer and activator of transcription (STAT)-3 is activated downstream of EGFR in SCCHN, and studies have demonstrated a role for STAT3 as an oncogene (Bromberg et al., 1998; Turkson et al., 1998). Constitutive activation of STAT3 has been detected in many cancers, including multiple myeloma, leukemia, lymphoma, prostate, breast, pancreas, lung, ovary, as well as SCCHN. A key downstream target of STAT3 is the gene encoding Bcl-XL, an antiapoptotic member of the Bcl-2 protein family. Overexpression of Bcl-XL has been reported in a majority of SCCHN, and it correlates with resistance to chemotherapy (Trask et al., 2002).
We previously demonstrated the feasibility of using a double-stranded deoxynucleotide transcription factor decoy to target activated STAT3, and we showed that the STAT3 decoy exhibited antitumor effects in SCCHN preclinical models, both alone and in combination with cytotoxic chemotherapy (Ahonen et al., 2003; Leong et al., 2003). The decoy binds to STAT3, abrogating its ability to bind to DNA response elements and induce transcription of target genes, resulting in decreased proliferation and increased apoptosis. To date, no STAT3 targeting strategy has been approved for the treatment of cancer. In this study, we investigated the antitumor efficacy of combining the STAT3 decoy with the tyrosine kinase inhibitor erlotinib (OSI-774; Tarceva), the negative enantiomer of gossypol (AT-101), or both, in preclinical models of SCCHN. Erlotinib has shown significant antitumor effects against SCCHN, and it is currently approved by the United States Food and Drug Administration (2004) for treatment of locally advanced or metastatic non–small-cell lung cancer after failure of at least one prior chemotherapy regimen and for use in combination with gemcitabine for the first-line treatment of patients with locally advanced, unresectable or metastatic pancreatic cancer (Pomerantz and Grandis, 2004). However, targeting of EGFR alone has only shown promise clinically when combined with standard cytotoxic approaches, including chemotherapy or radiation, in SCCHN (Burtness et al., 2005; Bonner et al., 2006). To date, no Bcl-XL inhibitors have been investigated in patients with SCCHN. Studies have shown that the negative enantiomer of gossypol binds to the Bcl-2 homology 3 domain of Bcl-XL and Bcl-2 to cause apoptosis through induction of DNA fragmentation; poly(ADP-ribose) polymerase cleavage; loss of mitochondrial membrane potential; cytochrome c release; and activation of caspases-3, -8, and -9 (Enyedy et al., 2001; Zhang et al., 2003; Dao et al., 2004; Oliver et al., 2004).
We hypothesized that combined targeting of the individual components of the EGFR-STAT3-Bcl-XL signaling pathway would result in increased antitumor effects. EGFR, STAT3, and Bcl-XL have each been implicated as important therapeutic targets in SCCHN. We observed enhanced antiproliferative effects when the STAT3 decoy was combined with either erlotinib or gossypol in vitro. When erlotinib and the STAT3 decoy were combined in an in vivo model of SCCHN, significant antitumor effects were achieved. The triple combination of erlotinib, the STAT3 decoy, and gossypol resulted in enhanced growth inhibition in vitro. These results suggest that combined targeting of the EGFR-STAT3-Bcl-XL signaling axis represents a potential treatment strategy for cancers characterized by activation of this signaling pathway, including SCCHN.
Materials and Methods
Antibodies and Reagents
Antibodies for p44/42 mitogen-activated protein kinase (MAPK), phospho-p44/42 MAPK, p70S6K, phospho-p70S6K, p-Akt, and Akt were purchased from Cell Signaling Technology Inc. (Danvers, MA). The cyclin D1 and VEGF antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The goat anti-rabbit IgG (H+L)-horseradish peroxidase conjugate secondary antibody was from Bio-Rad Laboratories (Hercules, CA). The β-tubulin primary antibody was from Abcam Inc (Cambridge, MA). Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) stain (for apoptosis) was purchased from Millipore Bioscience Research Reagents (Temecula, CA). Erlotinib (OSI-774; Tarceva) was provided by Genentech (South San Francisco, CA) (Moyer et al., 1997). (−)-Gossypol was a kind gift from Dr. Shaomeng Wang (University of Michigan, Ann Arbor, MI) (Yoo et al., 2004). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Sigma-Aldrich (St. Louis, MO). The enhanced chemiluminescence kit was purchased from Santa Cruz Biotechnology, Inc. The Annexin V-Cy3 Apoptosis Detection Kit Plus was from BioVision (Mountain View, CA).
Cell Culture
UM-22B and PCI-15B cell lines were derived from cervical lymph node metastases from patients with head and neck squamous cell carcinomas as described previously (Lin et al., 2006). UM-22B cells were provided by Dr. Thomas Carey (University of Michigan) (Ala-aho et al., 2004). The PCI-15B cell line was created at the University of Pittsburgh (Pittsburgh, PA) (Sacks et al., 1988). The 1483 cell line was derived from a primary tumor of the pharynx of a patient with SCCHN, and it was a kind gift from Dr. Gary Clayman (MD Anderson Cancer Center, Houston, TX) (Sacks et al., 1988). Cells were cultured in DMEM containing 10% heat-inactivated fetal bovine serum and 1× penicillin/streptomycin mix (both from Invitrogen, Carlsbad, CA) at 37°C with 5% CO2.
STAT3 Decoy Transfections
The STAT3 decoy sequence was 5′-CATTTCCCGTAAATC-3′ and 3′-GTAAAGGGCATTTAG-5′, and the mutant control sequence was 5′-CATTTCCCTTAAATC-3′ and 3′-GTAAAGGGAATTTAG-5′. Oligonucleotides were generated as described previously (Leong et al., 2003; Xi et al., 2005). The single-stranded sense and antisense oligonucleotides were synthesized by the DNA Synthesis Facility at the University of Pittsburgh, and they were purified by means of β-cyanothylphysphoramidite chemistry to minimize degradation of the oligonucleotides by endogenous nucleases. Equal amounts of sense and antisense oligonucleotides were combined and annealed as described previously (Xi et al., 2005). Decoy transfections were performed using Lipofectamine 2000 in Opti-MEMI media (both from Invitrogen) as follows: cells were plated to approximately 60 to 70% confluence and transfection media containing the STAT3 decoy or mutant control was added and incubated at 37°C with 5%CO2 for 4 h. Fresh DMEM containing 10%heat-inactivated fetal bovine serum and 1× penicillin/streptomycin mix was then added.
Dose-Response Experiments
Dose-response experiments were performed using increasing concentrations of each therapeutic reagent [erlotinib, (−)-gossypol, or STAT3 decoy] in DMEM containing 10%heat-inactivated fetal bovine serum and 1× penicillin/streptomycin mix. After 72 h, MTT assays were performed to determine the effects of drug treatment. Media were removed from the plates and replaced with 5 mg/ml MTT (Sigma-Aldrich). Then, the plates were incubated at 37°, 5%CO2 for 15 min. The MTT reagent was removed, and dimethyl sulfoxide was added to lyse the cells. The plate was then read at 595 nm in the μQuant microplate spectrophotometer (Bio-Tek Instruments, Winooski, VT) using KCjunior software. Data were normalized to untreated control cells, and the equation to calculate the percentage of killing is (ODuntreated − ODinhibitor)/ODuntreated × 100%. Curve-fit nonlinear regression analysis of sigmoidal dose-response curves with variable slope was performed using Prism version 4.03 for Windows (GraphPad Software Inc., San Diego, CA). MTT data were confirmed using trypan blue exclusion assays.
Combination Studies in Vitro to Determine Cell Viability
For combination experiments, UM-22B and PCI-15B cells were transfected with the IC50 concentrations of 12.6 or 38.3 nM, respectively, STAT3 decoy or mutant control decoy as described above. After 4 h, transfection media were removed, and DMEM (10% heat-inactivated fetal bovine serum, 1× penicillin/streptomycin mix) containing 5 or 0.16 µM erlotinib alone, 2.67 or 2.97 µM (−)-gossypol alone for UM-22B and PCI-15B cells, respectively, or a combination of both erlotinib and (−)-gossypol was added. Cell counts using trypan blue exclusion assay were performed after 72 h to assess cell viability.
Combination Studies in Vitro to Determine STAT3 Target Gene Expression by Immunoblotting
For the triple combination experiment with PCI-15B cells, 2.5 × 105 cells/well were seeded in a six-well plate in DMEM containing 10% FBS, and after 24 h the cells were transfected with 38.3 nM STAT3 decoy or mutant control, respectively, as described above. After 4 h, transfection media were removed, and DMEM (10% heat-inactivated FBS) containing 0.16 µM erlotinib alone, 2.97 µM (−)-gossypol alone, or a combination of both erlotinib and (−)-gossypol was added. For the cells transfected only with STAT3 decoy or the mutant control, only DMEM was added. After 24 h, cell lysate was extracted, and the protein content was quantitated using the Bradford reagent. Forty micrograms of whole cell protein lysate was run on 8% SDS-polyacrylamide gel electrophoresis gels and transferred onto Trans-Blot nitrocellulose membranes (Bio-Rad Laboratories) for 50 min at 21 V using a semidry transfer machine (Bio-Rad Laboratories). The membranes were blocked using 5% nonfat dry milk, 0.2% Tween 20 in 1× phosphate-buffered saline (TBST) for 2 h. Membranes were incubated in primary antibody diluted at 1:1000, except for phospho p70S6 kinase antibody at 1:500 dilution at 4°C overnight, and then they were washed five times with TBST (5 min/wash). However, for the β-tubulin antibody, a 1:5000 dilution was used. The membranes were then incubated with secondary antibody for 1 h (1:3000) at room temperature, followed by five washes in TBST. Blots were developed using enhanced chemiluminescence, according to the manufacturer’s instructions (Santa Cruz Biotechnology, Inc.). Densitometric analyses were performed using AlphaDigiDoc 1000 software (Alpha Innotech, San Leandro, CA).
In Vivo Tumor Xenograft Studies
All studies were approved by the institutional animal care and use committee, and they were carried out in accordance with institutional guidelines for animal care. Female athymic nude mice (4–6 weeks old) were injected with 2 × 106 1483 cells in the left and right dorsal flanks, resulting in two tumors per animal. Approximately 12 days later, after tumors were palpable, animals were assigned to two treatment groups by stratified randomization based on flank ratio (six mice in the STAT3 decoy or mutant control decoy group and five mice in the erlotinib plus STAT3 decoy or erlotinib plus mutant control decoy group). Daily intratumoral injections of STAT3 decoy or mutant control (35µg in 50µl) were delivered for 31 days. Erlotinib was dissolved in a 20% Trappsol (hydroxypropyl-β-cyclodextrin) solution (CTD, Inc., Cyclodextrin Resource, High Springs, FL), and it was delivered (90 mg/kg) by oral gavage daily, as described previously (Higgins et al., 2004). After 31 days, the mice were sacrificed.
In Vitro Apoptosis Assay
For the apoptosis assay, UM-22B and PCI-15B cells were seeded at a density of 3 × 105 cells/well in a six-well plate in DMEM containing 10% FBS. After 24 h, cells were transfected with either 12.6 or 38.3 nM, respectively, STAT3 decoy or mutant control decoy as described above. After 4 h, transfection media were removed, and DMEM plus 10% FBS containing 5 or 0.16µM erlotinib alone, 2.67 or 2.97 µM (−)-gossypol alone for UM-22B and PCI-15B cells, respectively, or a combination of both erlotinib and (−)-gossypol was added. After 24 h, cells were detached by trypsinization, counted, and pelleted at 1000 rpm for 5 min. Cell pellets were washed once with phosphate-buffered saline, pH 7.4, and then they were resuspended in 100 µl of annexin V binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2). Approximately 5 µl of annexin V-Cy3 was added per tube, and the suspension was allowed to incubate at room temperature for 15 min in the dark. Then, the stained cell suspension was dropped on the slides and covered with coverslips. The membrane of apoptotic cells is stained a bright orange when analyzed with a fluorescence microscope (TE 2000-E; Nikon, Melville, NY). The ratio (percentage) of apoptotic to total cells (apoptotic plus nonapoptotic cells) was calculated for each high-power field. For each treatment, 5 to 10 high-power fields of view were quantitated on each section using Meta-Morph software version 7.0 (Molecular Devices, Sunnyvale, CA).
Immunohistochemical Analyses
For Ki-67, the Ki-55 clone was purchased from Dako North America, Inc. (Carpinteria, CA), used at a dilution of 1:25. For VEGF, the 5C3.F8+ JH121 clone was purchased from LabVision Products (Fremont, CA), used at a 1:100 dilution. For immunohistochemical staining, 4-µm sections were cut from formalin-fixed paraffin-embedded tissue blocks, and they were deparaffinized and rehydrated using successive washes of xylene followed by ethanol. Heat-induced epitope retrieval was performed on the sections in a microwave oven (medium power for 10 min) using citrate buffer, pH 6.0. Endogenous peroxidase was blocked with 3% hydrogen peroxide. Sections were incubated in Universal Protein Blocker (Shandon Lipshaw, Pittsburgh, PA) for 20 min at room temperature.
For TUNEL analysis, ApopTag Plus Peroxidase In Situ Apoptosis Detection kit (Millipore Corporation, Billerica, MA) sections were deparaffinized, and then they were treated with 20 µg/ml proteinase K (Roche Diagnostics, Indianapolis, IN) solution at 37°C for 10 min to enhance the staining. Then, 3% hydrogen peroxide/methanol was used to block the endogenous peroxidase. Sections were then incubated with terminal deoxynucleotidyl transferase and a dinucleotide solution (digoxigenin-labeled dUTP and dATP) at 37°C for 25 min followed by a stop buffer step at 37°C for 30 min. Peroxidase-conjugated anti-dioxygenin antibody was then applied to the sections at room temperature for 30 min, and the reaction products were visualized by 0.03% 3,3′-diaminobenzidine tetrahydrochloride solution containing 2 mM hydrogen peroxide. Apoptosis using TUNEL was assessed as a percentage of all tumor, excluding areas of necrosis. Necrosis was determined as a percentage of total cross-sectional area of tumor. Histologic criteria for necrosis were zones of granular acellular debris either with or without neutrophilic infiltrates, or amorphous eosinophilic cellular outline “ghosts.”
For VEGF, staining was interpreted semiquantitatively as the product of intensity (0–3) and percentage of cells staining. For hematoxylin and eosin and TUNEL, only percentage of cells was noted, and only strong staining was considered positive. All slides were examined using a standard brightfield microscope (BX51; Olympus, Center Valley, PA). All photomicrographs were taken at 100× magnification using an attached digital camera (SPOT insight 4MP; Diagnostic Instruments, Inc., Sterling Heights, MI).
Statistical Analyses
For combination treatment experiments, to compare two treatment groups, p values were obtained by the nonparametric Wilcoxon-Mann-Whitney test, and p ≤ 0.05 was considered significant. Statistical analyses were performed using StatXact software (Cytel Software Corporation, Cambridge, MA).
Results
SCCHN Cell Lines Demonstrates Similar IC50 Values for Gossypol but Not for Erlotinib or the STAT3 Decoy
To determine the doses of inhibitors to use in our cell line models, we first performed dose-response experiments for erlotinib, STAT3 decoy, or gossypol. UM-22B, PCI-15B, and 1483 cells were treated with a range of doses of the inhibitors for 72 h, and MTT assays were performed to assess cell viability. The time point and range of doses used were selected based on kinetics experiments (data not shown) and previously published reports (Oliver et al., 2004; Seki et al., 2006; Sutter et al., 2006). Data were normalized to untreated control cells, and the percentage of cytotoxicity and IC50 values were calculated as described under Materials and Methods. The IC50 value for gossypol in all cell lines tested was approximately 3 µM, which is consistent with other reports of gossypol activity in SCCHN cell lines (Table 1) (Yoo et al., 2004). The IC50 value for erlotinib was 10, 0.33, and 7 µM for UM-22B, PCI-15B, and 1483 cell lines, respectively. The IC50 values for the STAT3 decoy varied between the cell lines examined, which was 12.6 nM for UM-22B, 38.3 nM for PCI-15B, or 2.05 nM for 1483. In the subsequent combination experiments, the IC50 concentrations of the STAT3 decoy and (−)-gossypol were used, whereas half of the IC50 concentration or the IC25 concentration was used for erlotinib. This is because the sigmoidal dose-response curves for the STAT3 decoy and (−)-gossypol in the SCCHN cell lines was quite steep, indicating that the IC25, IC50, and IC75 concentrations are within a narrow dose range (data not shown).
TABLE 1. IC50 values for erlotinib, STAT3 decoy, and gossypol in SCCHN cell lines.
UM-22B, PCI-15B, and 1483 cells were treated with a range of concentrations of erlotinib, STAT3 decoy, or gossypol for 72 h. MTT assays were then performed, and IC50 values were calculated. The experiment was performed three times with similar results.
| Cell Line | Erlotinib | Decoy | Gossypol |
|---|---|---|---|
| µM | nM | µM | |
| UM-22B | 10 | 12.6 | 2.669 |
| PCI-15B | 0.33 | 38.3 | 2.968 |
| 1483 | 7 | 2.05 | 2.135 |
Combining Erlotinib with the STAT3 Decoy Enhances Inhibition of Cell Viability in SCCHN Cell Lines
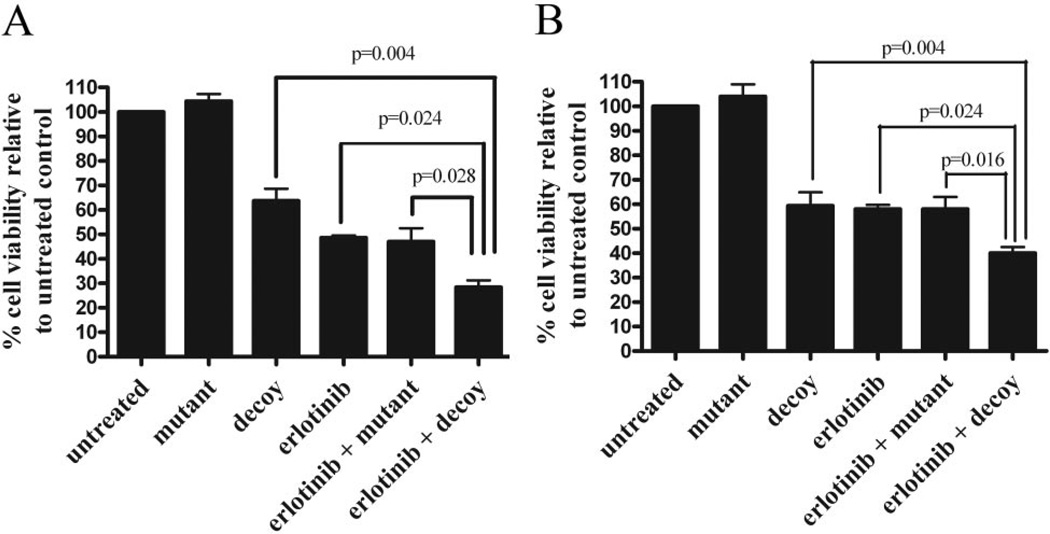
To assess the effect of simultaneous EGFR and STAT3 inhibition on cell viability, UM-22B cells were treated with 5 µM erlotinib and 12.6 nM STAT3 decoy. The mutant control decoy, which served as a control for the STAT3 decoy in all experiments, differs from the mutant control decoy by a single base-pair mutation, it is ineffective at binding and inhibiting activated STAT3, and it does not significantly decrease either cell viability or STAT3 target gene expression compared with an untreated control (Leong et al., 2003). Treatment of UM-22B cells with the STAT3 decoy alone resulted in 62.7% cell viability (Fig. 1A). Treatment with erlotinib alone resulted in 49.1% cell viability. Treatment with erlotinib plus the mutant control decoy resulted in 48.7% cell viability. Treatment with both the STAT3 decoy and erlotinib resulted in 29.6% cell viability, which was significantly different from cells treated with STAT3 decoy alone, erlotinib alone, or erlotinib plus the mutant control decoy (p = 0.004, p = 0.024, and p = 0.028, respectively) (Fig. 1A). Similar results were seen for PCI-15B cells where treatment with 38.3 nM STAT3 decoy resulted in 59.8% cell viability, treatment with erlotinib alone resulted in 59.5% cell viability, treatment with 0.16 µM erlotinib plus the mutant control decoy resulted in 59.44% cell viability, and treatment with both the STAT3 decoy and erlotinib resulted in 39.0% cell viability (Fig. 1B). In PCI-15B cells, the combination of the STAT3 decoy and erlotinib significantly decreased cell viability compared with the STAT3 decoy alone, erlotinib alone, or erlotinib and the mutant control decoy (p = 0.004, p = 0.024, and p = 0.016, respectively). These results indicate that combining erlotinib with the STAT3 decoy enhances antiproliferative effects.
Fig. 1.
Combining the STAT3 decoy with erlotinib enhances inhibition of cell viability in vitro. A, UM-22B cells were transfected with 12.6 nM STAT3 decoy or mutant control decoy for 4 h. Transfection media were removed, and DMEM containing 10% FBS and 5 µM erlotinib was added. Cells were then counted after 72 h using trypan blue dye exclusion. When the STAT3 decoy was combined with erlotinib, cell viability was significantly reduced compared with STAT3 decoy alone (p = 0.004), erlotinib alone (p = 0.024), or erlotinib plus mutant control decoy (p = 0.028). B, similar results were seen for PCI-15B cells (p = 0.004, p = 0.024, and p = 0.016) when treated with 0.1 µM erlotinib and 38.3 nM STAT3 decoy. Cumulative results are shown from five separate experiments.
Combining Erlotinib and the STAT3 Decoy Enhances in Vivo Antitumor Effects Compared with Either Treatment Alone
To determine the therapeutic efficacy of combining an EGFR inhibitor with the STAT3 decoy in vivo, a xenograft model of SCCHN was used. Studies have found that the maximum tolerated dose of erlotinib in nude mice is 100 mg/kg with daily administration (Higgins et al., 2004). The 1483 cell line is considered relatively resistant to erlotinib (the IC50 value is 7 µM); therefore, we chose to use 90 mg/kg erlotinib daily. The STAT3 decoy dose was based on previous in vivo studies; however, when 50 µg of STAT3 decoy was administered daily, the antitumor effects were so profound that we could not assess a benefit of the addition of erlotinib (90 mg/kg daily) (data not shown). This decrease in mean tumor volumes was apparent after 5 days of treatment, and animals were sacrificed after 14 days of treatment due to ulcerated tumors and weight loss. SCCHN xenografts frequently ulcerate, especially in the setting of repeated intratumoral inoculations. In the next experiment, nude mice bearing 1483 tumors were treated with the 35 µg of STAT3 decoy or mutant control decoy by intratumoral injection with 90 mg/kg erlotinib or the vehicle control by oral gavage for 31 days (five to six mice/group). In general, the combination of erlotinib with the STAT3 decoy inhibited tumor growth to a greater extent than either treatment alone (the mutant decoy was used as a control for the STAT3 decoy) (data not shown). Specifically, the tumor volumes were significantly reduced in the combined therapy group injected with erlotinib plus the STAT3 decoy compared with the monotherapy group injected with mutant control decoy alone at days 27, 29, and 31 (p = 0.01).
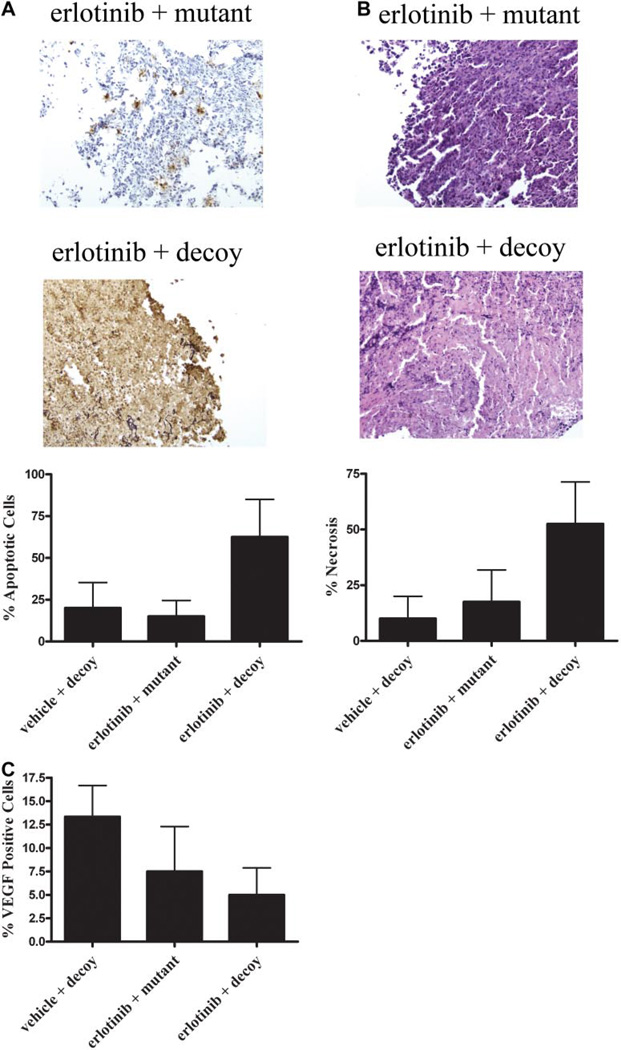
Tumors were harvested and immunohistochemical analysis was performed to assess induction of apoptosis and necrosis and also to quantitate VEGF, an angiogenesis marker and STAT3 target gene. The tumors that received erlotinib plus decoy also exhibited markedly increased apoptosis (62.5%) compared with vehicle plus decoy (20.0%) or erlotinib plus mutant (15.0%) (Fig. 2A). In addition, necrosis was increased in the erlotinib plus decoy-treated tumors (52.5%) compared with vehicle plus decoy (10.0%) and erlotinib plus mutant (17.5%) (Fig. 2B). The combination of the STAT3 decoy and erlotinib decreased the expression of VEGF by 8.3% compared with vehicle plus decoy and by 2.5% compared with erlotinib plus mutant (Fig. 2C). These results indicate that the combination of erlotinib plus STAT3 decoy increases apoptosis and induces necrosis, possibly through the down-modulation of VEGF, compared with the STAT3 decoy or erlotinib administered as single agents. The failure to consistently detect a difference in tumor volume between the mice that received both agents compared with monotherapy at each time point may be due to the necrosis that obscured an effect on gross tumor volumes, the relatively small number of mice per group, or both. The biologic significance of necrosis in a subcutaneous SCCHN xenograft model is unknown.
Fig. 2.
Combining the STAT3 decoy with erlotinib enhances cell growth inhibition in vivo. We inoculated 1483 cells (2 × 106) subcutaneously in the right and left flanks of 11 athymic nude mice. After 14 days, the tumors were palpable, and mice were randomized into two treatment groups. Tumors on the left flank were injected with the mutant control decoy, and tumors on the right flank were treated with the STAT3 decoy (35 µg) daily for 31 days. In addition, five mice received 90 mg/kg erlotinib, and six mice received vehicle control by oral gavage. Tumors were harvested at the end of treatment (day 31), and they were stained for apoptosis using a TUNEL assay (A) or for necrosis (B), by hematoxylin and eosin staining, and VEGF expression (C). The average percentage of apoptotic, necrotic, and VEGF-positive cells are represented. The percentage of apoptotic cells by TUNEL excluded the areas of necrosis as part of the denominator.
STAT3 Decoy in Combination with Gossypol Enhances Inhibition of Cell Viability in SCCHN Cell Lines
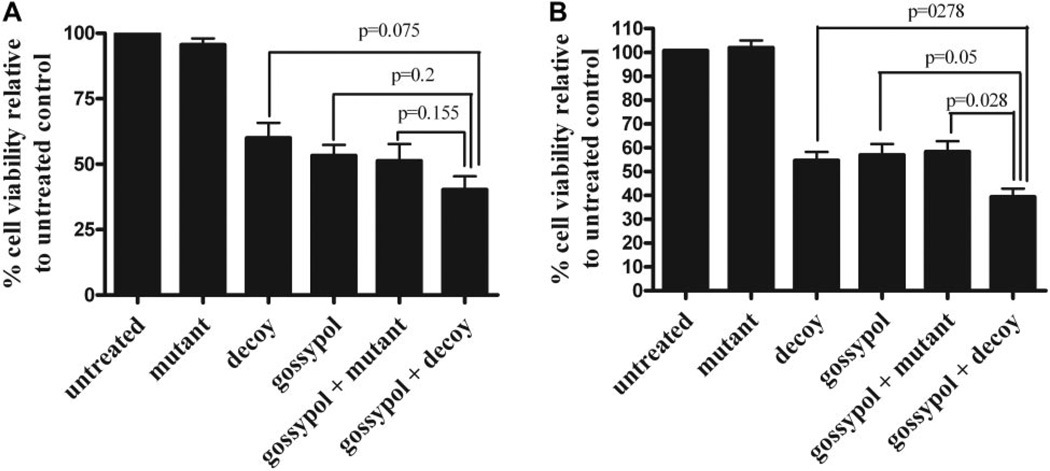
To determine the efficacy of combining a STAT3 inhibitor with a Bcl-XL inhibitor, the STAT3 decoy was combined with gossypol in UM-22B and PCI-15B cells. Cell viability was assessed using trypan blue dye exclusion assay after 72 h to determine the efficacy of the combination treatment. A trend was observed in UM-22B cells, with 40.3% cell viability after treatment with the STAT3 decoy plus gossypol, 61.8% cell viability with the STAT3 decoy alone (p = 0.075), 52.5% cell viability with gossypol alone, and 51.5% cell viability with the mutant control decoy plus gossypol, although it was not statistically significant (p = 0.155) (Fig. 3A). In PCI-15B cells, the combination of STAT3 decoy plus gossypol significantly enhanced inhibition of cell viability compared with either the STAT3 decoy alone, gossypol alone, or the mutant control decoy with gossypol (38.7% cell viability, 54.2% cell viability, 55.5% cell viability, and 56.4% cell viability, respectively; p = 0.0278, p = 0.05, and p = 0.0278) (Fig. 3B). These data indicate that the combination of the STAT3 decoy and gossypol resulted in enhanced inhibition of cell viability.
Fig. 3.
Combination of the STAT3 decoy with gossypol enhances inhibition of cell viability of head and neck cancer cells. A, UM-22B cells were treated with 12.6 nM STAT3 decoy or mutant control decoy for 4 h followed by treatment with 2.67 µM gossypol for 72 h. Cell counts were then performed using trypan blue dye exclusion. Combining the STAT3 decoy with gossypol augmented inhibition of cell viability compared with STAT3 decoy alone, gossypol alone, gossypol alone, or mutant control plus gossypol (p = 0.075, p = 0.2, and p = 0.155, respectively). B, similar results were seen when PCI-15B cells were treated with 38.3 nM STAT3 decoy and 2.97 µM gossypol (p = 0.0278, p = 0.5, and p = 0.0278). Cumulative results are shown from five separate experiments.
Combination of Erlotinib, STAT3 Decoy, and Gossypol Exhibits Enhanced Growth Inhibition of SCCHN Cells in Vitro
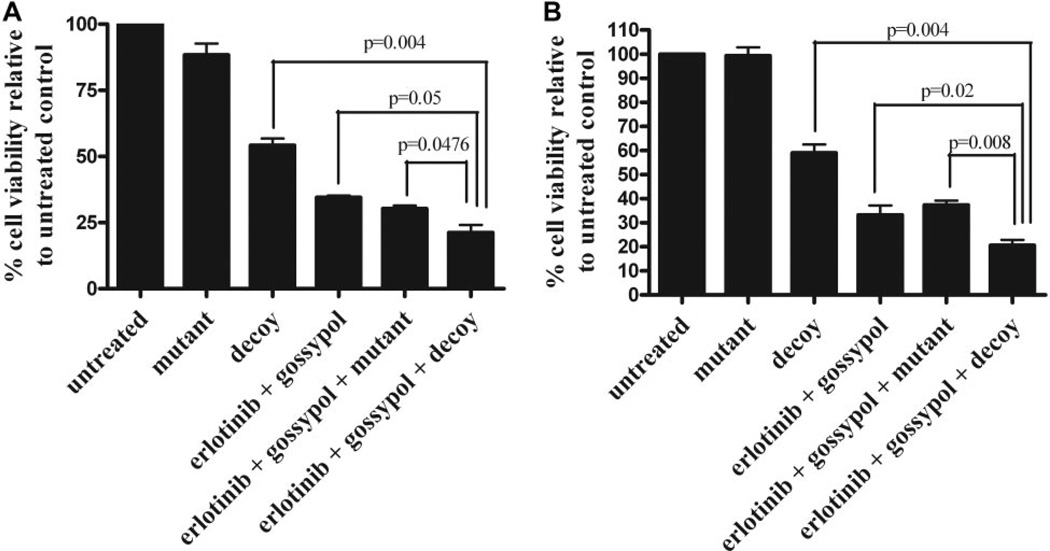
We next investigated the efficacy of combined inhibition of EGFR, STAT3, and Bcl-XL using a combination of erlotinib, the STAT3 decoy, and gossypol. UM-22B cells were treated with 5 µM erlotinib, 12.6 nM STAT3 decoy, and 2.67 µM gossypol, and then we compared these cells with cells treated with STAT3 decoy alone or the combination of erlotinib, the mutant control decoy, and gossypol. After 72 h of treatment, we found that the triple combination of erlotinib, the STAT3 decoy, and gossypol increased inhibition of cell viability compared with STAT3 decoy alone, erlotinib plus gossypol, or erlotinib plus mutant control decoy plus gossypol (21.3% cell viability, 54.3% cell viability, 34.1% cell viability, or 30.7% viability, respectively; p = 0.004, p = 0.05, and p = 0.0476) (Fig. 4A). Similar results were seen with PCI-15B cells, where the combination of erlotinib plus STAT3 decoy plus gossypol enhanced inhibition of cell viability compared with STAT3 decoy alone, erlotinib plus gossypol or erlotinib plus mutant control decoy, plus gossypol (19% cell viability compared with 59.6, 34.5, and 37.9% cell viability, respectively; p = 0.004, p = 0.02, and p = 0.008) (Fig. 4B).
Fig. 4.
Combining erlotinib, STAT3 decoy, and gossypol further enhances inhibition of cell viability in SCCHN cells. A, UM-22B cells were plated in 96-well plates, and then they were treated with 5 µM erlotinib, 12.6 nM STAT3 decoy, and 2.67 µM gossypol. Cell counts were performed by trypan blue dye exclusion at 72 h. The combination of erlotinib, STAT3 decoy, and gossypol enhanced inhibition of cell viability compared with STAT3 decoy alone (p = 0.004), erlotinib plus gossypol (p = 0.05), or the combination of erlotinib, mutant control, and gossypol (p = 0.0476). B, similar results were seen with PCI-15B treated with 0.16 µM erlotinib, 38.3 nM STAT3 decoy, and 2.97 µM gossypol (p = 0.004, p = 0.02, and p = 0.008, respectively). Cumulative results are shown from five separate experiments.
Combined Targeting of EGFR, STAT3, and Bcl-XL Increases SCCHN Apoptosis
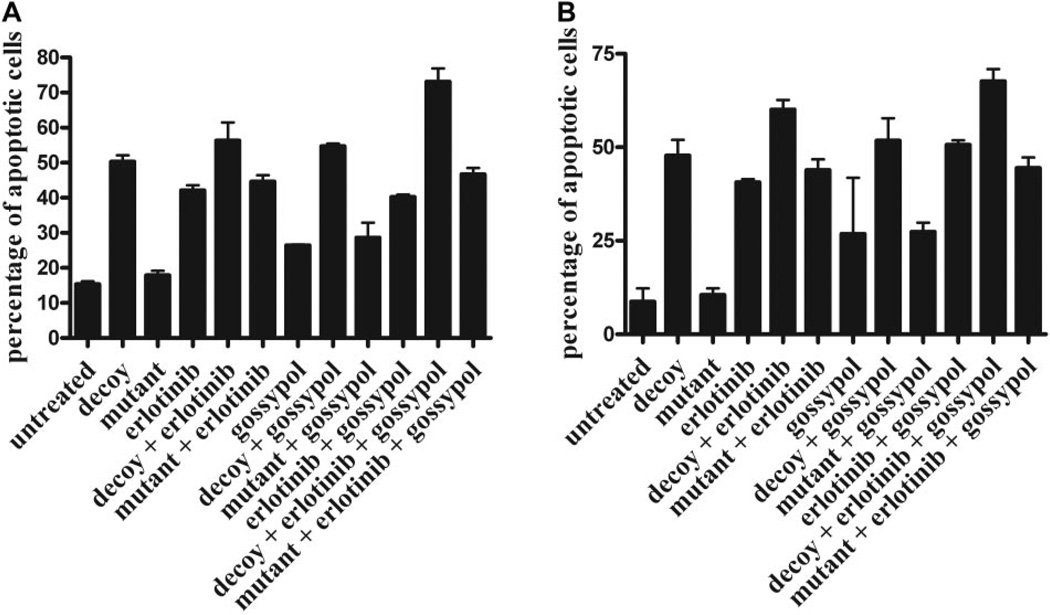
We examined apoptosis by annexin V analysis after STAT3 decoy, erlotinib, and gossypol treatment to determine whether the cytotoxic effects of combined EGFR, STAT3, and Bcl-XL targeting were due to increased apoptosis. In UM-22B cells (Fig. 5A), combined targeting enhanced apoptosis (74% apoptotic cells) at 24 h, compared with decoy alone (50% apoptotic cells), erlotinib alone (42% apoptotic cells), decoy plus erlotinib alone (57% apoptotic cells), gossypol alone (25% apoptotic cells), decoy plus gossypol (54% apoptotic cells), and erlotinib plus gossypol (39% apoptotic cells). Similar observations were seen in PCI-15B cells (Fig. 5B), where combined targeting enhanced apoptosis (66% apoptotic cells) at 24 h, compared with decoy alone (48% apoptotic cells), erlotinib alone (38% apoptotic cells), decoy plus erlotinib alone (60% apoptotic cells), gossypol alone (27% apoptotic cells), decoy plus gossypol (52% apoptotic cells), and erlotinib plus gossypol (50.5% apoptotic cells).
Fig. 5.
Combining erlotinib, STAT3 decoy, and gossypol induces apoptosis in vitro. UM-22B (A) and PCI-15B (B) cells were treated with STAT3 decoy/control decoy (12.6 and 38.3 nM, respectively), erlotinib (5 and 0.16 µM, respectively), and gossypol (2.67 and 2.97 µM, respectively) alone or in combination for 24 h, followed by annexin V assay. Percentage of apoptotic cells is expressed as mean ± S.E.M.
Combined Inhibition of EGFR, STAT3, and Bcl-XL Elicits Antitumor Effects by Inhibiting Target Gene Expression
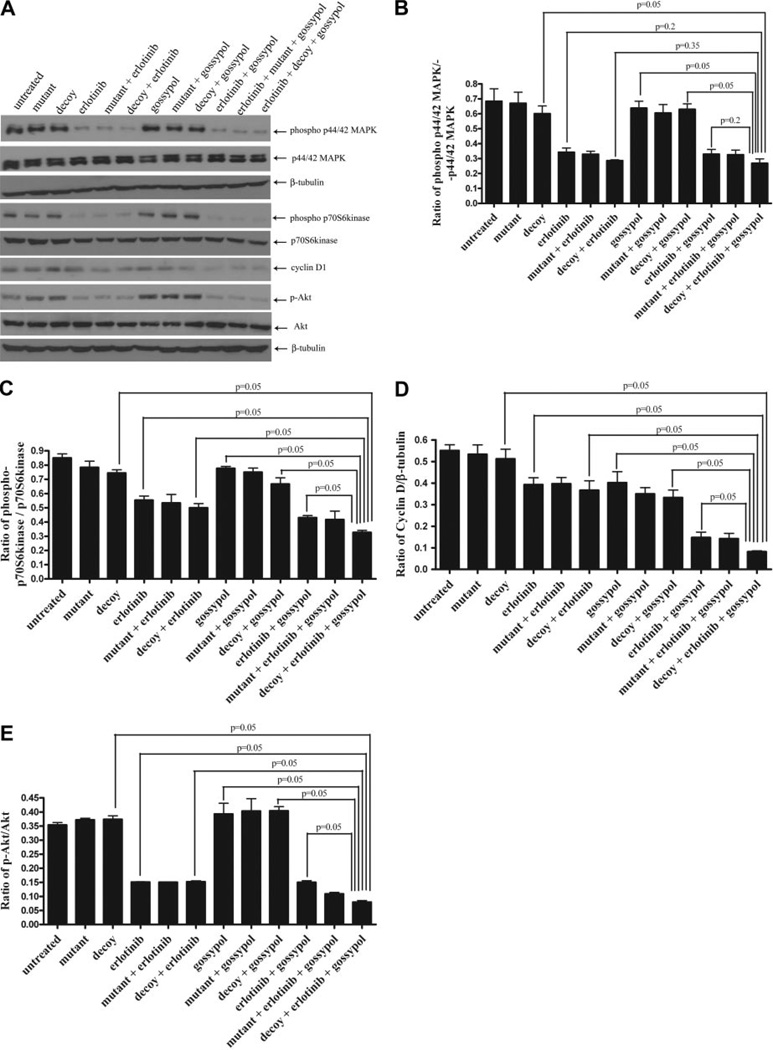
To investigate the effects of combined targeting of the EGFR-STAT3-Bcl-XL signaling pathway on the expression of selected proteins, PCI-15B cells were treated with IC50 concentrations of the STAT3 decoy (38.3 nM) and/or gossypol (2.97 µM) and the IC25 concentration oferlotinib (0.16 µM) for 24 h. Lysates were then prepared and subjected to immunoblotting for phospho-p44/42MAPK, p44/42 MAPK, cyclin D1, phospho-p70S6K, p70S6K, p-Akt, Akt, and β-tubulin (loading control) (Fig. 6A). Treatment with erlotinib alone or in combination with the decoy inhibited MAPK phosphorylation as shown by densitometric analysis compared with decoy alone (Fig. 6B). However, combining the decoy with erlotinib and gossypol did not further augment the down-modulation of p44/42 MAPK phosphorylation. P70S6K is downstream of EGFR and mammalian target of rapamycin, a protein that is up-regulated in SCCHN (Yokogami et al., 2000; Tibes et al., 2006). Likewise, we found that combination of the decoy with erlotinib and gossypol down-regulated phospho-p70S6 kinase compared with decoy alone (p = 0.05), decoy and gossypol in combination (p = 0.05), erlotinib alone (p = 0.05), or decoy plus erlotinib in combination (p = 0.05) (Fig. 6C). Combining the STAT3 decoy with erlotinib and gossypol resulted in significantly decreased expression of cyclin D1 compared with either the single or double combinations (p = 0.05) (Fig. 6D). In addition, combination of STAT3 decoy plus erlotinib plus gossypol down-regulated p-Akt compared with decoy alone (p = 0.05), erlotinib alone (p = 0.05), gossypol alone (p = 0.05), or erlotinib plus decoy (p = 0.05) (Fig. 6E). These results suggest that down-modulation of MAPK and p70S6 kinase are primarily mediated by erlotinib treatment in vitro. However, decreased expression of cyclin D1 and p-Akt seems to reflect the enhanced anti-proliferative effect of targeting the pathway at multiple points.
Fig. 6.
Combining erlotinib, STAT3 decoy, and gossypol inhibits the p44/42 MAPK and mammalian target of rapamycin pathways, as well as cyclin D1 and p-Akt protein expression in vitro. PCI-15B cells were treated with STAT3 decoy/control decoy (38.3 nM), erlotinib (0.16 µM), and/or gossypol (2.97 µM) alone or in combination for 24 h. A, lysates were collected, and 40 µg of protein/lane was immunoblotted for cyclin D1, phospho-p44/42 MAPK, p44/42 MAPK, phospho-p70S6 kinase, p70S6 kinase, p-Akt, Akt, and β-tubulin (loading control). Results shown are representative of three independent experiments. B to E are presentations of densitometry data of PCI-15B cells treated with decoy/control decoy, erlotinib, and gossypol (alone or in combination), and the data are expressed as a ratio with respect to β-tubulin or total protein. Protein levels are expressed as mean ± S.E.M. The results represent densitometry performed on Western blots from three independent experiments.
Discussion
Because of the complexity of signaling pathways and the multilevel cross-stimulation of parallel pathways in a cell, molecularly targeted inhibitors have not consistently performed satisfactorily in single-agent trials (Maione et al., 2006). Preclinical studies have focused on combining EGFR inhibitors or Bcl-XL inhibitors with standard treatments, either radiation or chemotherapy. Because no STAT3 inhibitor has reached the clinic to date, there are no clinical data on the therapeutic efficacy of a STAT3 inhibitor in combination with either standard treatments or experimental treatments such as EGFR or Bcl-XL inhibitors. Combined targeting of several molecules in a pathway whose component proteins are up-regulated in cancer is largely unexplored.
EGFR, STAT3, and Bcl-XL have each been implicated in cancer progression in a wide range of human tumors. Activation of EGFR by autocrine ligands leads to activation of STAT3 in SCCHN through direct interaction via SH2 domains with specific autophosphorylation sites in the cytoplasmic domain of the receptor. Activation of STAT3 leads to dimerization of STAT molecules, translocation into the nucleus and induction of critical target genes, including Bcl-XL. Previous studies have demonstrated that EGFR, STAT3, and Bcl-XL are constitutively activated in a variety of malignancies and that each of these proteins may serve as a therapeutic target in cancers such as SCCHN in which increased expression levels have been correlated with decreased survival, poor prognosis, and increased resistance to chemotherapy and radiation (Trask et al., 2002; Pomerantz and Grandis, 2004; Leeman et al., 2006).
To date, studies have primarily focused on targeting these proteins alone or in combination with established treatment modalities, such as chemotherapy or radiation. EGFR activates a variety of intracellular pathways and proteins that stimulate growth, proliferation, angiogenesis, metastasis, and survival, including the Ras/MAPK, phospholipase-Cγ phosphatidylinositol-3 kinase, and STATs. STAT3 represents a point of convergence for several upstream signaling pathways, including EGFR, platelet-derived growth factor receptor, Src, Bcr-Abl, and gp130/IL-6R where activation of STAT3 elicits expression of a variety of target genes, including Bcl-XL, cyclin D1, VEGF, MMP-2, and MMP-9 (Ruff-Jamison et al., 1994; Zhong et al., 1994; Darnell, 1997; Turkson et al., 1998; Shao et al., 2003; Sriuranpong et al., 2003). Therefore, the combined targeting of EGFR, STAT3, and Bcl-XL using erlotinib, STAT3 decoy, and gossypol in SCCHN may lead to enhanced antitumor activity without overlapping toxicities in a broader range of patients.
Dual-molecular targeting approaches are under active investigation in preclinical models of cancer and in selected early phase clinical trials. We found that combining the decoy with erlotinib enhanced cell growth inhibition in vitro. In addition, our data indicate that combining the STAT3 decoy and erlotinib in vivo may be an efficacious antitumor regimen. Immunohistochemical analysis of the tumors from this experiment indicate that the combination of erlotinib and the decoy increased TUNEL-positive cells and decreased expression of VEGF, leading us to hypothesize that the antitumor effects are the result of increased apoptosis and decreased angiogenesis. Reports of combined targeting of EGFR and STAT3 are few, but others have observed additive or synergistic growth inhibition, depending on the concentrations used, of an human cervical cancer cell line overexpressing EGFR when the EGFR tyrosine kinase inhibitor AG1478 and AG490 (a Janus tyrosine kinase 2 inhibitor) were combined in vitro (Dowlati et al., 2004). To date, there are no published reports investigating the effects of combining STAT3 and Bcl-XL inhibitors. We also found that combining the STAT3 decoy and gossypol to inhibit STAT3 and Bcl-XL resulted in enhanced antiproliferative effects in vitro.
To date, the antitumor effects of combining EGFR, STAT3, and Bcl-XL inhibitors have not been explored. When EGFR, STAT3, and Bcl-XL inhibitors were combined, we observed enhanced inhibition of cell viability in all three SCCHN cell lines tested. A study performed in both colon and breast cancer models combined an EGFR tyrosine kinase inhibitor (gefitinib; ZD1839), with protein kinase A antisense oligonucleotides and Bcl-2/Bcl-XL antisense oligonucleotides, and it found that combined targeting resulted in enhanced antiproliferative, proapoptotic, and antiangiogenic effects (Tortora et al., 2003a). The same group next studied the combination of the cyclooxygenase-2 inhibitor SC-236 with gefitinib and PKA antisense oligonucleotides in colon and breast preclinical models, and they also observed that combining three inhibitors resulted in enhanced antitumor effects compared with single or dual combinations of the inhibitors (Tortora et al., 2003b). Another group combined inhibitors targeting EGFR and VEGF receptor (AEE788; Traxler et al., 2004), and PDGF receptor, BCR-ABL, and c-Kit (imatinib; STI571) with chemotherapy in pancreatic cancer cell lines, and they found that the combination of small-molecule inhibitors with gemcitabine significantly inhibited tumor cell growth and prolonged survival in vivo (Yokoi et al., 2005). Our data provide further evidence that a combination of three inhibitors may be efficacious for cancer. Such a strategy may be particularly beneficial in view of the heterogeneity of most human tumors.
There are several potential mechanisms to explain the enhanced antitumor effects of combined targeting strategy. It is possible that each targeted molecule independently contributes to tumor progression and that effective strategies require simultaneous blockade of all three targets. Alternatively, cross-talk between signaling pathways provides a potential route to overcome blockade of a single target, which can be overcome with blockade of multiple targets. We found that the combination of erlotinib, STAT3 decoy, and gossypol caused down-modulation of cyclin D1, resulting in decreased cell cycle progression. Thus, inhibition of cell viability and proapoptotic effects observed indicate that combined targeting of EGFR, STAT3, and Bcl-XL inhibits the expression, phosphorylation, or both of several critical downstream signaling proteins, leading to apoptosis and necrosis of SCCHN tumors.
Acknowledgments
We acknowledge the Core facility (Department of Ophthalmology, University of Pittsburgh), which is supported by Core Grant for Vision Research EY08098, for assistance with the fluorescence microscope.
This work was supported by National Institutes of Health grants R01-CA101840, 2R01-CA77308, and P50-CA097190-01A1 (to J.R.G.).
ABBREVIATIONS
- SCCHN
squamous cell carcinoma of the head and neck
- EGFR
epidermal growth factor receptor
- STAT
signal transducer and activator of transcription
- MAPK
mitogen-activate protein kinase
- VEGF
vascular endothelial growth factor
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DMEM
Dulbecco’s modified Eagle’s medium
- OD
optical density
- FBS
fetal bovine serum
- TBST
0.2% Tween 20 in 1 × phosphate-buffered saline
- p-
phospho-
- AG1478
4-(3′-chloroanilino)-6,7-dimethoxy-quinazoline
- AG490
α-cyano-(3,4-dihydroxy)-N-benzylcinnamide
- SC-236
4-(5-(4-chlorophenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl)benzenesulfonamide
References
- Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, Rui H, Nevalainen MT. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–27292. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- Ala-aho R, Ahonen M, George SJ, Heikkila J, Grenman R, Kallajoki M, Kahari VM. Targeted inhibition of human collagenase-3 (MMP-13) expression inhibits squamous cell carcinoma growth in vivo. Oncogene. 2004;23:5111–5123. doi: 10.1038/sj.onc.1207678. [DOI] [PubMed] [Google Scholar]
- Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, Fu KK, Milas L. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–7356. [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol. 2005;23:8646–8654. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- Dao VT, Dowd MK, Martin MT, Gaspard C, Mayer M, Michelot RJ. Cytotoxicity of enantiomers of gossypol Schiff’s bases and optical stability of gossypolone. Eur J Med Chem. 2004;39:619–624. doi: 10.1016/j.ejmech.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Dowlati A, Nethery D, Kern JA. Combined inhibition of epidermal growth factor receptor and JAK/STAT pathways results in greater growth inhibition in vitro than single agent therapy. Mol Cancer Ther. 2004;3:459–463. [PubMed] [Google Scholar]
- Enyedy IJ, Ling Y, Nacro K, Tomita Y, Wu X, Cao Y, Guo R, Li B, Zhu X, Huang Y, et al. Discovery of small-molecule inhibitors of Bcl-2 through structure-based computer screening. J Med Chem. 2001;44:4313–4324. doi: 10.1021/jm010016f. [DOI] [PubMed] [Google Scholar]
- Higgins B, Kolinsky K, Smith M, Beck G, Rashed M, Adames V, Linn M, Wheeldon E, Gand L, Birnboeck H, et al. Antitumor activity of erlotinib (OSI-774, Tarceva) alone or in combination in human non-small cell lung cancer tumor xenograft models. Anticancer Drugs. 2004;15:503–512. doi: 10.1097/01.cad.0000127664.66472.60. [DOI] [PubMed] [Google Scholar]
- Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–241. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Jen YM, Cheng MF, Lin YS, Su WF, Hwang JM, Chang LP, Chao HL, Liu DW, Lin HY, et al. Squamous cell carcinoma of the buccal mucosa: an aggressive cancer requiring multimodality treatment. Head Neck. 2006;28:150–157. doi: 10.1002/hed.20303. [DOI] [PubMed] [Google Scholar]
- Maione P, Gridelli C, Troiani T, Ciardiello F. Combining targeted therapies and drugs with multiple targets in the treatment of NSCLC. Oncologist. 2006;11:274–284. doi: 10.1634/theoncologist.11-3-274. [DOI] [PubMed] [Google Scholar]
- Moyer JD, Barbacci EG, Iwata KK, Arnold L, Boman B, Cunningham A, DiOrio C, Doty J, Morin MJ, Moyer MP, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 1997;57:4838–4848. [PubMed] [Google Scholar]
- Oliver CL, Bauer JA, Wolter KG, Ubell ML, Narayan A, O’Connell KM, Fisher SG, Wang S, Wu X, Ji M, et al. In vitro effects of the BH3 mimetic, (−)-gossypol, on head and neck squamous cell carcinoma cells. Clin Cancer Res. 2004;10:7757–7763. doi: 10.1158/1078-0432.CCR-04-0551. [DOI] [PubMed] [Google Scholar]
- Pomerantz RG, Grandis JR. The epidermal growth factor receptor signaling network in head and neck carcinogenesis and implications for targeted therapy. Semin Oncol. 2004;31:734–743. doi: 10.1053/j.seminoncol.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- Ruff-Jamison S, Zhong Z, Wen Z, Chen K, Darnell JE, Jr, Cohen S. Epidermal growth factor and lipopolysaccharide activate Stat3 transcription factor in mouse liver. J Biol Chem. 1994;269:21933–21935. [PubMed] [Google Scholar]
- Sacks PG, Parnes SM, Gallick GE, Mansouri Z, Lichtner R, Satya-Prakash KL, Pathak S, Parsons DF. Establishment and characterization of two new squamous cell carcinoma cell lines derived from tumors of the head and neck. Cancer Res. 1988;48:2858–2866. [PubMed] [Google Scholar]
- Seki Y, Yamamoto H, Yee Ngan C, Yasui M, Tomita N, Kitani K, Takemasa I, Ikeda M, Sekimoto M, Matsuura N, et al. Construction of a novel DNA decoy that inhibits the oncogenic beta-catenin/T-cell factor pathway. Mol Cancer Ther. 2006;5:985–994. doi: 10.1158/1535-7163.MCT-05-0388. [DOI] [PubMed] [Google Scholar]
- Shao H, Cheng HY, Cook RG, Tweardy DJ. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63:3923–3930. [PubMed] [Google Scholar]
- Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- Sutter AP, Hopfner M, Huether A, Maaser K, Scherubl H. Targeting the epidermal growth factor receptor by erlotinib (Tarceva) for the treatment of esophageal cancer. Int J Cancer. 2006;118:1814–1822. doi: 10.1002/ijc.21512. [DOI] [PubMed] [Google Scholar]
- Tibes R, Keating MJ, Ferrajoli A, Wierda W, Ravandi F, Garcia-Manero G, O’Brien S, Cortes J, Verstovsek S, Browning ML, et al. Activity of alemtuzumab in patients with CD52-positive acute leukemia. Cancer. 2006;106:2645–2651. doi: 10.1002/cncr.21901. [DOI] [PubMed] [Google Scholar]
- Tortora G, Caputo R, Damiano V, Caputo R, Troiani T, Veneziani BM, De Placido S, Bianco AR, Zangemeister-Wittke U, Ciardiello F. Combined targeted inhibition of bcl-2, bcl-XL, epidermal growth factor receptor, and protein kinase A type I causes potent antitumor, apoptotic, and antiangiogenic activity. Clin Cancer Res. 2003a;9:866–871. [PubMed] [Google Scholar]
- Tortora G, Caputo R, Damiano V, Melisi D, Bianco R, Fontanini G, Veneziani BM, De Placido S, Bianco AR, Ciardiello F. Combination of a selective cyclo-oxygenase-2 inhibitor with epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 and protein kinase A antisense causes cooperative antitumor and antiangiogenic effect. Clin Cancer Res. 2003b;9:1566–1572. [PubMed] [Google Scholar]
- Trask DK, Wolf GT, Bradford CR, Fisher SG, Devaney K, Johnson M, Singleton T, Wicha M. Expression of Bcl-2 family proteins in advanced laryngeal squamous cell carcinoma: correlation with response to chemotherapy and organ preservation. Laryngoscope. 2002;112:638–644. doi: 10.1097/00005537-200204000-00009. [DOI] [PubMed] [Google Scholar]
- Traxler P, Allegrini PR, Brandt R, Brueggen J, Cozens R, Fabbro D, Grosios K, Lane HA, McSheehy P, Mestan J, et al. AEE788: a dual family epidermal growth factor receptor/ErbB2 and vascular endothelial growth factor receptor tyrosine kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2004;64:4931–4941. doi: 10.1158/0008-5472.CAN-03-3681. [DOI] [PubMed] [Google Scholar]
- Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Food and Drug Administration. Rockville, MD: United States Food and Drug Administration; 2004. Center for Drug Evaluation and Research new molecular entity (NME) drug and new biologic approvals in calendar year 2004. Available at http://www.fda.gov/cder/rdmt/nmecy2004.htm. [Google Scholar]
- Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–979. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- Yokoi K, Sasaki T, Bucana CD, Fan D, Baker CH, Kitadai Y, Kuwai T, Abbruzzese JL, Fidler IJ. Simultaneous inhibition of EGFR, VEGFR, and platelet-derived growth factor receptor signaling combined with gemcitabine produces therapy of human pancreatic carcinoma and prolongs survival in an orthotopic nude mouse model. Cancer Res. 2005;65:10371–10380. doi: 10.1158/0008-5472.CAN-05-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo GH, Piechocki MP, Oliver J, Lonardo F, Zumstein L, Lin HS, Kim H, Shibuya TY, Shehadeh N, Ensley JF. Enhancement of Ad-p53 therapy with docetaxel in head and neck cancer. Laryngoscope. 2004;114:1871–1879. doi: 10.1097/01.mlg.0000147914.51239.ed. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liu H, Guo R, Ling Y, Wu X, Li B, Roller PP, Wang S, Yang D. Molecular mechanism of gossypol-induced cell growth inhibition and cell death of HT-29 human colon carcinoma cells. Biochem Pharmacol. 2003;66:93–103. doi: 10.1016/s0006-2952(03)00248-x. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]