Abstract
CRM1 (or exportin 1, Xpo1) transports proteins out of the cell nucleus through the nuclear pore complex. In the cytoplasm, GTP hydrolysis and consequent dissociation of Ran from CRM1 releases low-affinity substrates, while additional factors facilitate release of high-affinity substrates. Here we provide a model for human CRM1 export complex assembly and disassembly through structural and biochemical analyses of CRM1 bound to the substrate snurportin 1 (SNUPN, also called snuportin 1).
Like all members of the karyopherin-β (Kap-β; also known as importin and exportin) family of nuclear transport factors, the chromosome region maintenance 1 protein (CRM1) binds transport substrates and nucleoporins to target substrates to the nuclear pore complex. The Ran GTPase regulates CRM1-substrate interactions and transport directionality1. Ran exists mostly in the GTP state in the nucleus and mostly in the GDP state in the cytoplasm. CRM1 binds RanGTP and export substrate with positive cooperativity, enabling substrates to be loaded on CRM1 in the nucleus and released in the cytoplasm as GTP is hydrolyzed on Ran2–5.
We recently solved the 2.9-Å-resolution X-ray structure of the binary CRM1–SNUPN complex (PDB 3GB8)6, which is an intermediate in the assembly and disassembly steps of nuclear export7. SNUPN-bound CRM1 is a ring-shaped protein comprising 20 HEAT repeats, but the CRM1 ring is not planar, as HEAT repeats 2–7 (H2–H7; most of H1 is disordered) twist away from the plane of the ring with an ~45° rotation (Fig. 1a,b and Supplementary Fig. 1 online)6. The 27-residue C-terminal C-helix (residues 1,031–1,057) lies across the plane of the ring and connects helix H20B to the B helices of H8–H12 (Fig. 1a and Supplementary Fig. 2 online). SNUPN binds the convex side of the CRM1 ring, with a leucine-rich nuclear export signal (LR-NES) in its Kap-β (importin-β)-binding domain (known as sIBB) interacting with H11A and H12A and its nucleotide-binding domain (NBD) contacting H12A, H13A and H14B6.
Figure 1.
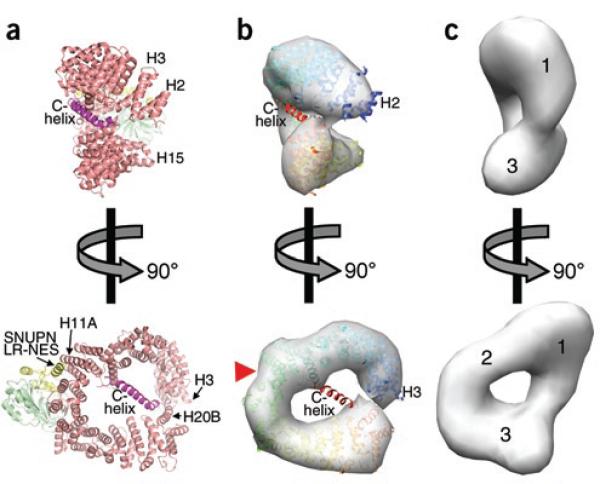
Comparison of unliganded and SNUPN-bound states of CRM1. (a) Ribbon diagram of the CRM1–SNUPN complex (PDB 3GB8)6. CRM1 is shown in pink and its C-extension in magenta, and several of the 20 HEAT repeats (H1–H20) are labeled. The sIBB and NBD domains of SNUPN are yellow and green, respectively. (b,c) Orthogonal views of the 22-Å-resolution density map calculated from coordinates of SNUPN-bound CRM1 (b) and the EM map of unliganded CRM1 (c)8. Ribbon diagram of SNUPN-bound CRM1 (blue-red color ramp from N to C terminus) is also shown in b, and a red triangle marks the LR-NES–binding site. N-terminal, central and C-terminal portions of CRM1 in the EM map are numbered in c.
Binary interactions between CRM1 and LR-NES or RanGTP are weak (dissociation constant (KD) > 8 μM in each case)7–9. However, the ligands bind CRM1 with positive cooperativity: each increases the other's affinity by ~500-fold7–9. To understand the structural basis of this cooperativity, we compared SNUPN-bound CRM1 with the 22-Å EM map of unliganded CRM1 (Fig. 1 and Supplementary Methods online)8. The ring curvatures and relative N- and C-terminus orientations of the two CRM1 structures are different. Previous analysis of the EM map suggested that repeat H1 lies next to H18–H20 at the C terminus, thus occluding the RanGTP-binding site at H1–H3 (ref. 8). In contrast, the N-terminal third of SNUPN-bound CRM1 is rotated ~45° out of the plane of the ring, exposing the Ran-binding B helices of repeats H1–H3 (Fig. 1 and Supplementary Fig. 1).
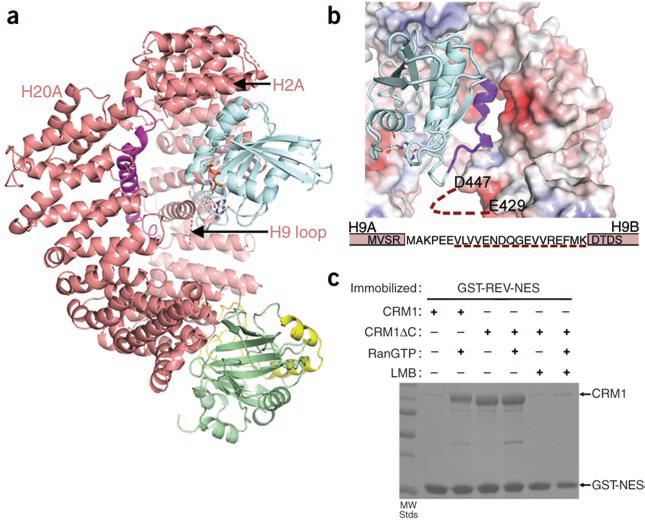
We docked RanGTP onto the CRM1–SNUPN complex based on alignment with the structure of the export-Kap-β protein CSE1 (also known as CSE1L) in complex with its substrate Kap60 (Saccharomyces cerevisiae homolog of SRP1, KPNA1 and importin-α) complex and Ran (Fig. 2a)10. Superposition of repeats H2–H7 resulted in an r.m.s. deviation of 1.78 Å for the closest 125 Cα atoms (Supplementary Fig. 3 online; CRM1 and CSE1 structures are compared in Supplementary Discussion and Supplementary Fig. 4 online). The CRM1–SNUPN complex accommodates RanGTP with minimal steric clashes (contacts <2.5 Å: Ran is ~2.2 Å from Val125 and the side chains of Lys367 and Asp313 of CRM1). The modeled Ran is oriented with its basic patch (residues 129–142) located next to a CRM1 acidic patch at H7B and H8B (Fig. 2b). The disordered H9 acidic loop, which was previously suggested to bind Ran8, is also nearby (previous mutagenesis of repeat H9 (ref. 8) is discussed in the Supplementary Discussion). It appears that little conformational change is needed for SNUPN-bound CRM1 to bind RanGTP, explaining CRM1's high affinity for the GTPase (KD ~15 nM)7,8. Complementary electrostatics at the modeled interface further support the idea that CRM1 conformation in the binary complex is poised for Ran binding and is likely to resemble the fully active CRM1 conformation of the ternary CRM1–substrate–Ran complex. Thus, we propose that cooperativity between Ran and substrate ultimately derives from equilibrium of CRM1 between the free structure observed in EM, in which the Ran and most probably the substrate sites are unavailable, and the structure observed in the SNUPN complex. Both ligands shift the equilibrium to the latter conformation, thus reciprocally enhancing each other's affinity.
Figure 2.
Positive cooperativity of substrate and Ran binding to CRM1. (a) The CRM1–SNUPN binary intermediate can accommodate RanGTP. Repeats H2–H7 of SNUPN-bound CRM1 were superimposed with H2–H7 of the CSE1–Kap60– RanGTP complex10 (PDB 1WA5). CRM1 and SNUPN are colored as in Figure 1 and RanGTP is in blue. CSE1 and Kap60 are omitted for clarity. (b) The electrostatic surface potential of SNUPN-bound CRM1 is shown with RanGTP from the superimposed CSE1–Kap60–Ran complex. The basic patch of Ran (residues 129–142) is in purple, the H9 loop sequence is shown and a dashed line indicates the disordered portion of this loop. (c) Pulldown assays with immobilized GST-NES of HIV1-REV and CRM1 proteins in the presence or absence of RanGTP and/or leptomycin B (LMB) were visualized by SDS-PAGE and Coomassie blue staining.
Because the C-helix bisects the CRM1 ring, we speculated that it might have a role in governing the global rearrangement of the protein between the inactive and active states6. Therefore, we compared substrate binding of wild-type CRM1 and the C-terminally truncated CRM1ΔC, from which residues 1,026–1,071 were removed. The deleted C-extension (which includes the C-helix and the extended portion that follows) did not contact binding sites for LR-NES, the SNUPN NBD or Ran and did not participate in crystal contacts (Fig. 1a, Supplementary Fig. 2 and Supplementary Methods). CRM1ΔC bound the LR-NES of HIV1-REV even in the absence of Ran (Fig. 2c). Thus, the C-extension appears to be important in allosteric coupling of substrate and Ran, stabilizing unliganded CRM1 in an inactive conformation that is incompatible with substrate binding. It was unclear whether the extension directly contacts the Ran and/or substrate sites because it could not be located in the low-resolution EM map of unliganded CRM1. However, its importance in stabilizing the inactive state suggests that it may be oriented differently in unliganded as compared to substrate-bound CRM1, perhaps toward the unusually large lobe of density at the N terminus of the former (region 1 in Fig. 1c). In summary, positive cooperativity in Ran and substrate binding appear to be achieved through global conformational changes in the CRM1 ring that may be aided by the large trans-ring C-extension.
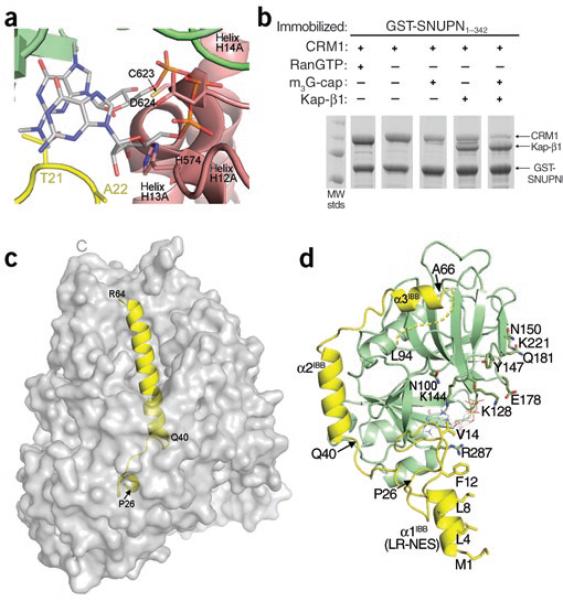
Upon the entrance of the CRM1 nuclear export complex into the cytoplasm, RanGAP-catalyzed GTP hydrolysis results in RanGDP dissociation from CRM1, shifting CRM1 equilibrium toward the inactive conformation and decreasing substrate affinity2–5. Low-affinity substrates such as LR-NESs are released into the cytoplasm, but high-affinity substrates such as SNUPN remain stably associated with CRM1 even after Ran dissociation6,7,11. In the latter cases, other factors in the cytoplasm must participate to complete nuclear export. Cytoplasmic SNUPN binds the trimethylguanosine (m3G)-cap structure of U snRNPs and the nuclear import factor Kap-β1 (ref. 12). The m3G-cap oligonucleotide has been reported to decrease CRM1-SNUPN affinity7. The NBDs of SNUPN are virtually identical in CRM1–SNUPN and m3G-cap–SNUPN complexes13 (Cα r.m.s. deviation 0.62 Å). Notably, the m3G-cap site is part of a nuclear export signal (NES) epitope that interacts with CRM1 (ref. 6), and that site in CRM1-bound SNUPN is occupied by CRM1 H12A and H13A helices and by sIBB residues 21TAA23 (Fig. 3a). Thus, CRM1 binding and m3G-cap binding are mutually incompatible, explaining the low affinity of m3G-cap–bound SNUPN for CRM1.
Figure 3.
Comparison of CRM1-, Kap-β1- and m3G-cap-nucleotide-bound SNUPN proteins. (a) The m3G-cap binding site in the CRM1–SNUPN complex. The m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD is shown as sticks (PDB 1XK5)13. CRM1 is shown as a pink ribbon and the sIBB and NBD of SNUPN as yellow and green ribbons, respectively. (b) Pulldown assays using immobilized GSTSNUPN and CRM1 were performed in the presence of RanGTP, m3G-cap nucleotide or Kap-β1. Bound proteins were separated by SDS-PAGE and visualized with Coomassie blue staining. (c) The Kap-β1–sIBB complex (PDB 2P8Q) is shown with the molecular surface of Kap-β1 in gray and the sIBB as a yellow ribbon. (d) Ribbon diagram of CRM1-bound SNUPN, showing that most of the sIBB (yellow) interacts with its NBD (green). m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD13 is shown as sticks. The structure is drawn with residues 40–52 of the sIBB in the same orientation as in c.
Kap-β1, like m3G-cap, also decreases CRM1-SNUPN affinity, and both factors together effectively dissociate the complex (Fig. 3b and Supplementary Methods). Kap-β1 binds sIBB with high affinity (KD = 2 nM), and in the Kap-β1–sIBB structure, sIBB residues 26–64 form a small 310-helix linked to a long α-helix, placing the termini 50 Å apart (Fig. 3c)14. Although sIBB residues 1–24 were not in the Kap-β1–sIBB structure, they contain additional binding determinants that are predicted to bind the N-terminal arch of Kap-β1 (ref. 14), further distancing the SNUPN N terminus and NBD. CRM1-bound sIBB adopts a very different structure: residues 1–16 (α1IBB and a short extended segment) form a LR-NES that binds CRM1 (ref. 6), and the rest of the sIBB wraps around the NBD, consistent with suggestions of intramolecular interactions between the domains (Fig. 3d)15. Residue Ser15 of the LR-NES is only 8 Å away from the NBD, allowing the LR-NES and the NES epitope on the NBD to bind adjacent CRM1 NES sites (Fig. 1a; LR-NES and NBD bind H11–H12 and H12–H14, respectively)6. Both the m3G-cap nucleotide and CRM1 bind SNUPN with KD values of ~1 μM6,13. Beside disrupting NBD-CRM1 interactions, m3G-cap binding would also disrupt sIBB-NBD intramolecular interactions (Fig. 3a), thus increasing sIBB accessibility to Kap-β1. Once sIBB binds Kap-β1, the probability of SNUPN re-binding to CRM1 is low given the high affinity of the Kap-β1–sIBB interaction and the physical separation of the two NES epitopes of SNUPN by Kap-β1. Such a multifactor dissociation mechanism is likely to ensure greater precision of nuclear export and coordination with subsequent nuclear import. In conclusion, the dissociation of SNUPN is driven by a combination of Ran-mediated changes to the tertiary conformation of CRM1 with structural changes to SNUPN and steric clashes induced by cytoplasmic SNUPN ligands.
Supplementary Material
ACKNOWLEDGMENTS
We thank Q. Jiang for help with EM map analyses and M. Rosen for critical reading of the manuscript. This work is funded by US National Institutes of Health grants R01GM069909, R01GM069909-03S1 and 5-T32-GM008297, Welch Foundation grant I-1532 and the University of Texas Southwestern Endowed Scholars Program. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US Department of Energy, Office of Energy Research, under contract no. W-31-109-ENG-38.
Footnotes
Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Gorlich D, Kutay U. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 2.Stade K, Ford CS, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 3.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 5.Fornerod M, Ohno M, Yoshida M, Mattaj IW. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 6.Dong X, et al. Nature. 2009;458 advance online publication, doi:10.1038/nature07975. April. [Google Scholar]
- 7.Paraskeva E, et al. J. Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petosa C, et al. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. J. Biol. Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 10.Matsuura Y, Stewart M. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 11.Engelsma D, Bernad R, Calafat J, Fornerod M. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber J, et al. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strasser A, Dickmanns A, Luhrmann R, Ficner R. EMBO J. 2005;24:2235–2243. doi: 10.1038/sj.emboj.7600701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. J. Biol. Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 15.Ospina JK, et al. Mol. Biol. Cell. 2005;16:4660–4671. doi: 10.1091/mbc.E05-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.