Abstract
IL-33, a member of the IL-1 family of cytokines, is produced by many cell types, including macrophages, yet its regulation is largely unknown. Treatment of primary murine macrophages with a panel of Toll-like receptor (TLR) (e.g., TLR2, TLR3, TLR4, and TLR9) agonists and non-TLR (e.g., MDA5, RIG-I) agonists revealed a pattern of gene and protein expression consistent with a role for IRF-3 in the expression of IL-33. Accordingly, induction of IL-33 mRNA was attenuated in IRF-3−/− macrophages and TBK1−/− mouse embryonic fibroblasts. Despite the fact that all IL-33 agonists were IRF-3-dependent, LPS-induced IL-33 mRNA was fully inducible in IFN-β−/− macrophages, indicating that IL-33 is not dependent on IFN-β as an intermediate. Epinephrine (EPI) and Bordetella pertussis adenylate cyclase toxin (ACT), cAMP-activating agents, activate CREB and greatly synergized with LPS to induce IL-33 mRNA in macrophages. Both LPS-induced and ACT/LPS-enhanced expression of IL-33 mRNA was partially, but significantly, inhibited by the Protein Kinase A (PKA) inhibitor, H-89, but not by tyrosine kinase or PKC inhibitors. Two IL-33 mRNA species derived from two alternative promoters encode full-length IL-33; however, the shorter “A” species is preferentially induced by all IL-33-inducing agonists except Newcastle Disease Virus (NDV), a RIG-I agonist, that induced expression of both “A” and “B” transcripts. Together, these studies greatly extend what is currently known about the regulation of IL-33 induction in macrophages stimulated by bacterial and viral agonists that engage distinct innate immune signaling pathways.
Keywords: Macrophages, murine, IL-33, TLR, gene regulation, cytokines, virus, signal transduction, molecular biology
Introduction
IL-33 was originally identified by Girard and colleagues (1) as a member of the IL-1 family of cytokines based on bioinformatics analysis that revealed that the IL-33 sequence was similar to that of IL-1 family members. IL-33 has also been referred to as IL-1F11, C90RF26, DVS27, and NF-HEV (2). IL-33 was first identified and cloned from canine vasospastic arteries after subarachnoid hemorrhage and it was shown to be highly inducible by IL-1α, IL-1β, and to some extent, by IFN-γ in cultured smooth muscle cells (1). Like IL-1α, the sequence of IL-33 was predicted to have a nuclear localization sequence that would be expected to facilitate its translocation to the nucleus. Nuclear IL-33 was also cloned from human high endothelial venule (HEV) cells and its N-terminus has a homeodomain-like helix-turn helix configuration that is similar to the Drosophila homeodomain transcription factor, engrailed (1). Like IL-1β, IL-33 can be processed by caspase-1 in vitro (2). However, in contrast to IL-1β, cleavage of IL-33 is not required for its biological activity (3).
The receptor for IL-33 is ST2. An orphan receptor, ST2 is conserved across species with homologs in the genomes of mouse, rat, and fruit flies. In humans, there are four ST2 isoforms: soluble sST2 (IL1RL-a), that lacks the transmembrane and cytoplasmic domains and is largely inducible by various immune disorders (4, 5); and a transmembrane ST2L, which is similar to the IL-1 Receptor (6, 7), binds IL-33 on cells and is expressed in a tissue-specific manner on the surface of Th2 cells and mast cells, but not on Th1 cells (8, 9). ST2V is a splice variant of ST2 that lacks the third immunoglobulin motif and C-terminal portion of ST2, and, another ST2 splice variant, ST2LV, lacks the transmembrane domain (10, 11). Soluble ST2 directly binds IL-33 and suppresses activation of NF-κB in EL-4 cells, that stably express ST2L, suggesting that it acts as a decoy receptor (12).
The C-terminus of IL-33 is important for binding to membrane-bound ST2L. The IL-33/ST2L complex subsequently associates with IL-1 receptor accessory protein (IL-1RAcP) to enable IL-33-dependent activation of NF-κB and MAP kinases (JNK, ERK1/2, and p38) (2, 13, 14). As observed for IL-1-mediated signaling, IL-33-receptor interaction recruits the adapter molecule, MyD88, to the receptor complex that, in turn, recruits IRAK1, IRAK4, and TRAF6, leading to MAP kinase activation and NF-κB translocation (2). When this occurs in differentiated Th2 cells, IL-33-mediated signaling can enhance induction of cytokines typically associated with Th2 responses (e.g., IL-4, IL-5, and IL-13) (2). However, secretion of proinflammatory cytokines, e.g., IL-6, IL-1β, TNF-α, and MCP-1 has also been observed in primary mouse-derived mast cells after treatment with IL-33 (15–17). IL-33 also activates basophils, eosinophils, NK, NKT cells, cardiomyocytes, and cultured glial cells (18–22). IL-33 promotes survival and adhesion of human mast cells, as well as production of IL-8 and IL-13 by human umbilical cord blood-derived mast cells (23). In mice, administration of anti-ST2L antibody enhances the Th1 cytokine response and inhibits allergic airway inflammation (24, 25). Thus, although IL-33 can elicit induction of proinflammatory mediators, it favors development of a Th2-biased immune response.
Active secretion of IL-33 from cells has not been reported. Like, HMGB1, IL-1α, or IL-1β, the IL-33 gene sequence does not contain a classical secretory leader sequence (26). Also, like IL-1α, IL-33 exhibits dual functions as both a cytokine and nuclear chromatin modulator (2, 27). IL-33 has been found to be chromatin-associated in the nucleus of endothelial cells and has the capacity to regulate transcription (27). IL-33 binds to chromatin in the surface of nucleosome by docking to the pockets of histone H2A-H2B dimer (28). Despite the fact that IL-33 has been detected in multiple tissues (2), very little is known about how IL-33 production is regulated. Therefore, in the studies described herein, we measured the steady-state levels of IL-33 mRNA or IL-33 protein in macrophages and fibroblasts after exposure to different Toll-like receptor (TLR) ligands, as well as non-TLR ligands including viral pathogens. Our studies revealed that IL-33 is regulated by two transcription factors, Interferon Regulatory Factor-3 (IRF-3) and cAMP response element binding (CREB) protein, but not by Protein Kinase C (PKC) or tyrosine kinases, as supported by pharmacologic inhibition studies. Importantly, while two mRNA species encoding full-length IL-33 are transcribed, the shorter “A” transcript is preferentially induced by all IL-33 inducers identified except the RIG-I agonist, NDV, that induced both “A” and “B” transcripts. These studies provide new insights into the regulation of IL-33 gene expression in macrophages.
Materials and Methods
Reagents
Protein-free E. coli K235 LPS (<0.008% protein) was prepared by modification of the phenol-water extraction method described previously (29). The synthetic lipoprotein S-[2,3-Bis(palmitoyloxy)-(2–RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-Lys4-OH, trihydrochloride (P3C) was purchased from EMC Microcollections (Tuebingen, Germany). Polyinosinic:polycytidylic acid [p(I:C)] was purchased from Amersham Biosciences (Pittsburgh, PA). 5,6-dimethylxanthenone-4-acetic acid (DMXAA) was purchased from Sigma-Aldrich. Anti-phospho IRF-3, anti-β-actin, anti-pSTAT1, anti-p-tyrosine mouse mAb (P-tyr-100 #9411) and anti-p-CREB (Ser 133), that also detects phosphorylation of the CREB-related protein, ATF-1(#9198), were purchased from Cell Signaling, (Beverly, MA, USA). Anti-total IRF3 antibody was obtained from Invitrogen (Carlsbad, CA, USA). Adenylate cyclase toxin (ACT), a potent inducer of cyclic AMP, was the kind gift of Dr. Erik Hewlett (University of Virginia, Charlottesville, VA). Tyrosine kinase inhibitors, PP2 (Src family of protein tyrosine kinases) and EGF/FGF/PDGF Receptor Tyrosine kinase Inhibitor (RTKi), the PKC inhibitor, Go 6983, Epinephrine, and the PKA inhibitor, H-89, were purchased from Calbiochem, EMD Chemicals, Inc. (Gibbstown, NJ).
Cell culture
Primary murine peritoneal macrophages were obtained by peritoneal lavage from 6 to 8-wk old C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME), IFN-β−/−, and IRF-3−/− mice 4 days after i.p. injection with sterile thioglycollate as described previously (30). IFN-β−/− mice (backcrossed ≥N8 onto a C57BL/6 background) (31) were bred homozygously at the University of Maryland, Baltimore. IRF-3−/− mice (backcrossed N≥15 onto a C57BL/6 background) were bred homozygously at University of Massachusetts Medical School and thioglycollate-elicited macrophages were kindly provided by Dr. Katherine Fitzgerald. Macrophages were cultured in RPMI supplemented with 2% FCS, 2 mM glutamine, penicillin, and streptomycin as described previously (30). Mouse embryonic fibroblasts (MEFs) from TBK1+/+ and TBK1−/− mice were a gift of Dr. W.-C. Yeh (University of Toronto, Toronto, Canada). RIG-I and RIG-I−/− mouse embryonic fibroblasts were the kind gift of Dr. S. Akira (32). Embryonic fibroblasts were cultured in DMEM (BioWhittaker), supplemented with 10% (vol/vol) FBS (HyClone Laboratories), glutamate (2 mM), penicillin (10, 000 U/ml), and streptomycin (10,000 µg/ml) at 37° C in 5% CO2 in air.
Cell stimulation
Primary murine macrophages and MEFs were cultured (4 × 106 cells/well) in 6-well plates. After overnight incubation, culture medium was replaced with fresh medium and cells were stimulated with medium only, LPS (100 ng/ml), polyI:C (100 µg/ml), P3C (1 µg/ml), or CpG DNA (1 µg/ml), by transfection of polyI:C (Tfp(I:C))10µg/ml (1 µl Lipofectin was complexed with 1 µg of polyI:C and transfected into the macrophages (32), or by infection with Newcastle Disease Virus (NDV) or Vesicular Stomatitis Virus (VSV) (multiplicity of infection (MOI) =10) for the times indicated in the figures.
Western analysis
Macrophages were washed with PBS and then lysed in buffer (1% Triton X-100, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 10 mM TRIS with protease inhibitor cocktail (Roche) and 1 mM sodium vanadate), and boiled for 5 min with Laemmli lysis buffer for SDS-PAGE and subsequent Western analysis. Twenty microgram of total protein in Laemmli buffer was boiled for 5 min, resolved by 10% SDS-PAGE in Tris/glycine/SDS buffer (25 mM Tris, 250 mM glycine, 0.1% SDS) from Bio-Rad (Hercules, CA), and then electrotransferred onto Immobilon-P transfer membranes (Millipore, Bedford, MA) at 100 V for 1.5 h (4° C). After blocking for 1 h in TBS-T (20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween 20) containing 5% nonfat milk, membranes were washed 3 times in TBS-T and probed for 20 h at 4° C with the respective antibodies, according to manufacturer’s instructions. Following washing in TBS-T, membranes were incubated with secondary HRP-conjugated, anti-rabbit IgG from Cell Signaling (1: 2,000 dilution) for 1 h at room temperature, washed three times in TBS-T, and bands were detected using ECL plus reagents (Amersham Pharmacia Biotech, Piscataway, NJ). Densitometric signals from Western blots were quantified using the ImageJ program from the NIH (http://rsbweb.nih.gov/ij/).
Measurement of steady-state mRNA by quantitative real-time PCR
Total RNA was isolated by using Trizol reagent from Invitrogen (Carlsbad, CA, USA) as specified by the manufacturer’s instructions and quantified by spectrophotometric analysis. The cDNA was prepared from 1 µg of total RNA using iScript reverse transcriptase (Bio-Rad, Hercules, CA, USA) and both poly-oligo dT and random primer mix, as recommended by the manufacturer’s instructions. The resulting cDNA was quantified by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and ABI Prism 7900HT cycler as described previously (33). Primers for detection of IL-33, IFN-β, IL-12 p40, Elam, ISG-56, IP-10, and GAPDH mRNAs were designed using the Primer Express 2.0 program (Applied Biosystems). The IL-33 sense primer 5’-TGAGACTCCGTTCTGGCCTC-3’ and anti-sense primer 5’-CTCTTCATGCTTGGTACCCGAT-3’ were used for most of the experiments in this study.
Full-length IL-33 is encoded by 2 mRNA species, “A” and “B,” that are transcribed from different promoters. In order to ascertain which mRNA species was induced in murine macrophages, we designed two specific forward primers that correspond to the two respective 5’UTRs, (5’-GGGGCTCACTGCAGGAAAGTA-3’ (AK075849.1) and 5’- CAGCTGCAGAAGGGAGAAAT-3’ (AK163464.1), and one common reverse primer from the 3rd exon (5’- CTTATGGTGAGGCCAGAACG-3’). PCR products were cloned into the TA cloning vector (pDrive) and further sequenced by the Biopolymer and Genomics Core Laboratory at the University of Maryland, Baltimore. The real time PCR primers, “A” mRNA (AK075849.1) forward primer (5’-GGGCTCACTGCAGGAAAGTA-3’), “B” mRNA (AK163464.1) forward primer (5’-CAGCTGCAGAAGGGAGAAAT-3’), and the common reverse primer (5’-GGACCAGGGCTTCGCCT-3’) were used to amplify specific IL-33 transcripts produced in vitro and in vivo.
Flow cytometry analysis
To preclude the need for scraping cells for flow cytometric studies, thioglycollate-elicited peritoneal macrophages from C57BL/6J mice were cultured on 6-well low cluster and low adhesion plates (Corning, Inc.). Washes were carried out in centrifuge tubes and the cells were re-plated and treated with medium alone, polyI:C transfection (10 µg/ml) or infected with NDV (MOI = 10) for 14 h. Cells were harvested for analysis by gentle shaking, washed with PBS, and then fixed with 4% p-formaldehyde (PFA) for 10 min at room temperature. Cells were blocked and permeabilized for 30 min with PBST (PBS, 1% BSA, 1% normal donkey serum, 0.3 % Triton X-100) at room temperature. IL-33 was detected using a goat polyclonal antibody (R&D Systems) directed against the protein, followed by secondary and tertiary staining with biotin-conjugated donkey anti-goat IgG and Cy3-conjugated streptavidin, respectively (Jackson ImmunoResearch Laboratories). Cells were washed in PBST and suspended in PBS for immediate analysis using a FACSCalibur. Analytic gates were set to exclude cellular debris and aggregates. CELLQuest software (Becton Dickinson) was used to analyze the data.
Results
Induction of IL-33 mRNA in mouse peritoneal macrophages by TLR and non-TLR agonists
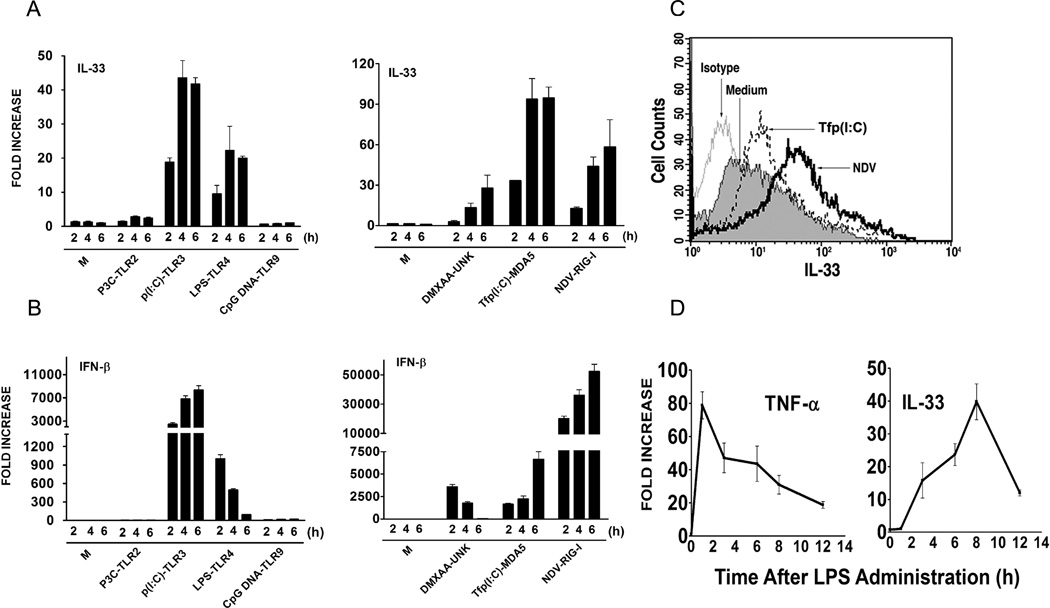
While IL-33 has been shown to be produced by macrophages (34), little is known about its transcriptional regulation by TLR and non-TLR stimuli. Therefore, we initially measured steady-state IL-33 mRNA levels in mouse primary macrophages after treatment with TLR2, TLR3, TLR4, and TLR9 agonists, e.g., P3C, poly I:C (p(I:C)), LPS, and CpG DNA, respectively. As shown in Figure 1A (left panel), stimulation by TLR3 and TLR4 agonists strongly up-regulated IL-33 mRNA expression in a time-dependent manner; however, there was no IL-33 mRNA induced when macrophages were stimulated with TLR2 (P3C) or TLR9 (CpG DNA) ligands. The ability of the TLR agonists to induce IL-33 mRNA correlated with their ability to induce IFN-β (Figure 1B, left panel). Thus, we hypothesized that non-TLR ligands that are strong inducers of IFN-β would also be strong inducers of IL-33 gene expression. Therefore, IL-33 mRNA was measured in macrophages treated with the anti-cancer drug, DMXAA, that uses an unknown (UNK), non-TLR, non-RIG-I, non-MDA5 sensor (35), transfected polyI:C (Tfp(I:C)), an MDA5 ligand (32), or NDV infection, a RIG-I ligand (32, 36), all of which are potent inducers of IFN-β (32, 35, 36). Figure 1A (right panel) shows that IL-33 mRNA was potently induced by Tfp(I:C) and NDV, and to a lesser extent, by DMXAA, in primary macrophage cultures with kinetics similar to the active TLR agonists, and that each of these agents also induced IFN-β mRNA (Figure 1B, right panel). These findings were confirmed by measuring intracellular IL-33 protein in macrophages by FACS analysis after treatment with Tfp(I:C) or NDV infection for 14 h (Figure 1C). Thus, we found a strong correlation between agents that are known IFN-β inducers and the ability of these agents to induce IL-33.
Figure 1. Induction of IL-33 in peritoneal macrophages by TLR and non-TLR agonists.
(A) IL-33 mRNA or (B) IFN-β mRNA was measured by quantitative real-time PCR at the indicated times following stimulation with LPS (100 ng/ml), P3C (1 µg/ml), CpG DNA (1 µM), DMXAA (100 µg/ml), p(I:C) (100 µg/ml), transfection (Tf) of p(I:C) (10 µg/ml) or NDV infection (MOI=10) in peritoneal macrophages. (C) Intracellular IL-33 protein was measured in macrophages by staining with anti-IL-33 antibody and analyzed by FACS, 14 h after stimulation of cells with Tfp(I:C) (10 µg/ml) or NDV infection (MOI=10). M; medium (unstimulated). (D) TNF-α and IL-33 mRNA were measured by real-time PCR in liver samples at the indicated times after lethal injection of LPS (35 mg/kg body weight). M; medium (unstimulated control). Data shown are representative of two separate experiments.
We also examined the induction of IL-33 in vivo, in the livers of mice after lethal challenge with LPS. IL-33 mRNA levels increased over 8 h in liver samples and then diminished, in contrast to TNF-α mRNA levels that peaked after only 1 h (Figure 1D). Thus, IL-33 mRNA is induced relatively late in response to LPS in vivo.
Induction of IL-33 mRNA in LPS-stimulated murine peritoneal macrophages is independent of IFN-β production
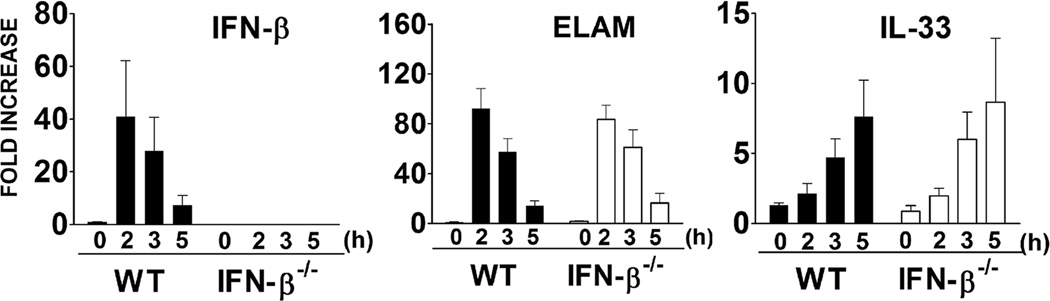
IFN-β is the primary Type I IFN induced by TLR stimulation in macrophages and often functions in an autocrine/paracrine manner through the IFN-α/β receptor to activate transcription factors, e.g., STAT1, STAT2, that induce expression of IFN-β-dependent genes including MCP-5 and iNOS (37), as well as anti-inflammatory genes such as IL-10, IL-4 (38), and SOCS-1 (35). To determine if LPS-induced IL-33 was dependent on the production and autocrine action of IFN-β, macrophages from WT and IFN-β−/− mice were stimulated by LPS and induction of IFN-β (control), endothelial leukocyte adhesion molecule (ELAM) (IFN-β-independent (39)), and IL-33 mRNA were compared (Figure 2). We confirmed that IFN-β−/− macrophages did not produce IFN-β mRNA in response to LPS, in contrast to WT macrophages (left panel). Expression of the gene that encodes ELAM is strongly regulated by transcription factor NF-κB (39) and was comparably induced by LPS in WT and IFN-β-deficient macrophages (middle panel). Similar to ELAM mRNA, IL-33 mRNA was comparably induced by LPS in WT and IFN-β−/− macrophages (right panel). These data indicate that induction of IL-33 mRNA by LPS is IFN-β-independent.
Figure 2. IL-33 mRNA induction in macrophages is independent of IFN-β.
IFN β, ELAM, and IL-33 mRNAs were measured by quantitative real-time PCR at the indicated times after stimulation with LPS (100 ng/ml) in WT and IFN-β−/− macrophages. Data shown are representative of two separate experiments.
TLR- and non-TLR-induced IL-33 mRNA is TBK1-dependent in MEFs
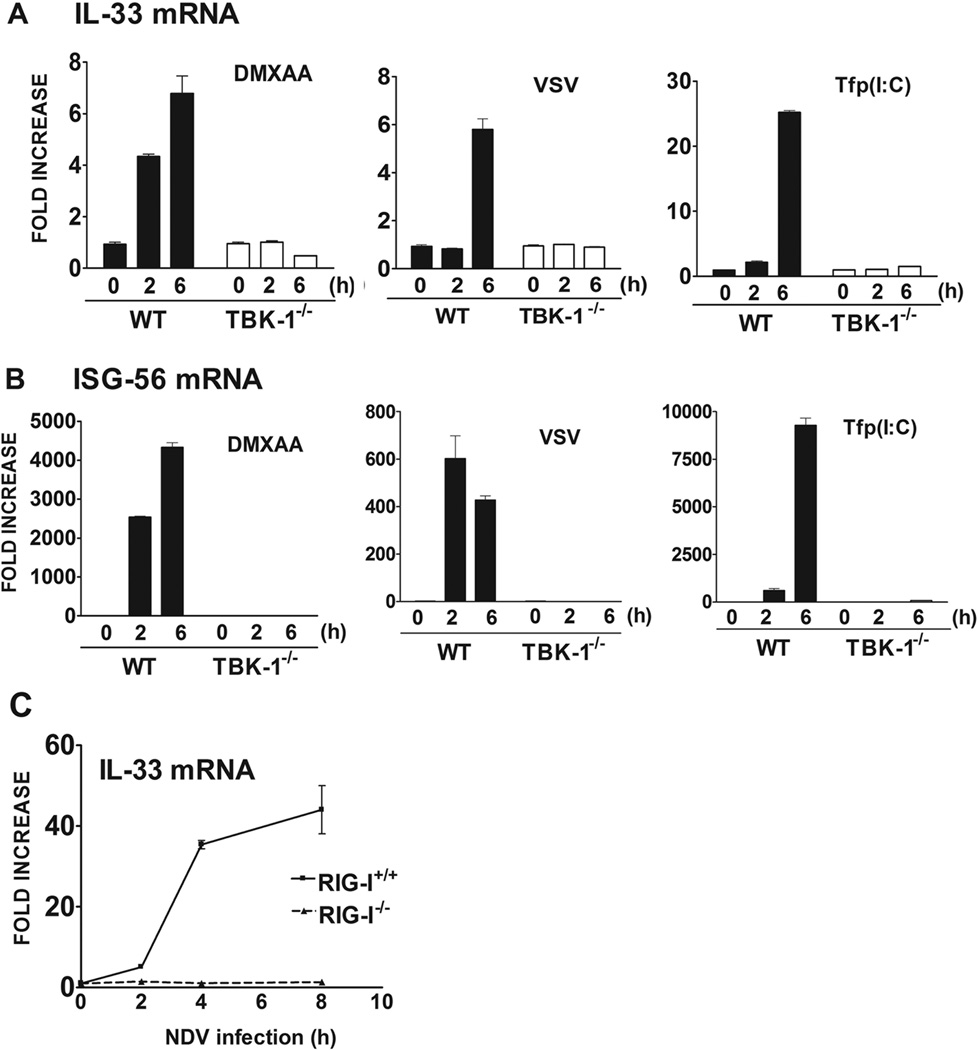
Activated TRIF, MDA5, and RIG-I recruit TBK-1, leading ultimately to activation of IRF-3, a key transcription factor that is required for induction of IFN-β and other IRF-3-dependent gene products, e.g., RANTES (32, 36, 40). Since, the induction of IL-33 was IFN-β-independent, but inducible by stimuli that share the capacity to induce IFN-β through activation of IRF-3, we next sought to determine if IL-33 induction was also dependent on TBK-1, the kinase that leads to IRF-3 activation. Since TBK-1−/− mice do not survive (35), we utilized TBK-1−/− MEFs for these experiments. Figure 3A shows that in contrast to WT MEFs, IL-33 mRNA failed to be induced in TBK-1−/− MEFs after stimulation with DMXAA (stimulates IRF-3-induced IFN-β through an UNK sensor), VSV infection (RIG-I), or Tfp(I:C) (MDA5), while ELAM mRNA expression was induced by these three agonists in both in WT and TBK-1−/− MEFs (data not shown). IL-33 mRNA was also not induced in TBK-1−/− MEFs when compared to WT MEFs after stimulation by LPS (data not shown). As an additional control, ISG-56 mRNA, an IFN-β-inducible gene, was also measured in these same samples. Induction of ISG-56 mRNA by these agonists (Figure 3B) was also abrogated in TBK-1−/− MEFs when compared to WT MEFs. Thus, IL-33 mRNA induction by these stimuli in MEFs is TBK-1-dependent.
Figure 3. IL-33 mRNA induction in mouse embryonic fibroblasts is dependent on TBK-1.
(A) IL-33 and (B) ISG-56 mRNA were measured by quantitative real-time PCR at the indicated times after stimulation with DMXAA (100 µg/ml), VSV infection (10 MOI), and Tf p(I:C) (10 µg/ml) in WT and TBK 1−/− mouse embryonic fibroblasts. (C) IL-33 mRNA was measured by quantitative real-time PCR at the indicated times after NDV-infection (MOI=10) in WT and RIG-I−/− mouse embryonic fibroblasts. Data shown are representative of two separate experiments.
NDV-induced IL-33 mRNA is RIG-I-dependent in MEFs
Previous studies have shown that NDV infection activates the RIG-I signaling pathway and uses IPS/MAVS, rather than TRIF, to activate TBK-1, IRF-3, and NF-κB (41). Since RIG-I−/− mice also fail to survive (35), we utilized WT and RIG-I−/− MEFs to determine whether induction of IL-33 mRNA is dependent upon RIG-I. Figure 3C shows a significant, time-dependent increase in IL-33 mRNA levels after NDV infection of WT MEFs that was completely eliminated in RIG-I−/− MEFs. There was no difference in induction of IL-33 mRNA in RIG-I+/+ and RIG-I−/− MEFs when stimulated with DMXAA, Tfp(I:C), or LPS, ligands that do not activate RIG-I (35) (data not shown).
Induction of IL-33 is IRF-3-dependent in macrophages
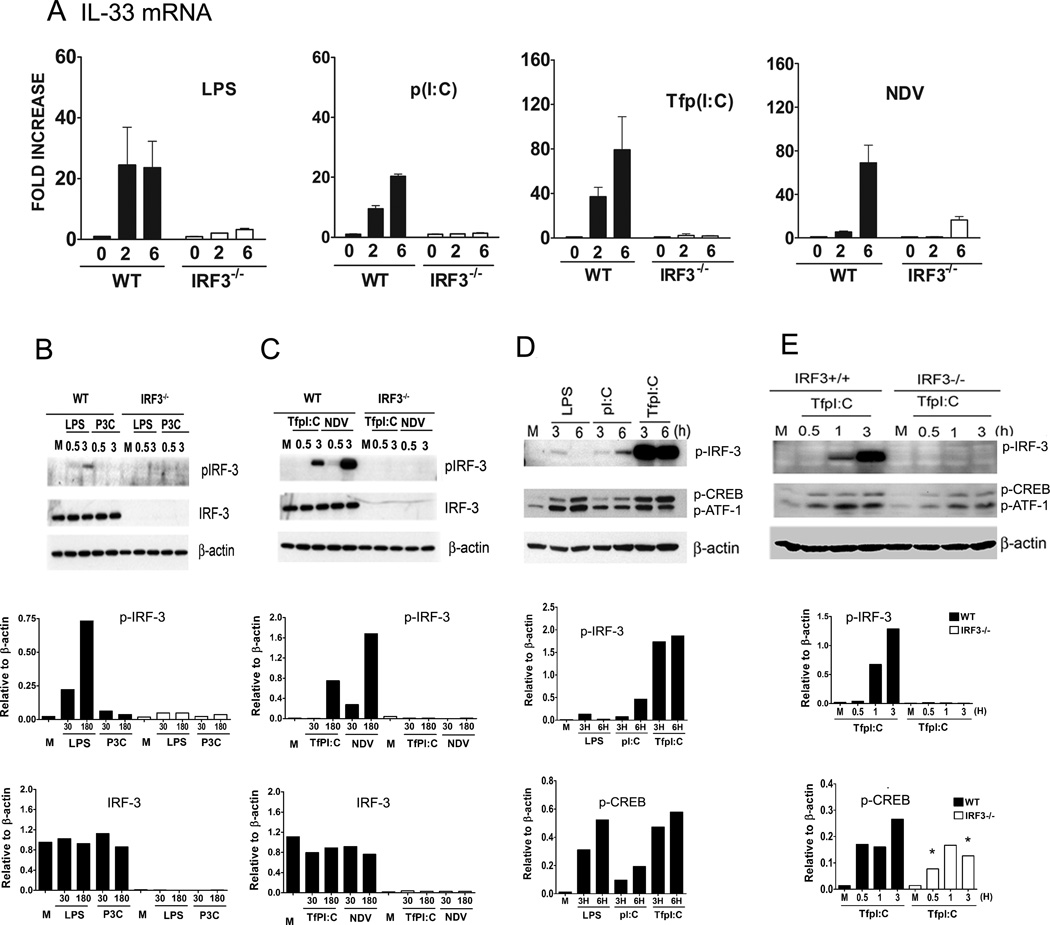
Since the induction of IL-33 could not be attributed to the autocrine/paracrine action of IFN-β (Figure 2), but was dependent on TBK1 (Figure 3), we next hypothesized that IL-33 induction was IRF-3-dependent. Therefore, IL-33 mRNA was measured in WT and IRF3−/− peritoneal macrophages that were stimulated with LPS, P3C, p (I:C), or Tfp(I:C), or infected with NDV. Figure 4A shows that induction of IL-33 mRNA was significantly inhibited in IRF3−/− macrophages compared to WT macrophages after stimulation with these TLR and non-TLR agonists that activate TBK-1 and induce IFN-β expression (Figures 1 and 3A). IL-33 mRNA failed to be induced in WT or IRF3−/− macrophages after treatment with P3C (data not shown), consistent with the failure of this TLR2 agonist to activate the “MyD88-independent,” TRIF-dependent pathway (37, 42, 43). However, MyD88-dependent expression of TNF-α mRNA was comparably induced in P3C-treated WT or IRF-3−/− macrophages (data not shown).
Figure 4. IRF-3 is required for induction of IL-33 in macrophages by various agonists.
(A) IL-33 mRNA was measured by real-time PCR at the indicated times after stimulation with LPS (100 ng/ml), p(I:C) (100 µg/ml), Tf p(I:C) (10 µg/ml), and NDV infection (MOI=10) in WT and IRF3−/− macrophages. (B) and (C). Whole cell lysates were prepared from macrophages after stimulation with LPS (100 ng/ml), P3C (1 µg/ml), Tfp(I:C) (10 µg/ml), or NDV infection (MOI=10) at the indicated times and were subjected to Western analysis using anti-phospho-IRF-3 (p-IRF-3), anti-IRF-3, and anti-β-actin antibodies. β-actin was used as a loading control. The panels below each Western blot represent the denstitometry data quantified after normalization to β-actin. (D) Whole cell lysates were prepared after stimulation with LPS (100 ng/ml), p(I:C) (100 µg/ml), or Tfp(I:C) (10 µg/ml) at the indicated times in macrophages and whole cell lysates were subjected to Western analysis using anti-phospho-IRF-3 (p-IRF-3), anti-phospho-CREB (p-CREB), and β-actin antibodies. Panels below each Western blot represent the densitometry data quantified after normalization to β-actin. Note that the anti-CREB antibody used also detects p-ATF-1 (see Materials and Methods) which is similarly modulated. (E) Whole cell lysates were obtained from the WT and IRF-3−/− macrophages after stimulation with Tfp(I:C) (10 µg/ml) and these lysates were analyzed for phospho-IRF-3 and phospho-CREB by Western blot as described for (D). M; medium (unstimulated control). Data shown are representative of two separate experiments.
Total cell lysates were extracted from WT and IRF3−/− macrophages treated with LPS, P3C, Tfp(I:C), and NDV infection and were subjected to Western analysis for detection of phosphorylated-IRF-3 (pIRF-3), total IRF-3, and β-actin, the latter being used as a protein loading control. These Western blots were subjected to scanning densitometry and quantified after normalization to the β-actin signal and these data are presented below each Western blot. As seen in Figures 4B and 4C, relative levels of total IRF-3 protein were comparable in WT macrophages regardless of treatment, and as expected, IRF-3 protein was not detected in IRF-3−/− macrophages. Phosphorylated IRF-3 was detected in WT macrophages after stimulation with LPS (Fig. 4B), Tfp(I:C), and NDV infection (Fig. 4C) for 3 h. NDV and TfpI:C were more potent activators of p-IRF-3 than LPS at this time point. No phosphorylation of IRF-3 was detected in samples treated with the TLR2 agonist, P3C, in WT or IRF3−/− macrophages (Fig. 4B), consistent with the failure of this agonist to induce IL-33 or IFN-β mRNA (Figure 1). Thus, TBK-1-dependent phosphorylation of IRF-3 is required for induction of IL-33 in macrophages.
We next measured phosphorylation of IRF-3 in macrophages after stimulation with TLR and non-TLR agonists for longer periods of time, i.e., 3 or 6 hours. Figure 4D confirms and extends the data in Figure 4B that LPS- and polyI:C-induced phosphorylation of IRF-3 was transient and relatively weak compared to that induced by TfpI:C: LPS-induced phosphorylation of IRF-3 was detected at 3 h, but no longer at 6 h, whereas p(I:C)-induced phosphorylation of IRF-3 was detectable at 3 h and continued to increase at 6 h. Robust phosphorylation of IRF-3 by Tfp(I:C) was observed at both 3 h and 6 h.
Role of cAMP-CREB pathway in the transcriptional regulation of IL-33
The cAMP response element binding (CREB) proteins are transcription factors that bind to DNA sequences called cAMP response elements (CRE) and thereby regulate expression of certain genes. cAMP-activating drugs produce second messengers, such as cAMP or Ca2+, that, in turn, activate protein kinase A (PKA), that translocates to the nucleus where it activates CREB (44, 45). In addition, CREB activation can be driven by TLR or non-TLR ligands that activate MAPKs pathways, involving ERK and p38 MAPK signaling (46). Activated CREB binds to a CRE and is then bound by CREB binding protein that co-activates it, leading to increased or decreased transcription (47). Figure 4D shows that in addition to IRF-3 being phosphorylated, CREB phosphorylation was also up-regulated by LPS, p(I:C), and Tfp(I:C); however, the relative degree of CREB activation induced by these agonists differed from their relative abilities to activate IRF-3: LPS- and Tfp(I:C)-induced phosphorylation of CREB was only slightly greater than that induced by p(I:C). Stimulation of IRF-3−/− macrophages with Tfp(I:C), the agonist shown in Fig. 4D to be the most potent of the non-infectious IRF-3 activating agents, resulted in reduced phosphorylation of CREB compared to WT macrophages (Figure 4E), suggesting that IRF-3 may contribute partially to the activation of CREB by Tfp(I:C).
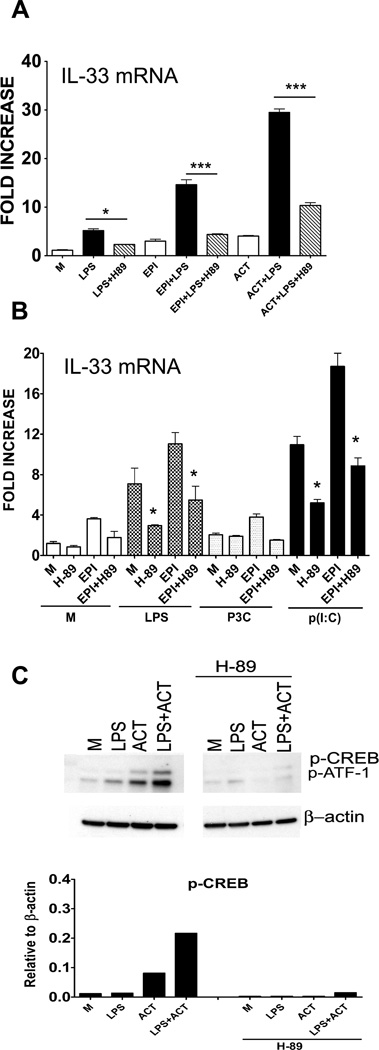
In Th1 cells, cAMP inhibits Th1 cytokine production, whereas cAMP markedly increases Th2 cytokine production in GATA3-expressing cells (48). cAMP also activates p38, that in turn, activates GATA3, leading to Th2 cytokine production (49, 50). Since IL-33 has been associated with a Th2 bias, we hypothesized that cAMP might also be involved in regulation of IL-33 production. Therefore, we measured the effects of cAMP agonists on LPS-induced IL-33 mRNA in peritoneal macrophages. Figure 5A illustrates that LPS-induced IL-33 mRNA was enhanced ~5-fold compared to untreated macrophages, but in the presence of potent cAMP activating drugs, epinephrine (EPI) or adenylate cyclase toxin (ACT), a synergistic induction of LPS-induced IL-33 mRNA expression was observed. Both LPS- and LPS + EPI/ACT-induced IL-33 mRNA was significantly inhibited in the presence of the PKA inhibitor, H-89 (Figure 5A). A similar trend was observed when macrophages were treated with LPS and p(I:C), but not P3C, in the absence or presence of EPI (Figure 5B). These data support the possible involvement of the PKA-CREB pathway in IL-33 mRNA induced by LPS and other IL-33-inducing agonists in macrophages.
Figure 5. Evidence for the cAMP-CREB pathway in regulation of IL-33 mRNA in peritoneal macrophages.
(A) IL-33 mRNA was measured by real-time PCR at the indicated times following stimulation with LPS (100 ng/ml), without or with cAMP-activating drugs, epinephrine (EPI; 10 µM) or ACT (20 nM), in the absence or presence of the PKA inhibitor, H-89 (10 µM). Data shown are the average ± SEM of three separate experiments (*, p≤ 0.05; **, p≤ 0.01; ***, p≤ 0.001). (B) IL-33 mRNA was measured as described in (A) after stimulation of macrophages with LPS (100 ng/ml), P3C (1 µg/ml), or p(I:C) (100 µg/ml), without or with cAMP-activating drug, epinephrine (EPI; 10 µM), in the absence or presence of the PKA inhibitor, H-89 (10 µM). Data shown are the average ± SEM of three separate experiments (*, p≤ 0.05; **, p≤ 0.01; ***, p≤ 0.001). (C) Whole cell lysates were prepared after stimulation of macrophages for 2 h with LPS (100 ng/ml), without or with ACT (20 nM), in the absence or presence of the PKA inhibitor, H-89 (10 µM). Whole cell lysates were analyzed by Western blot using anti-phospho-CREB and anti-β-actin antibodies. β-actin was used as loading control. The panel below the Western blot represents the densitometry data quantified after normalization to β-actin. M; medium (unstimulated control). Data shown are representative of two separate experiments.
In contrast to the synergy observed between LPS and either cAMP-activating agent at the level of IL-33 mRNA expression, LPS-induced CREB phosphorylation was only slightly enhanced by the presence ACT. Nonetheless, H-89 also inhibited phosphorylation of CREB induced by LPS alone, ACT alone, or both (Figure 5C), as indicated by decreased signal in the blots and in the corresponding normalized densitometric scan. Consistent with a role for PKA in the induction of IL-33 by LPS, H-8, a PKA inhibitor that is a ~10-fold less potent than H-89 (51–53), inhibited LPS-, ACT-, and LPS/ACT-induced p-CREB much less than H-89 at the same concentration (data not shown).
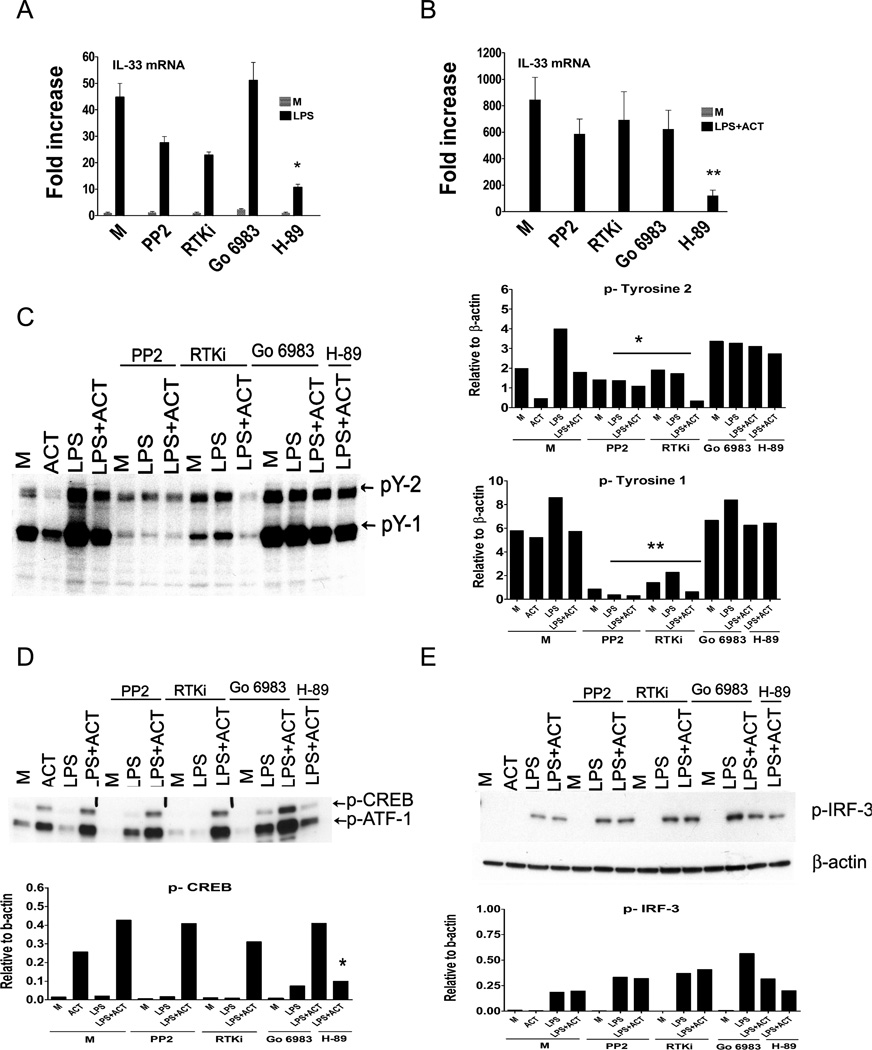
To determine if tyrosine kinases or Protein Kinase C (PKC) were involved in signaling leading to IL-33 mRNA expression, we next compared induction of IL-33 mRNA by LPS alone (Figure 6A) or LPS + ACT (Figure 6B), in the presence of tyrosine kinase inhibitors (PP2 or RTKi), the PKC inhibitor, Go 6983, and as well the PKA inhibitor H-89 (as a control). LPS-induced IL-33 mRNA was not statistically inhibited in the presence of tyrosine kinase inhibitors, PP2 or RTKi, or the PKC inhibitor, under conditions where H-89, the PKA inhibitor, significantly inhibited LPS- and LPS + ACT-induced IL-33 mRNA in macrophages as was observed in Figure 5A. We confirmed that PP2 and RTKi were active at concentrations used in Figure 6A and 6B by showing inhibition of tyrosine phosphorylation (p-Y) induced by LPS or LPS + ACT by inclusion of either of the two tyrosine kinase inhibitors in the cultures, while neither the PKC or PKA inhibitors had any effect on phospho-tyrosine induction (Figure 6C; densitometric analyses of the two major phospho-tyrosine species, pY-2 and pY-1, are presented to the right of each Western blot). As was observed at the level of IL-33 mRNA, there was no inhibition of p-CREB in the presence of the receptor tyrosine kinase or PKC inhibitors, under conditions where phosphorylation of CREB was significantly inhibited by H-89 (Figure 6D; densitometric analysis is below the blot). Finally, there was no significant inhibition of LPS-induced phospho-IRF-3 in presence of any of these inhibitors (Figure 6E; densitometric analysis is below the blot).
Figure 6. Role of receptor tyrosine kinase, PKC, and PKA in the induction of IL-33 mRNA in macrophages.
(A) IL-33 mRNA was measured by quantitative real-time PCR following stimulation with LPS (100 ng/ml), in the absence or presence of receptor tyrosine kinase inhibitors (PP2 (2.5 µM) or RTKi (1 µM), the PKC inhibitor, Go 6983 (5 µM), or the PKA inhibitor, H-89 (10 µM). (B) IL-33 mRNA was measured by quantitative real-time PCR following stimulation with LPS (100 ng/ml) + ACT (20 nM), in the absence or presence of receptor tyrosine kinase inhibitors (PP2 (2.5 µM) or RTKi (1 µM)), the PKC inhibitor ,Go 6983 (5 µM), or the PKA inhibitor, H-89 (10 µM). (C) Whole cell lysates were prepared after stimulation of macrophages following stimulation with LPS (100 ng/ml), without or with ACT (20 nM), in the absence or presence of receptor tyrosine kinase inhibitors (PP2 (2.5 µM) or RTKi (1 µM)), the PKC inhibitor, Go 6983 (5 µM), or the PKA inhibitor, H-89 (µM) for 1 h. The whole cell lysates were analyzed by Western blot using anti-phospho-Tyrosine (pY) antibody. The panels to the right represent the densitometry data quantified after normalization of the two major bands, pY-2 and pY-1, to β-actin. Data shown are the average ± SEM of three separate experiments (*, p≤ 0.05; **, p≤ 0.01; ***, p≤ 0.001). (D) The same gel shown in (C) was re-probed with anti-phospho-CREB antibody and (E) anti-phospho-IRF-3 antibody. The panels below (D) and (E) represent the densitometry data quantified after normalization to β-actin. M; medium (unstimulated control). The data presented represent individual experiments of at least 2 experiments that were similar in design and outcome.
Mouse gene IL-33 organization and induction of specific IL-33 transcripts
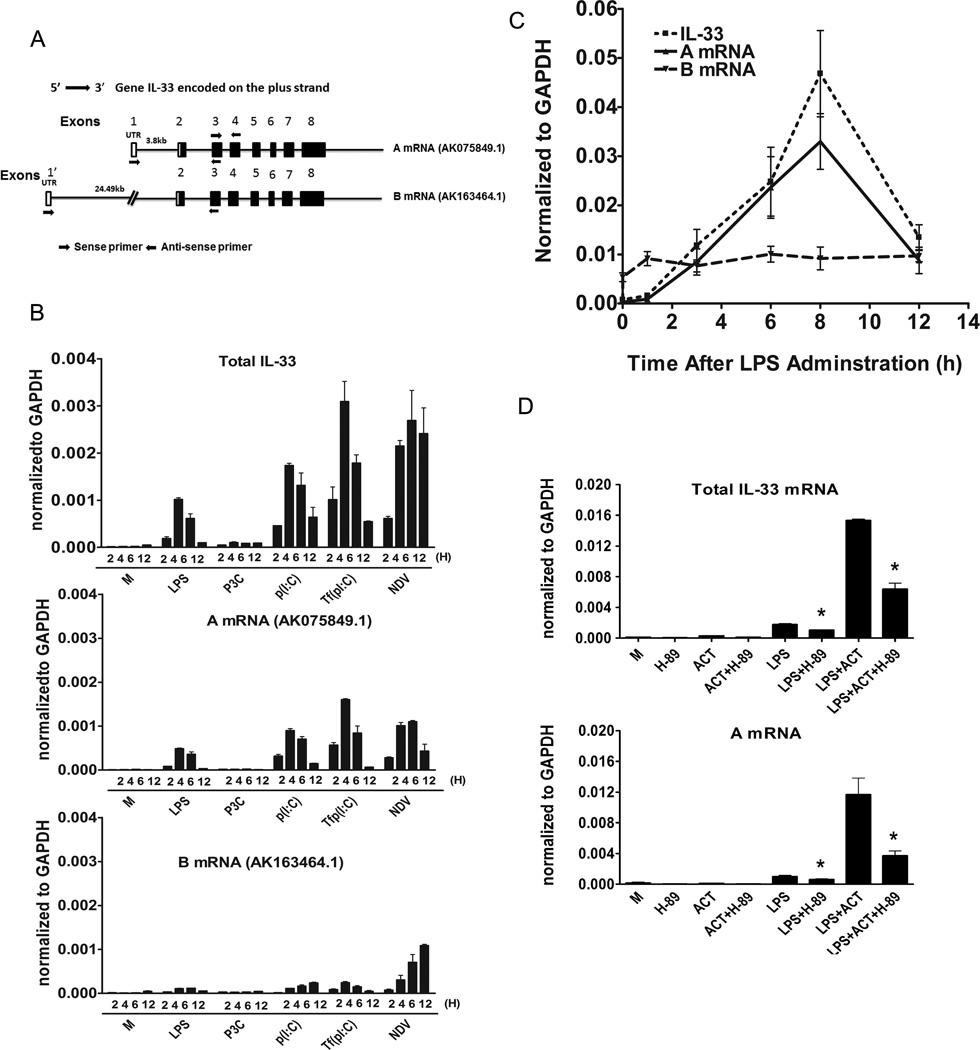
Originally, Baekkevold et al. (1) published that the mouse IL-33 genomic structure was comprised of seven exons. The major difference between human and mouse IL-33 genes was the size of the first intron that was estimated to be 9 kb in the human IL-33 gene, but only 2 kb in mouse ortholog. However, recent advances in sequencing analysis and aceView (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=mouse&term=IL-33&submit=Go) showed that the first exon identified in the paper by Baekkevold et al. (1) is actually exon 2. The IL-33 gene is localized on mouse chromosome 19 and the sequence of the mouse IL-33 gene has been defined by 188 gene bank accessions from 185 cDNA clones from different mouse tissues. The IL-33 gene contains 11 different GT-AG introns and is transcribed into seven different mRNA species, with five alternatively spliced variants and two unspliced forms. Thus, there are 3 possible alternative promoters and four validated altered polyadenylation sites. Two of these mRNAs, transcripts “A” and “B” (see Figure 7A), are derived from two alternative promoters, and are both predicted to encode a full-length IL-33 protein encoded by eight exons. The other splice variants are not predicted to result in a full-length protein and were therefore not studied further.
Figure 7. Mouse gene IL-33 organization and induction of alternative IL-33 transcripts.
(A) Mouse IL-33 genomic structure. Open boxes indicate non-translated exon sequences and black boxes are coding sequences. Both coding and untranslated sequences are distributed over eight exons. IL-33 pre-mRNA is comprised of species that vary between 35,609 bp (AK163464.1) and 14,915 bp (AK075849.1), flanked by two different non-translated regions at the 5’ end (5’UTR). The “A” mRNA (AK075849.1) and the “B” mRNA (AK163464.1) species are as shown in the figure. Total IL-33 mRNA was measured by quantitative real-time PCR using the primers that flank the region between exons 3 and 4; the “A” mRNA species was measured by using primers in first exon and second exon and whereas “B” mRNA was measured by using the primers derived from sequences in exon 1’ and exon 2. (B) IL-33 mRNA (total IL-33, “A” mRNA and “B” mRNA) was measured by real-time PCR at the indicated times following stimulation with LPS (100 ng/ml), P3C (1 µg/ml), p(I:C) (100 µg/ml), Tfp(I:C) (10 µg/ml), or NDV infection (MOI=10) of peritoneal macrophages. (C) IL-33 mRNA (total IL-33, “A” mRNA, and “B” mRNA) was measured by real-time PCR in liver samples, at the indicated times, after lethal injection of LPS (35 mg/kg body weight). M; medium (unstimulated control). (D) Total IL-33 and “A” mRNA was measured by quantitative real-time PCR at the indicated times following stimulation with LPS (100 ng/ml), without or with ACT (20 nM), in absence or presence of the PKA inhibitor, H-89 (10 µM). M; medium (unstimulated control). Data shown are the average ±SEM of three separate experiments (*, p≤ 0.05).
The shorter transcript, “A,” is 14,915 bp (AK075849.1) and longer transcript, “B,” comprises a pre-mRNA length of 35,609 bp (AK163464.1). Transcripts A and B are identical with the exception of the 5’ untranslated region (5’ UTR). We designed two specific forward primers that correspond to the two respective 5’UTRs, (5’-GGGGCTCACTGCAGGAAAGTA-3’ (AK075849.1) and 5’- CAGCTGCAGAAGGGAGAAAT-3’ (AK163464.1), and one common reverse primer from the 3rd exon (5’- CTTATGGTGAGGCCAGAACG-3’) to identify these two mRNA species in murine macrophages. These two sets of primers amplified two predominant (i.e., each primer set amplified a single PCR product) PCR products from LPS-stimulated macrophage cDNA. These two PCR products were gel purified and cloned into the TA cloning vector (pDrive). Sequence analyses confirmed that both “A” and “B” IL-33 mRNAs are expressed in murine macrophages. The inducibility of these distinct mRNA species by TLR and non-TLR ligands was next analyzed by real-time PCR using primers specific for “A” or “B” mRNA. As shown in Figure 7B, LPS, p(I:C), and Tfp(I:C) induced “A” mRNA (AK075849.1) and the induction paralleled that of total IL-33 mRNA, whereas “B” mRNA (AK163464.1) was only poorly induced when stimulated with these three agonists. However, after infection of macrophages with NDV, both “A” and “B” IL-33 mRNA species were expressed. We also observed “A” mRNA was predominately induced in liver samples after sub-lethal dose of LPS administration, whereas “B” IL-33 mRNA remained at basal levels, indicating that “A” mRNA is preferentially induced by LPS in vivo and in vitro (Figure 7C). Finally, LPS-induced IL-33 total mRNA and “A” mRNA were greatly augmented in the presence of the cAMP-activating agent, ACT (Figure 7D), whereas “B” mRNA was not significantly increased (data not shown). LPS/ACT-induced total IL-33 mRNA and “A” mRNA were partially, but significantly, inhibited in presence the PKA inhibitor, H-89 (Figure 7D).
Discussion
Relatively little is known about the molecular regulation of IL-33 gene expression. To approach this problem, we studied the expression of IL-33 mRNA in murine macrophages and fibroblasts after stimulation with a diverse panel of TLR and non-TLR ligands. Although these various stimuli elicit signaling through distinct membrane-associated, endosome-associated, or cytosolic receptors, our data suggest that a common pathway for IL-33 gene expression is the activation of TBK1 that leads to activation of the transcription factor, IRF-3 (Figures 3A and 4A). IRF-3 is a key transcriptional activator and is central to the transcriptional upregulation of IFN-β and other genes (54). Once produced, IFN-β acts in an autocrine/paracrine fashion to activate the IFN-α/β receptor (IFNAR), leading to transcriptional induction of >300 IFN-stimulated genes (55). While induction of IL-33 in macrophages was entirely IRF-3-dependent (Figure 4A), the failure to observe any difference in LPS-induced IL-33 mRNA in LPS-stimulated WT and IFN-β−/− macrophages indicates that induction of IL-33 is not IFN-β-dependent. This suggests the possibility that activated IRF-3 might bind directly to regulatory regions within the IL-33 gene to drive IL-33 transcription.
Given the multiple reports of the association of IL-33 with asthma, we explored the possibility that other transcription factors were associated with IL-33 gene expression. NF-κB, AP-1, NF-AT, CREB, and STATs have been strongly implicated in genes expressed in allergic asthma (56). Phosphorylated CREB was significantly increased in patients with bronchial asthma and its induction correlated with the inflammatory status (57). Augmentation of phospho-CREB was suppressed upon inhaled glucocorticoid treatment in an experimental model of asthma (57). Both TLR and non-TLR ligands have been reported to activate second signals that ultimately activate PKA and its subsequent translocation to the nucleus where it activates CREB (44, 45). Therefore, the effects of cAMP-activating agents and the PKA inhibitor, H-89, were tested for their effects on inducible IL-33 mRNA expression. Both EPI and ACT, known cAMP agonists (58, 59), had little effect on IL-33 mRNA levels alone, but synergistically increased levels LPS-induced IL-33 mRNA. Both LPS-induced and LPS + EPI/ACT-induced IL-33 mRNA were sensitive to pharmacologic inhibition of PKA, as was p(I:C) and p(I:C) + EPI-induced IL-33 mRNA (Figure 5A, B). This data supports the role of the transcription factor, CREB, in the induction of IL-33 mRNA. It is interesting to note that in IRF-3−/− macrophages, CREB phosphorylation induced by TfpI:C was diminished (Fig. 7C), suggesting yet another mechanism by which IRF-3 may regulate IL-33 gene expression.
Transcription of IL-33 is complex, with seven different mRNA species, five alternatively spliced variants, and two unspliced forms (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=mouse&term=IL-33&submit=Go). Only two transcripts, “A” and “B” (Figure 7A) encode the full-length IL-33 protein. While our initial cloning experiments revealed that both “A” and “B” mRNA variants are produced in stimulated primary murine macrophages, all of the stimuli tested, except for NDV, resulted in preferential induction of the shorter IL-33 “A” mRNA variant (Figure 7B). In contrast, NDV infection induced a mixture of both “A” and “B” transcripts. These data confirm and significantly extend a very recent report by Talabot-Ayer et al. (60) who also found expression of IL-33 gene from two alternative promoters (i.e., “IL33a” and “IL-33b” mRNA; designated in our study as “B” and “A,” respectively, consistent with the nomenclature used by the data base (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=mouse&q=Il33), whose expression in the mouse were organ-specific- and inducible by LPS and pI:C. In this report, we have significantly extended their findings by showing that additional TLR and non-TLR agonists that share the ability to induce TBK-1 and IRF-3 by distinct pathways elicit the shorter “A” mRNA species preferentially in primary murine macrophages, with the exception of NDV that induces both “A” and “B” transcripts. In addition, we provide evidence that this preferential induction of the “A” mRNA species is reflected in vivo as well, in liver samples after administration of a lethal dose of LPS to mice. Finally, our data indicate that the cAMP activating drug, ACT, enhanced LPS-induced “A” mRNA (Figure 7D), but not the “B” mRNA (data not shown), and that this induction was PKA-dependent. The significance of these two mRNA species, and of preferential induction of the “A” species by all agonists but NDV (RIG-I), will await a complete elucidation of regulatory elements in the promoter and possible intronic regulatory sequences in this gene.
Thymic Stromal Lymphopoietin (TSLP) is produced by epithelial cells in response to certain microbial products (e.g., peptidoglycan, lipoteichoic acid, and double stranded RNA), injury, or inflammatory cytokines (e.g., IL-1β and TNF-α) (61). TSLP is detected in the airways of asthmatic patients and the level of TSLP mRNA was shown to be proportional to the severity of the disease (62). Both IL-33 and TSLP activate Th2 cells to produce Th2 cytokines (63, 64). These findings suggest the possibility that viral infection and recruitment of Th2 cytokine-producing cells may amplify Th2 inflammation through the production of IL-33 and TSLP (65) in asthmatic airways. Viral infection of epithelial cells ultimately results in tissue damage and, eventually, release of full-length IL-33 from necrotic cells (66). In chronic asthma, continual inflammation also results in tissue disruption and release of IL-33 (64). This released IL-33 acts on different cells to produce Th2 cytokines as well as proinflammatory cytokines to increase severity of asthma (64). In Th1 cells, cAMP inhibits Th1 cytokine production, whereas cAMP markedly increases Th2 cytokine production in GATA3-expressing cells (48). cAMP also activates p38, that in turn, activates GATA3, leading to Th2 cytokine production (49, 50). Since IL-33 has been associated with a Th2 bias, increased cAMP may be involved in regulation of IL-33 production. These observations are strengthened by our findings showing that cAMP agonists significantly enhanced LPS-induced IL-33 mRNA in macrophages through a PKA-dependent process.
Lastly, IL-33 has also been shown to be a chromatin-associated in the nuclei of endothelial cells and has the capacity to regulate transcription (27). IL-33 binds to chromatin in the surface of nucleosome by docking to the pockets of the histone H2A-H2B dimer (28). Therefore, IL-33 is a dual function protein, in that it acts as both a cytokine and an intracellular nuclear protein. IL-33 may be an important cytokine involved in initiation and perpetuation of inflammation in the case of asthma (64). Depending on the tissue type, IL-33 may contribute to the resolution of inflammation-associated arthrosclerosis and cardiac function (21, 67), expulsion of the intestinal-dwelling nematode, or infection-induced tissue damage (66, 68). Thus, depending on the immune mechanism underlying the pathogenesis of each disease condition, IL-33 has the potential as a therapeutic target in various diseases.
Acknowledgments
We acknowledge Drs. Leah Cole and Vladimir Toshchakov for their help and advice during these studies.
This work was supported by NIH grant (AI189797) (SNV).
Abbreviations used in this paper
- IRF-3
interferon regulatory factor-3
- P3C
S-[2-3-Bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-Lys4-OH trihydrochloride
- DMXAA
5,6-dimethylxanthenone-4-acetic acid
- PKA
Protein Kinase A
- MDA5
Melanoma Differentiation Associated gene 5
- RIG-I
retinoic-acid-inducible protein I
- IRAK
interleukin-1 receptor-associated kinase
- H2A-H2B
histones H2A, H2B
- poly-(I:C)
polyinosinic:polycytidylic acid
- TBK1
TANK-binding kinase 1
- CpG DNA
cytidine-phosphateguanosine DNA
- CHX
cycloheximide
- TSLP
Thymic Stromal Lymphopoietin
- RTKi
EGF/FGF/PDGF receptor tyrosine kinase inhibitor
Footnotes
Conflict of interest statement: The authors have no conflict of interest.
References
- 1.Baekkevold ES, Roussigne M, Yamanaka T, Johansen FE, Jahnsen FL, Amalric F, Brandtzaeg P, Erard M, Haraldsen G, Girard JP. Molecular characterization of NF-HEV, a nuclear factor preferentially expressed in human high endothelial venules. Am J Pathol. 2003;163:69–79. doi: 10.1016/S0002-9440(10)63631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Cayrol C, Girard JP. The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc Natl Acad Sci U S A. 2009;106:9021–9026. doi: 10.1073/pnas.0812690106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oshikawa K, Kuroiwa K, Tago K, Iwahana H, Yanagisawa K, Ohno S, Tominaga SI, Sugiyama Y. Elevated soluble ST2 protein levels in sera of patients with asthma with an acute exacerbation. Am J Respir Crit Care Med. 2001;164:277–281. doi: 10.1164/ajrccm.164.2.2008120. [DOI] [PubMed] [Google Scholar]
- 5.Rossler U, Thomassen E, Hultner L, Baier S, Danescu J, Werenskiold AK. Secreted and membrane-bound isoforms of T1, an orphan receptor related to IL-1-binding proteins, are differently expressed in vivo. Dev Biol. 1995;168:86–97. doi: 10.1006/dbio.1995.1063. [DOI] [PubMed] [Google Scholar]
- 6.Iwahana H, Yanagisawa K, Ito-Kosaka A, Kuroiwa K, Tago K, Komatsu N, Katashima R, Itakura M, Tominaga S. Different promoter usage and multiple transcription initiation sites of the interleukin-1 receptor-related human ST2 gene in UT-7 and TM12 cells. Eur J Biochem. 1999;264:397–406. doi: 10.1046/j.1432-1327.1999.00615.x. [DOI] [PubMed] [Google Scholar]
- 7.Bergers G, Reikerstorfer A, Braselmann S, Graninger P, Busslinger M. Alternative promoter usage of the Fos-responsive gene Fit-1 generates mRNA isoforms coding for either secreted or membrane-bound proteins related to the IL-1 receptor. Embo J. 1994;13:1176–1188. doi: 10.1002/j.1460-2075.1994.tb06367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu D, Chan WL, Leung BP, Huang F, Wheeler R, Piedrafita D, Robinson JH, Liew FY. Selective expression of a stable cell surface molecule on type 2 but not type 1 helper T cells. J Exp Med. 1998;187:787–794. doi: 10.1084/jem.187.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meisel C, Bonhagen K, Lohning M, Coyle AJ, Gutierrez-Ramos JC, Radbruch A, Kamradt T. Regulation and function of T1/ST2 expression on CD4+ T cells: induction of type 2 cytokine production by T1/ST2 cross-linking. J Immunol. 2001;166:3143–3150. doi: 10.4049/jimmunol.166.5.3143. [DOI] [PubMed] [Google Scholar]
- 10.Tominaga S, Kuroiwa K, Tago K, Iwahana H, Yanagisawa K, Komatsu N. Presence and expression of a novel variant form of ST2 gene product in human leukemic cell line UT-7/GM. Biochem Biophys Res Commun. 1999;264:14–18. doi: 10.1006/bbrc.1999.1469. [DOI] [PubMed] [Google Scholar]
- 11.Iwahana H, Hayakawa M, Kuroiwa K, Tago K, Yanagisawa K, Noji S, Tominaga S. Molecular cloning of the chicken ST2 gene and a novel variant form of the ST2 gene product, ST2LV. Biochim Biophys Acta. 2004;1681:1–14. doi: 10.1016/j.bbaexp.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa H, Hayakawa M, Kume A, Tominaga S. Soluble ST2 blocks interleukin-33 signaling in allergic airway inflammation. J Biol Chem. 2007;282:26369–26380. doi: 10.1074/jbc.M704916200. [DOI] [PubMed] [Google Scholar]
- 13.Chackerian AA, Oldham ER, Murphy EE, Schmitz J, Pflanz S, Kastelein RA. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179:2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 14.Palmer G, Lipsky BP, Smithgall MD, Meininger D, Siu S, Talabot-Ayer D, Gabay C, Smith DE. The IL-1 receptor accessory protein (AcP) is required for IL-33 signaling and soluble AcP enhances the ability of soluble ST2 to inhibit IL-33. Cytokine. 2008;42:358–364. doi: 10.1016/j.cyto.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Moulin D, Donze O, Talabot-Ayer D, Mezin F, Palmer G, Gabay C. Interleukin (IL)-33 induces the release of pro-inflammatory mediators by mast cells. Cytokine. 2007;40:216–225. doi: 10.1016/j.cyto.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Ali S, Huber M, Kollewe C, Bischoff SC, Falk W, Martin MU. IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc Natl Acad Sci U S A. 2007;104:18660–18665. doi: 10.1073/pnas.0705939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allakhverdi Z, Smith DE, Comeau MR, Delespesse G. Cutting edge: The ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J Immunol. 2007;179:2051–2054. doi: 10.4049/jimmunol.179.4.2051. [DOI] [PubMed] [Google Scholar]
- 18.Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol. 2008;121:1484–1490. doi: 10.1016/j.jaci.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson CA, Christophi GP, Gruber RC, Wilmore JR, Lawrence DA, Massa PT. Induction of IL-33 expression and activity in central nervous system glia. J Leukoc Biol. 2008;84:631–643. doi: 10.1189/jlb.1207830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pecaric-Petkovic T, Didichenko SA, Kaempfer S, Spiegl N, Dahinden CA. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood. 2009;113:1526–1534. doi: 10.1182/blood-2008-05-157818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanada S, Hakuno D, Higgins LJ, Schreiter ER, McKenzie AN, Lee RT. IL-33 and ST2 comprise a critical biomechanically induced and cardioprotective signaling system. J Clin Invest. 2007;117:1538–1549. doi: 10.1172/JCI30634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol. 2008;20:1019–1030. doi: 10.1093/intimm/dxn060. [DOI] [PubMed] [Google Scholar]
- 23.Iikura M, Suto H, Kajiwara N, Oboki K, Ohno T, Okayama Y, Saito H, Galli SJ, Nakae S. IL-33 can promote survival, adhesion and cytokine production in human mast cells. Lab Invest. 2007;87:971–978. doi: 10.1038/labinvest.3700663. [DOI] [PubMed] [Google Scholar]
- 24.Coyle AJ, Lloyd C, Tian J, Nguyen T, Erikkson C, Wang L, Ottoson P, Persson P, Delaney T, Lehar S, Lin S, Poisson L, Meisel C, Kamradt T, Bjerke T, Levinson D, Gutierrez-Ramos JC. Crucial role of the interleukin 1 receptor family member T1/ST2 in T helper cell type 2-mediated lung mucosal immune responses. J Exp Med. 1999;190:895–902. doi: 10.1084/jem.190.7.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez-Ramos JC, Levinson D, Radbruch A, Kamradt T. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci U S A. 1998;95:6930–6935. doi: 10.1073/pnas.95.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith DE. IL-33: a tissue derived cytokine pathway involved in allergic inflammation and asthma. Clin Exp Allergy. 2009 doi: 10.1111/j.1365-2222.2009.03384.x. [DOI] [PubMed] [Google Scholar]
- 27.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussel L, Erard M, Cayrol C, Girard JP. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A-H2B acidic pocket. EMBO Rep. 2008;9:1006–1012. doi: 10.1038/embor.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntire FC, Sievert HW, Barlow GH, Finley RA, Lee AY. Chemical, physical, biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry. 1967;6:2363–2372. doi: 10.1021/bi00860a011. [DOI] [PubMed] [Google Scholar]
- 30.Salkowski CA, Kopydlowski K, Blanco J, Cody MJ, McNally R, Vogel SN. IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J Immunol. 1999;163:1529–1536. [PubMed] [Google Scholar]
- 31.Deonarain R, Alcami A, Alexiou M, Dallman MJ, Gewert DR, Porter AC. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J Virol. 2000;74:3404–3409. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Ohno T, Oboki K, Kajiwara N, Morii E, Aozasa K, Flavell RA, Okumura K, Saito H, Nakae S. Caspase-1, caspase-8, and calpain are dispensable for IL-33 release by macrophages. J Immunol. 2009;183:7890–7897. doi: 10.4049/jimmunol.0802449. [DOI] [PubMed] [Google Scholar]
- 35.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, Williams BR, Major J, Hamilton TA, Fenton MJ, Vogel SN. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 38.Kozovska ME, Hong J, Zang YC, Li S, Rivera VM, Killian JM, Zhang JZ. Interferon beta induces T-helper 2 immune deviation in MS. Neurology. 1999;53:1692–1697. doi: 10.1212/wnl.53.8.1692. [DOI] [PubMed] [Google Scholar]
- 39.Whelan J, Ghersa P, Hooft van Huijsduijnen R, Gray J, Chandra G, Talabot F, DeLamarter JF. An NF kappa B-like factor is essential but not sufficient for cytokine induction of endothelial leukocyte adhesion molecule 1 (ELAM-1) gene transcription. Nucleic Acids Res. 1991;19:2645–2653. doi: 10.1093/nar/19.10.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 41.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 42.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, Freudenberg M, Beutler B. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Muhlradt PF, Akira S. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J Immunol. 2000;164:554–557. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 44.Delghandi MP, Johannessen M, Moens U. The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell Signal. 2005;17:1343–1351. doi: 10.1016/j.cellsig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Rehfuss RP, Walton KM, Loriaux MM, Goodman RH. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991;266:18431–18434. [PubMed] [Google Scholar]
- 46.Wiggin GR, Soloaga A, Foster JM, Murray-Tait V, Cohen P, Arthur JS. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol Cell Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 48.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CH, Zhang DH, LaPorte JM, Ray A. Cyclic AMP activates p38 mitogen-activated protein kinase in Th2 cells: phosphorylation of GATA-3 and stimulation of Th2 cytokine gene expression. J Immunol. 2000;165:5597–5605. doi: 10.4049/jimmunol.165.10.5597. [DOI] [PubMed] [Google Scholar]
- 50.Klein-Hessling S, Bopp T, Jha MK, Schmidt A, Miyatake S, Schmitt E, Serfling E. Cyclic AMP-induced chromatin changes support the NFATc-mediated recruitment of GATA-3 to the interleukin 5 promoter. J Biol Chem. 2008;283:31030–31037. doi: 10.1074/jbc.M805929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 52.Meja KK, Catley MC, Cambridge LM, Barnes PJ, Lum H, Newton R, Giembycz MA. Adenovirus-mediated delivery and expression of a cAMP-dependent protein kinase inhibitor gene to BEAS-2B epithelial cells abolishes the anti-inflammatory effects of rolipram, salbutamol, and prostaglandin E2: a comparison with H-89. J Pharmacol Exp Ther. 2004;309:833–844. doi: 10.1124/jpet.103.060020. [DOI] [PubMed] [Google Scholar]
- 53.Johannes FJ, Prestle J, Dieterich S, Oberhagemann P, Link G, Pfizenmaier K. Characterization of activators and inhibitors of protein kinase C mu. Eur J Biochem. 1995;227:303–307. doi: 10.1111/j.1432-1033.1995.tb20389.x. [DOI] [PubMed] [Google Scholar]
- 54.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 55.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnes PJ, Adcock IM. Transcription factors and asthma. Eur Respir J. 1998;12:221–234. doi: 10.1183/09031936.98.12010221. [DOI] [PubMed] [Google Scholar]
- 57.Chiappara G, Chanez P, Bruno A, Pace E, Pompeo F, Bousquet J, Bonsignore G, Gjomarkaj M. Variable p-CREB expression depicts different asthma phenotypes. Allergy. 2007;62:787–794. doi: 10.1111/j.1398-9995.2007.01417.x. [DOI] [PubMed] [Google Scholar]
- 58.Yasuda G, Umemura S, Jeffries WB. Effect of epinephrine on cAMP accumulation in cultured rat inner medullary collecting duct cells. Am J Physiol. 1997;272:F192–F197. doi: 10.1152/ajprenal.1997.272.2.F192. [DOI] [PubMed] [Google Scholar]
- 59.Perkins DJ, Gray MC, Hewlett EL, Vogel SN. Bordetella pertussis adenylate cyclase toxin (ACT) induces cyclooxygenase-2 (COX-2) in murine macrophages and is facilitated by ACT interaction with CD11b/CD18 (Mac-1) Mol Microbiol. 2007;66:1003–1015. doi: 10.1111/j.1365-2958.2007.05972.x. [DOI] [PubMed] [Google Scholar]
- 60.Talabot-Ayer D, Calo N, Vigne S, Lamacchia C, Gabay C, Palmer G. The mouse interleukin (Il)33 gene is expressed in a cell type- and stimulus-dependent manner from two alternative promoters. J Leukoc Biol. 2011;91:119–125. doi: 10.1189/jlb.0811425. [DOI] [PubMed] [Google Scholar]
- 61.Allakhverdi Z, Comeau MR, Jessup HK, Yoon BR, Brewer A, Chartier S, Paquette N, Ziegler SF, Sarfati M, Delespesse G. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J Exp Med. 2007;204:253–258. doi: 10.1084/jem.20062211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ying S, O'Connor B, Ratoff J, Meng Q, Mallett K, Cousins D, Robinson D, Zhang G, Zhao J, Lee TH, Corrigan C. Thymic stromal lymphopoietin expression is increased in asthmatic airways and correlates with expression of Th2-attracting chemokines and disease severity. J Immunol. 2005;174:8183–8190. doi: 10.4049/jimmunol.174.12.8183. [DOI] [PubMed] [Google Scholar]
- 63.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 10:103–110. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]
- 65.Kato A, Favoreto S, Jr, Avila PC, Schleimer RP. TLR3- and Th2 cytokine-dependent production of thymic stromal lymphopoietin in human airway epithelial cells. J Immunol. 2007;179:1080–1087. doi: 10.4049/jimmunol.179.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luthi AU, Cullen SP, McNeela EA, Duriez PJ, Afonina IS, Sheridan C, Brumatti G, Taylor RC, Kersse K, Vandenabeele P, Lavelle EC, Martin SJ. Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity. 2009;31:84–98. doi: 10.1016/j.immuni.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Miller AM, Xu D, Asquith DL, Denby L, Li Y, Sattar N, Baker AH, McInnes IB, Liew FY. IL-33 reduces the development of atherosclerosis. J Exp Med. 2008;205:339–346. doi: 10.1084/jem.20071868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Humphreys NE, Xu D, Hepworth MR, Liew FY, Grencis RK. IL-33, a potent inducer of adaptive immunity to intestinal nematodes. J Immunol. 2008;180:2443–2449. doi: 10.4049/jimmunol.180.4.2443. [DOI] [PubMed] [Google Scholar]