Abstract
Total chemical synthesis was used to site-specifically 13C-label active site Asp25 and Asp25’ residues in HIV-1 protease and in several chemically synthesized analogues of the enzyme molecule. 13C NMR measurements were consistent with a monoprotonated state for the catalytic dyad formed by the interacting Asp25, Asp25’ side chain carboxyls.
Introduction
Direct experimental determination of the ionization states of the side chain carboxyl’s of catalytic residues Asp25 and Asp25’ in the HIV-1 protease is of particular importance for understanding the catalytic mechanism of this important enzyme, and for designing new inhibitors. All potent inhibitors interact with the catalytic carboxyls of Asp25/25’.1 X-ray crystallography is not a suitable technique for determining the ionization states of these side chains because of the very weak electron density for hydrogen atoms. Neutron diffraction crystallography on the other hand provides unambiguous solution for the location of the hydrogens for a macromolecule in the crystal state.2 However, the methodology is severely limited because of difficulties in obtaining the large single crystals required for neutron diffraction (at least 1 mm).3 An alternative method for determining the ionization state of the catalytic carboxyl moieties is NMR measurements of site specifically isotope-labelled proteins.
NMR measurements have previously been performed for complexes of HIV-1 protease with the C2-symmetric DMP-323 inhibitor4 and with the asymmetric KNI-272 inhibitor.5 In the case of DMP-323, it was found that both Asp25 and Asp25’ side chains are protonated over the pH range extending from 2.2 to 7.0 (13C δ, 176.4 ppm), whereas for KNI-272 one aspartate side chain Asp25 is charged (13C δ, 177.4 ppm) while the other side chain carboxyl Asp25’ is protonated (13C δ, 175.8 ppm, deuterium isotope effect 0.17 ppm). Interestingly, the 13C-chemical shifts in the presence of the inhibitor KNI-272 were found to be essentially independent of pH in the range 2.5 – 7.0, indicating extreme pKa values for the side chain carboxyl’s of Asp25 and Asp25’ in the HIV-1 protease- KNI-272 complex.
In a contemporaneous report, researchers used total chemical synthesis to prepare site-specifically 13Cγ-Asp25/25’-labelled enzyme and used 13C-NMR measurements to establish the ionization states of the Asp25/25’ side chains in a complex of HIV-1 protease with pepstatin.6 Two 13C-resonances at 178.8 ppm and 172.4 ppm were detected, with only one of them (178.8 ppm) demonstrating significant (~0.2 ppm upfield) isotope effect when H2O solvent was replaced by D2O. Again, as in the complex with KNI-272, these resonances did not titrate in the pH range of 2.5 – 6.5. In the same paper, these authors reported a study of unliganded HIV-1 protease and concluded that both Asp25 and Asp25’ are deprotonated at pH 6.0. Their interpretation was based on the observation of a single 13C NMR 13Cγ–carboxylate resonance, at ~180.2 ppm, for HIV-1 protease bearing 13C-label only at Cγ-atom of both Asp25 and Asp25’. To make an assignment, a model 11-residue peptide (corresponding to residues 20–30 of the HIV-1 protease polypeptide) was used, where the 13Cγ–aspartate resonated at ~180.2 ppm when charged and at much lower chemical shift when protonated (pKa of the Asp side chain in the model peptide was found to be near 4.0). However, detailed biochemical mechanistic studies by Meek and co-workers indicated a monoprotonated state (i.e. one carboxylate, one carboxyl) for the side chain carboxyls of Asp25/25’ in the unliganded HIV-1 protease.7 More recent theoretical studies also suggested a monoprotonated state.8 To resolve this discrepancy, we decided to revisit the published experiments6 using the more sensitive equipment available at the present day, and to extend our studies to analogues of the HIV-1 protease prepared by total chemical synthesis.
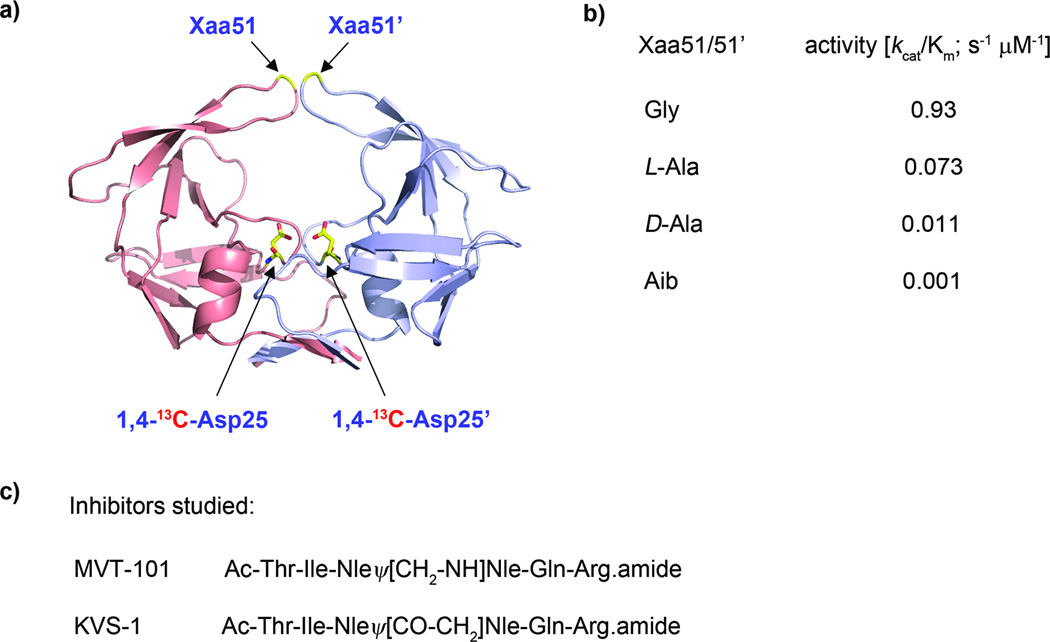
In previous work, we have shown that substitutions of residue Gly51, at the tip of the flaps in the HIV-1 protease, with L-Ala, D-Ala, or Aib strongly affect the catalytic activity of the enzyme despite the large distance between residue 51 and catalytic site (Fig. 1).9 Furthermore, by applying a combination of experimental methods (protein NMR, pulse-EPR, X-ray crystallography) and molecular dynamics (MD) simulations we have shown that catalytic rate reduction in these analogues is caused by alteration of dynamic and conformational properties leading to lower concentrations of enzyme states preorganized for catalysis.9
Fig. 1.
(a) Catalytic dimer form (2 × 99 amino acids) of HIV-1 protease. The (1,4)-13C-labelled Asp25/25’ residues and Xaa51/51’ (where Xaa = Gly, L-Ala, D-Ala, Aib) are indicated with arrows. (b) Catalytic efficiencies are listed for the wild-type HIV-1 protease (Xaa = Gly) and for three chemical analogues. (c) Inhibitors studied in this report include the reduced isostere MVT-101 and ketomethylene isostere KVS-1.
Results and discussion
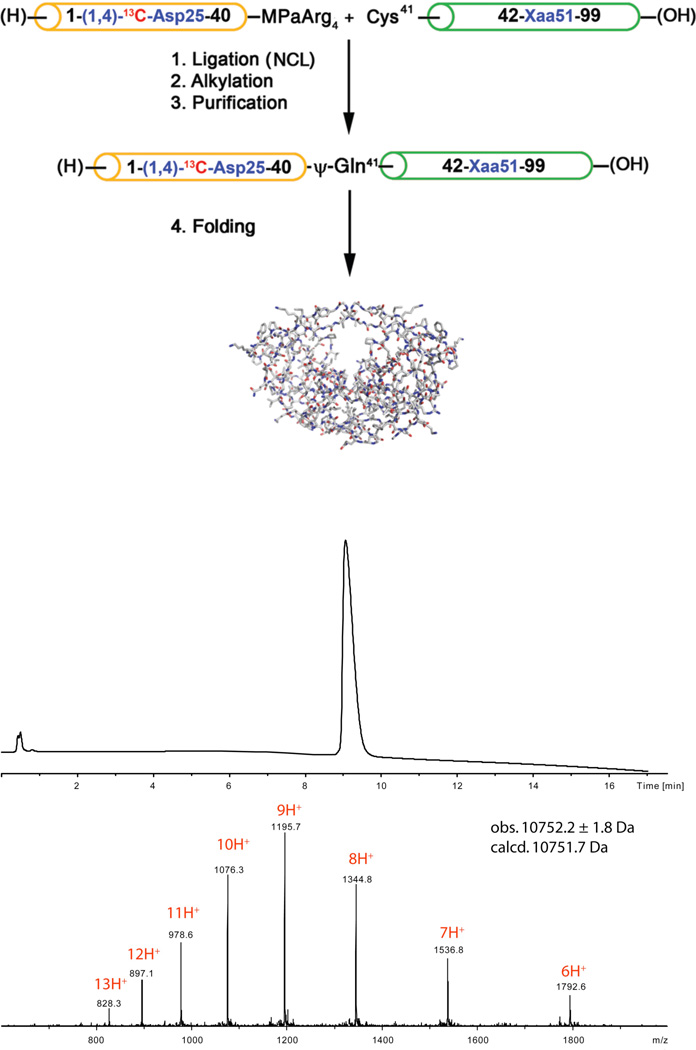
In this study, we used dual site specific labelling with 13C, in order to have a reference signal against which to measure 13C chemical shifts of the side chain carboxyls in the active site. (1,4)-13C-Asp25/25’ wild-type [1–99]HIV-1 protease and selected chemical analogues Gly51Xaa (Xaa = L-Ala, D-Ala, Aib, where Aib is α-aminoisobutyric acid) were prepared by total chemical synthesis based on a two segment native chemical ligation.10,11 Peptides (1-(1,4)-13C-Asp25-40). αthioester and (Cys41-Xaa51-99) were synthesized by stepwise Boc chemistry solid phase peptide synthesis (SPPS), in which the 13C-labelled Asp25 residue was introduced as Boc-(1,4)-13C-Asp(γCO2Allyl). Synthetic peptides were cleaved and deprotected using anhydrous HF, and were purified by reverse phase HPLC. After native chemical ligation under standard conditions,10 residue Cys41 was alkylated with 2-bromoacetamide to form a ψ-homo-Gln41 residue. After removal of the formyl groups from Trp6 and Trp42, the full length (1–99)-polypeptides were purified by reverse-phase HPLC and folded by two-step dialysis [analytical HPLC and MS data are provided in the Supplementary Information]. A representative synthesis is shown in Figure 2.
Fig. 2.
Chemical synthesis of the wild-type HIV-1 protease and analogues was based on the native chemical ligation of two synthetic peptide segments.11 (Upper) Synthetic scheme. In the Scheme, MPaArg4 = 3-mercaptopropionate tetraarginine.amide (i.e. the thioester leaving group). (Lower) Analytical LCMS data for the synthetic enzyme with Gly51/51’.
In the present study, we employed three analogue enzymes with reduced catalytic rates (and which thus undergo autoproteolysis to a lesser extend) to carefully measure 13C NMR chemical shifts of the catalytic Asp25 and Asp25’ residue side chain carboxyls in HIV-1 protease, unliganded and – for [L-Ala51/51’]HIV-1 protease – complexed with inhibitors. We used a 21.1 Tesla NMR spectrometer (226 MHz for 13C and 900 MHz for 1H) equipped with a super-cooled preamplifier for the 13C-channel, which gave enhanced sensitivity for direct detection of NMR of the 13C-nucleus (National Magnetic Resonance Facility at Madison, Wisconsin). Spectra were collected for samples of unliganded enzymes and, in addition, for complexes of [L-Ala51/51’]HIV-1 protease with MVT-101 and KVS-1 inhibitors.
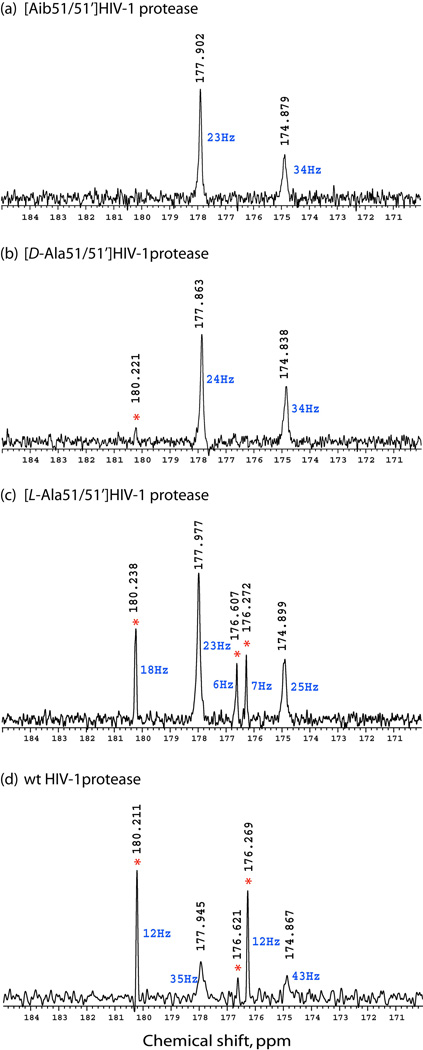
1H-decoupled 13C-spectra are displayed in Fig. 3 for the unliganded enzymes at pH 5.7. For the essentially inactive [Aib51/51’]HIV-1 protease (see Fig. 1 b),9 only two peaks at 177.9 ppm (side chain carboxyl(ate)) and 174.9 ppm (backbone amide) were observed (Fig. 3 a). In the case of the [D-Ala51/51’]HIV-1 protease,9 which has only poor proteolytic activity, an additional small peak at 180.2 ppm was observed (Fig. 3 b). For the [L-Ala51/51’]HIV-1 protease (~10-times lower activity than that of the wild-type enzyme),9 additional peaks at ~ 180.2 ppm, 176.6 ppm and 176.3 ppm were observed (Fig. 3 c). For the wild-type HIV-1 protease, these additional peaks dominated the spectrum (Fig. 3 d), with the 180.2 ppm peak being the most dominant as in the previously reported data.6 It should be noted that peaks at 177.9 ppm and 174.9 ppm had much greater linewidth (2–3 times) than the sharp additional peaks at 180.2 ppm, 176.6 ppm and 176.3 ppm. Recording spectra for the same samples in D2O showed no significant isotope effect (30 ppb for carboxyl(ate)s and 50 ppb for amides) for signals at 177.9 ppm and 174.9 ppm at pH 5.7 (pD 6.1).
Fig. 3.
226 MHz 13C-{1H} NMR spectra for unliganded HIV-1 protease and three chemically synthesized analogue enzymes. (a) [Aib51/51’]HIV-1 protease, (b) [D-Ala51/51’]HIV-1 protease, (c) [L-Ala51/51’]HIV-1 protease, (d) wild-type HIV-1 protease. Red asterisk indicates peaks originating from peptide autoproteolysis products. Linewidths of the peaks are in blue. All samples were prepared in 18.9 mM Na.phosphate buffer (pH 5.7), containing 5.4 % (v/v) D2O and 100 µM DSS-d6. Concentrations of protein were 0.29 – 0.41 mM.
pH titration experiments on the unliganded enzymes showed a significant effect on chemical shift for resonances at 180.2 ppm (Δδ of 1.3 ppm upfield when pH was reduced from 5.7 to 3.9, pKa 4.5), but only a very slight effect on the resonances at 177.9 ppm (Δδ of 0.23 ppm upfield when pH was reduced from 5.7 to 3.9, pKa ~ 5). For the resonances at 180.2 ppm, the observed pKa 4.5 (typical pKa for γ-carboxylate of aspartic acid) in combination with the narrower linewidths suggests that they might be coming from unstructured peptide fragments resulting from autoproteolytic activity of HIV-1 protease.12 Inspection of samples by analytical HPLC-MS prior to and after the NMR measurements showed that products of autoproteolysis did not exceed 10% in the most active wild-type HIV-1 protease.
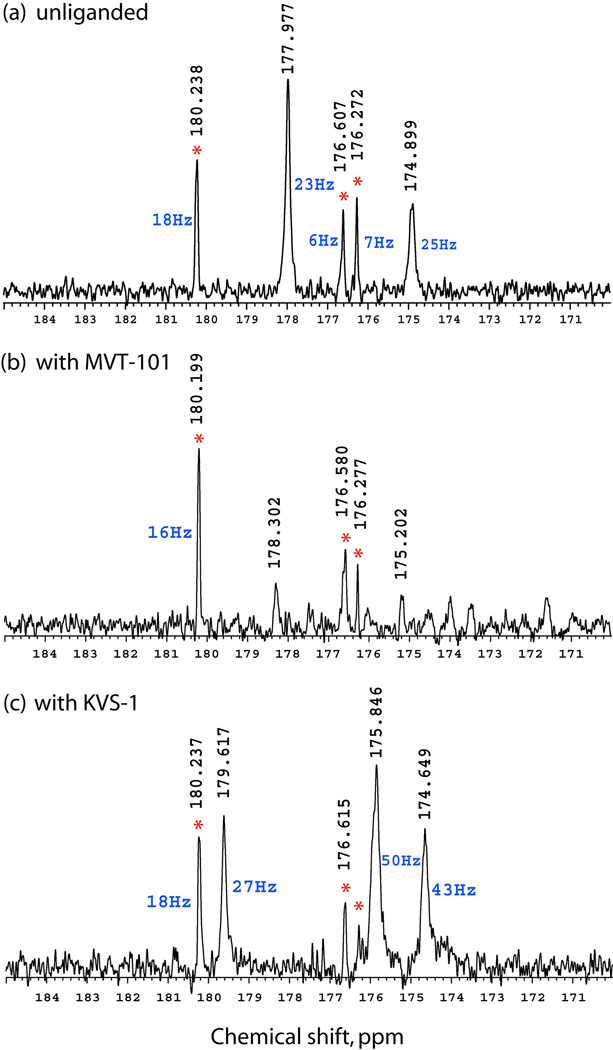
We then performed 13C-NMR measurements on the complex of the [L-Ala51/51’]analogue with the reduced isostere inhibitor MVT-101 and the ketomethylene isostere inhibitor KVS-1 (the enzyme was folded in the absence of an inhibitor and each of the inhibitors was separately added prior to the NMR measurements). These inhibitor molecules bind HIV-1 protease and form H-bonds with the catalytic Asp25 and Asp25’ side chain carboxyls, thus altering their 13C chemical shifts. With the MVT-101 inhibitor, we observed sharp peaks of high intensity at 180.2 ppm, 176.6 ppm and 176.3 ppm, the same as for the wild-type HIV-1 protease and its [L-Ala51]analogue (Fig. 4 b). In addition, two broader peaks of lower intensity were observed at 178.3 ppm and 175.2 ppm. Additional experiments, where inter-pulse delay was varied from 2 s to 5 s, showed much greater intensity for these peaks at 178.3 ppm and 175.2 ppm with the 5 s inter-pulse delay (Supplementary Fig. 1), suggesting a much longer T1-longitudinal relaxation time constant for these resonances.
Fig. 4.
226 MHz 13C-{1H} NMR spectra for [L-Ala51/51’]HIV-1 protease: (a) unliganded; (b) complexed with the reduced isostere inhibitor MVT-101; and (c) with the ketomethylene isostere inhibitor KVS-1. Red asterisk indicates peaks originating from unfolded peptidic autoproteolysis products. [L-Ala51/51’]HIV-1 protease was folded by dialysis without inhibitors present; products of autoproteolysis did not exceed 5 % by HPLC-analysis. Inhibitors were added after dialysis. In (a) acquisition time was set at 0.5 s and inter-pulse delay was 1 s, while in (b) and (c) inter-pulse delay was extended to 5 s to obtain better S/N ratio spectra.
In the complex with inhibitor KVS-1, again we have observed three sharp resonances at 180.2 ppm, 176.6 ppm and 176.3 ppm and, in addition, broader peaks at 179.6 ppm, 175.9 ppm and 174.6 ppm (Fig. 4 c). A synthetic model 11-residue peptide (residues 20–30 of HIV-1 protease sequence) showed 13C-NMR signals at 180.2 ppm and 176.6 ppm; the same resonances have been observed for synthetic precursor peptide (1–40).thioester at pH 5.7 when aspartate Asp25 is deprotonated.
Taken in total, these results suggest that the previously detected 180.2 ppm 13C-resonance6 was the product(s) of autoproteolysis,12 and that the signal came from short unfolded peptide(s) and did not originate from Asp25/25’ in the intact, folded HIV-1 protease. This interpretation is supported by the observation that the intensity of the signal at 180.2 ppm is apparently a function of the catalytic efficiency of a given chemical analogue of HIV-1 protease: for unliganded enzymes, the more active the enzyme, the more intense the observed resonance at 180.2 ppm was.
To unambiguously rule out the possibility that different peaks might be coming from different conformational states of HIV-1 protease, we performed 13C-EXSY (exchange spectroscopy) for unliganded [L-Ala51/51’]HIV-1 protease with mixing times of 0.2 s and 1 s and did not find any exchange cross-peaks. Additional support for the 180.2 ppm peak originating from shorter peptide fragments comes from the observed narrower linewidth of these resonances (6–18 Hz) in comparison with broader peaks coming from folded enzymes and enzyme/inhibitor complexes (23–43 Hz). This indicates slower T2*-transverse relaxation for the sharper peaks, and is consistent with the unfolded nature of short peptides.
Conclusions
We conclude that in the unliganded HIV-1 protease enzyme the ionization state of the catalytic dyad made up of Asp25/Asp25’ side chain carboxyls is likely to be monoprotonated, as elucidated in biochemical experiments by Meek and co-workers.7 The true chemical shift for 13Cγ-Asp25/Asp25’ in the unliganded enzyme is δ 177.9 ppm – which is midway between the two extremes for charged (181 ppm) and protonated aspartates (175 ppm)4 – and is in agreement with a monoprotonated state for the catalytic dyad. Most probably, rapid exchange on the NMR time scale makes it impossible to detect individual 13C-resonances for protonated Asp25 and charged Asp25’.
Recently we have demonstrated that conformational dynamics of the HIV-1 protease protein molecule are correlated with the dynamics of the catalytic aspartates.9 The pKa of the active site aspartates is likely to be affected in different conformers. Such complexity may account for the absence in our measurements of titration behavior (protein dynamics will change with pH) and the absence of any significant deuterium isotope effect. We suggest that Fourier transform infrared spectroscopy may be better suited to unambiguously resolve this conundrum.13
Supplementary Material
Acknowledgments
We acknowledge funding from the DOE “Genomics to Life” program (grant no. DE-PS02-07ER07-14). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture. We acknowledge Klaas Hallenga, Marco Tonelli and William M. Westler at National Magnetic Resonance Facility at Madison for help with NMR experiments.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental procedures for the NMR measurements, analytical HPLC and mass-spectrometry characterization of studied proteins. See DOI: 10.1039/b000000x/
Notes and references
- 1.Wlodawer A, Vondrasek J. Annu. Rev. Biophys. Biomol. Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 2.Adachi M, Ohhara T, Kurihara K, Tamada T, Honjo E, Okazaki N, Arai S, Shoyama Y, Kimura K, Matsumura H, Sugiyama S, Adachi H, Takano K, Mori Y, Hidaka K, Kimura T, Hayashi Y, Kiso Y, Kuroki R. Proc. Natl. Acad, Sci. USA. 2009;106:440–444. doi: 10.1073/pnas.0809400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu N, Sugiyama S, Maruyama M, Takahashi Y, Adachi M, Tamada T, Hidaka K, Hayashi Y, Kimura T, Kiso Y, Adachi H, Takano K, Murakami S, Inoue T, Kuroki R, Mori Y, Matsumura H. Cryst. Growth Des. 2010;10:2990–2994. [Google Scholar]
- 4.Yamazaki T, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Eyermann CJ, Hodge CN, Lam PYS, Ru Y, Jadhav PK, Chang C-H, Weber PC. J. Am. Chem. Soc. 1994;116:10791–10792. [Google Scholar]
- 5.Wang Y-X, Freedberg DI, Yamazaki T, Wingfield PT, Stahl SJ, Kaufman JD, Kiso Y, Torchia DA. Biochemistry. 1996;35:9945–9950. doi: 10.1021/bi961268z. [DOI] [PubMed] [Google Scholar]
- 6.Smith R, Brereton IM, Chai RY, Kent SBH. Nature Struct. Biol. 1996;3:946–950. doi: 10.1038/nsb1196-946. [DOI] [PubMed] [Google Scholar]
- 7.Hyland LJ, Tomaszek T, Meek TD. Biochemistry. 1991;30:8454–8463. doi: 10.1021/bi00098a024. [DOI] [PubMed] [Google Scholar]
- 8.Piana S, Carloni, P P. Proteins: Struct. Func. Genet. 2000;39:26–36. doi: 10.1002/(sici)1097-0134(20000401)39:1<26::aid-prot3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 9.Torbeev VY, Raghuraman H, Hamelberg D, Tonelli M, Westler WM, Perozo E, Kent SBH. Proc Natl Acad Sci USA. 2011;108:20982–20987. doi: 10.1073/pnas.1111202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson PE, Kent SBH. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 11.Torbeev VY, Mandal K, Terechko VA, Kent SBH. Bioorg. Med. Chem. Lett. 2008;18:4554–4557. doi: 10.1016/j.bmcl.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 12.Rose JR, Salto R, Craik CS. J. Biol. Chem. 1993;268:11939–11945. [PubMed] [Google Scholar]
- 13.Hellwig P, Barquera B, Gennis RB. Biochemistry. 2001;40:1077–1082. doi: 10.1021/bi002154x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.