Abstract
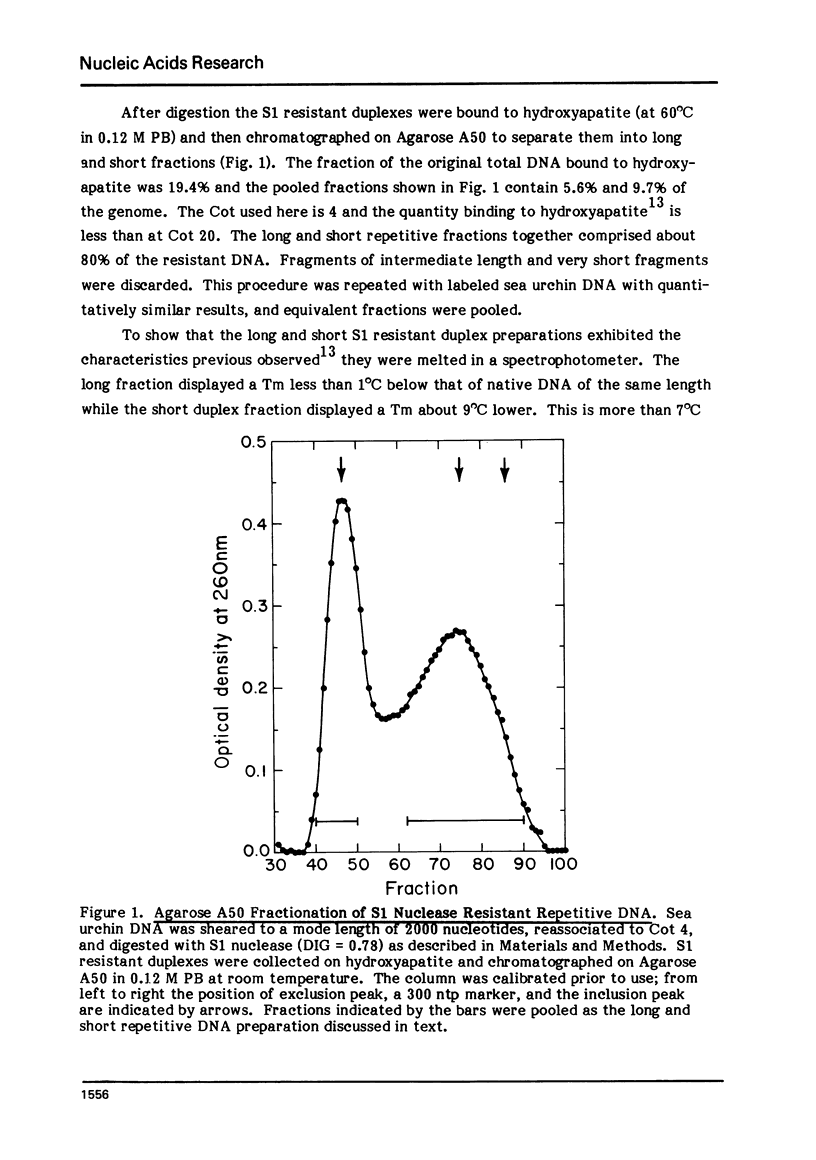
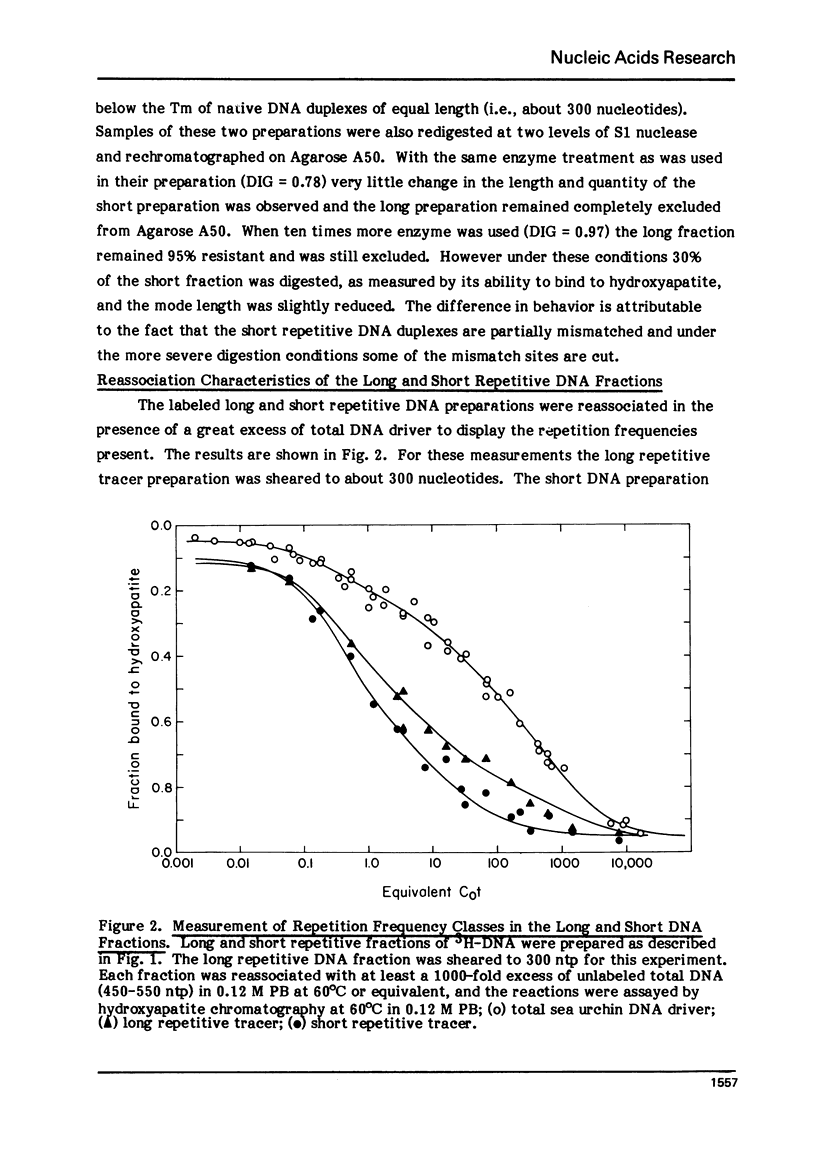
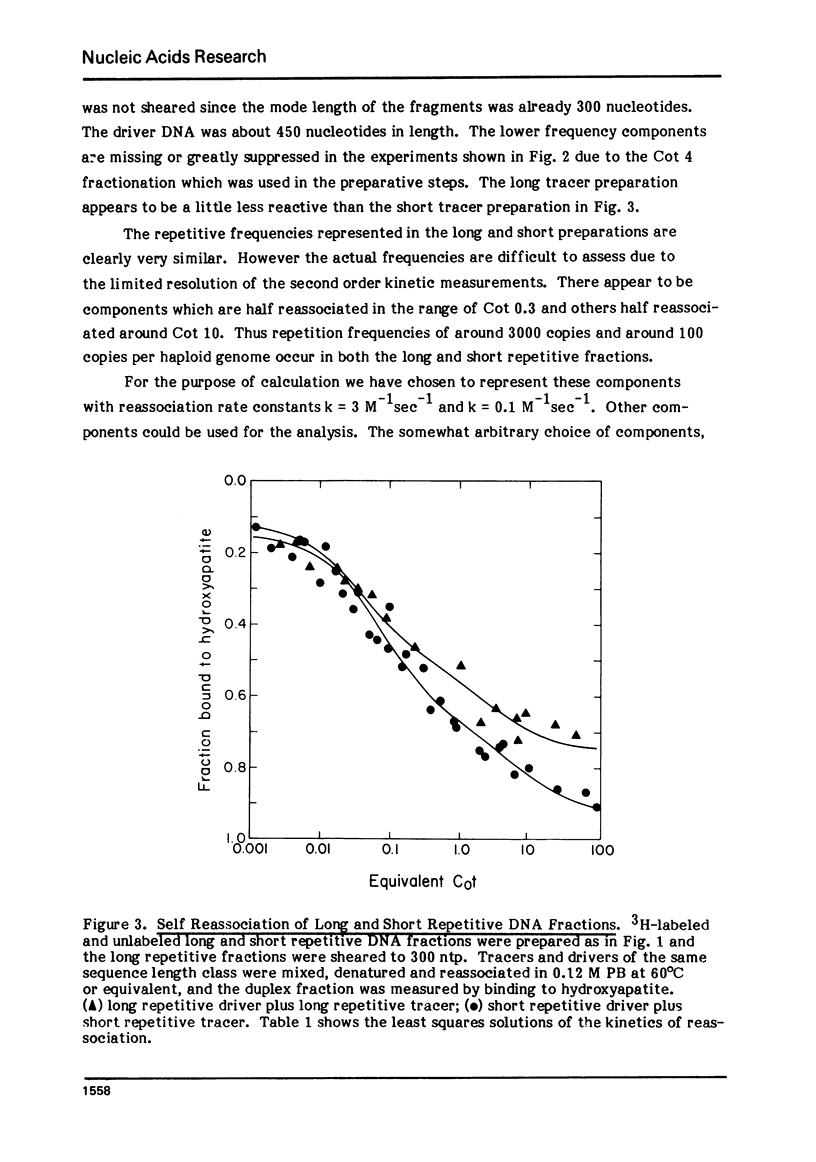
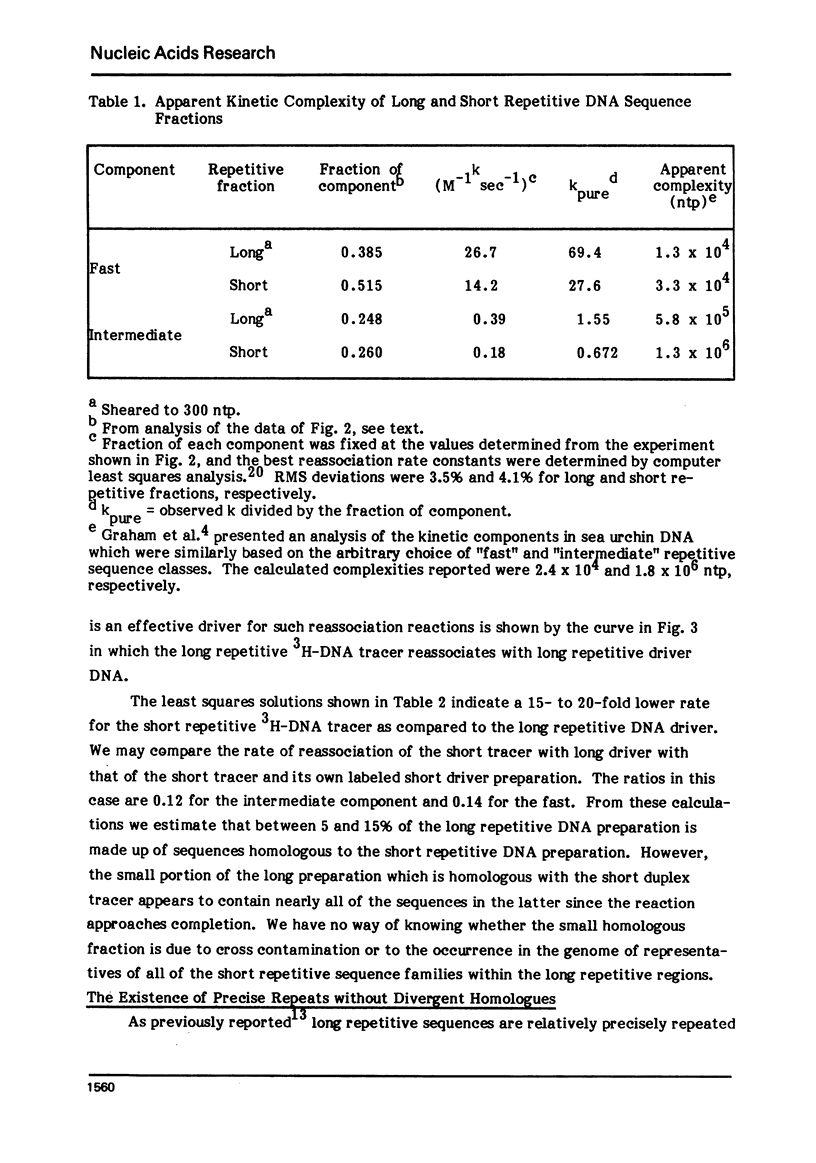
Long and short repetitive sequences of sea urchin DNA were prepared by reassociation of 2000 nucleotide long fragments to Cot 4 and digestion with the single strand specific nuclease S1. The S1 resistant duplexes were separated into long repetitive and short repetitive fractions on Agarose A50. The extent of shared sequences was studied by reassociating a labeled preparation of short repetitive DNA with an excess of unlabeled long repetitive DNA. Less than 10% of the long repetitive DNA preparation was able to reassociate with the short repetitive DNA. Thus the long and short repetitive elements appear to be principally independent sequence classes in sea urchin DNA. Precisely reassociating repetitive DNA was prepared by four successive steps of reassociation and thermal chromatography on hydroxyapatite. This fraction (3% of the genome) was reassociated by itself or with a great excess of total sea urchin DNA. The thermal stability of the products was identical in both cases (Tm=81 degrees C), indicating that precisely repeated sequences do not have many imprecise copies in sea urchin DNA.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim Biophys Acta. 1966 Jan 18;114(1):158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- Angerer R. C., Davidson E. H., Britten R. J. DNA sequence organization in the mollusc Aplysia californica. Cell. 1975 Sep;6(1):29–39. doi: 10.1016/0092-8674(75)90070-7. [DOI] [PubMed] [Google Scholar]
- Birnstiel M., Telford J., Weinberg E., Stafford D. Isolation and some properties of the genes coding for histone proteins. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2900–2904. doi: 10.1073/pnas.71.7.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. DNA sequence arrangement and preliminary evidence on its evolution. Fed Proc. 1976 Aug;35(10):2151–2157. [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Gene regulation for higher cells: a theory. Science. 1969 Jul 25;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971 Jun;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3175–3179. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M. E., Britten R. J., Davidson E. H. Sequence organization in Xenopus DNA studied by the electron microscope. J Mol Biol. 1975 Aug 5;96(2):317–333. doi: 10.1016/0022-2836(75)90351-4. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Purdom I. F. Clustering of transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):411–429. doi: 10.1016/0022-2836(73)90014-4. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Kurer V. Isolation and some properties of DNA coding for tRNA1met from Xenopus laevis. Cell. 1976 Jun;8(2):183–195. doi: 10.1016/0092-8674(76)90002-7. [DOI] [PubMed] [Google Scholar]
- Crain W. R., Eden F. C., Pearson W. R., Davidson E. H., Britten R. J. Absence of short period interspersion of repetitive and non-repetitive sequences in the DNA of Drosophila melanogaster. Chromosoma. 1976 Jul 30;56(4):309–326. doi: 10.1007/BF00292953. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Graham D. E., Neufeld B. R., Chamberlin M. E., Amenson C. S., Hough B. R., Britten R. J. Arrangement and characterization of repetitive sequence elements in animal DNAs. Cold Spring Harb Symp Quant Biol. 1974;38:295–301. doi: 10.1101/sqb.1974.038.01.033. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Crain W. R., Britten R. J., Davidson E. H., Kafatos F. C. DNA sequence organization in the lepidopteran Antheraea pernyi. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2289–2293. doi: 10.1073/pnas.73.7.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsheit A. B., Davidson N., Brown D. D. An electron microscope heteroduplex study of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri. J Mol Biol. 1974 Dec 5;90(2):301–314. doi: 10.1016/0022-2836(74)90375-1. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Klein W. H., Davis M. M., Wold B. J., Britten R. J., Davidson E. H. Structural gene sets active in embryos and adult tissues of the sea urchin. Cell. 1976 Apr;7(4):487–505. doi: 10.1016/0092-8674(76)90200-2. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Rochaix J. D. The amplified ribosomal DNA of dytiscid beetles. Proc Natl Acad Sci U S A. 1974 May;71(5):1819–1823. doi: 10.1073/pnas.71.5.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., Crain W. R., Ruderman J. V., Moore G. P., Barnett T. R., Higgins R. C., Gelfand R. A., Galau G. A., Britten R. J., Davidson E. H. DNA sequence organization in the genomes of five marine invertebrates. Chromosoma. 1975 Jul 21;51(3):225–251. doi: 10.1007/BF00284817. [DOI] [PubMed] [Google Scholar]
- Kedes L. H., Cohn R. H., Lowry J. C., Chang A. C., Cohen S. N. The organization of sea urchin histone genes. Cell. 1975 Nov;6(3):359–369. doi: 10.1016/0092-8674(75)90185-3. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Schmid C. W., Davidson N. Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell. 1975 Feb;4(2):141–155. doi: 10.1016/0092-8674(75)90121-x. [DOI] [PubMed] [Google Scholar]
- Noll H. Characterization of macromolecules by constant velocity sedimentation. Nature. 1967 Jul 22;215(5099):360–363. doi: 10.1038/215360a0. [DOI] [PubMed] [Google Scholar]
- Patterson J. B., Stafford D. W. Characterization of sea urchin ribosomal satellite deoxyribonucleic acid. Biochemistry. 1971 Jul 6;10(14):2775–2779. doi: 10.1021/bi00790a019. [DOI] [PubMed] [Google Scholar]
- Schachat F. H., Hogness D. S. Repetitive sequences in isolated Thomas circles from Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1974;38:371–381. doi: 10.1101/sqb.1974.038.01.040. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Gross K., Telford J., Birnstiel M. Molecular analysis of the histone gene cluster of psammechinus miliaris: II. The arrangement of the five histone-coding and spacer sequences. Cell. 1976 Aug;8(4):471–478. doi: 10.1016/0092-8674(76)90214-2. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Unequal crossover and the evolution of multigene families. Cold Spring Harb Symp Quant Biol. 1974;38:507–513. doi: 10.1101/sqb.1974.038.01.055. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H. A comparison of the structural organization of amplified ribosomal DNA from Xenopus mulleri and Xenopus laevis. J Mol Biol. 1975 May 15;94(2):151–161. doi: 10.1016/0022-2836(75)90074-1. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H., Carroll D., Brown D. D., Deutch A., Higashinakagawa T., Dawid I. B. Amplified ribosomal DNA from Xenopus laevis has heterogeneous spacer lengths. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2823–2827. doi: 10.1073/pnas.71.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]



