Abstract
We have demonstrated label-free optical detection of viral nucleoprotein binding to a polyvalent anti-influenza aptamer by monitoring the surface-enhanced Raman (SERS) spectra of the aptamer-nucleoprotein complex. The SERS spectra demonstrated that selective binding of the aptamer-nucleoprotein complex could be differentiated from that of the aptamer alone based solely on the direct spectral signature for the aptamer-nucleoprotein complex. Multivariate statistical methods, including principal components analysis, hierarchical clustering, and partial least squares, were used to confirm statistically significant differences between the spectra of the aptamer-nucleoprotein complex and the spectra of the unbound aptamer. Two separate negative controls were used to evaluate the specificity of binding of the viral nucleoproteins to this aptamer. In both cases, no spectral changes were observed that showed protein binding to the control surfaces, indicating a high degree of specificity for the binding of influenza viral nucleoproteins only to the influenza-specific aptamer. Statistical analysis of the spectra supports this interpretation. AFM images demonstrate morphological changes consistent with formation of the influenza aptamer-nucleoprotein complex. These results provide the first evidence for the use of aptamer-modified SERS substrates as diagnostic tools for influenza virus detection in a complex biological matrix.
Introduction
Influenza remains a major source of morbidity and mortality worldwide. Severe outbreaks, such as the 1918 and 1957 flu pandemics caused by the highly pathogenic avian influenza (HPAIV) H5N1 virus, resulted in a ~60% mortality rate.1 More recently, the 2009 influenza A (H1N1) outbreak reached a high pandemic alert level. As a result, surveillance methods are being implemented to track outbreaks of current and emerging strains, as well as aid in the development of vaccine and disease intervention strategies.2
Current diagnostic tools available for routine laboratory diagnosis of influenza rely on the detection of viral particles or typing of the viral genome.3–5 However, these assays suffer from a lack of sensitivity and reproducibility, or often require synthetic labeling or species-specific reagents. As a result, new approaches for rapid, reliable, and sensitive means of detecting influenza virus in clinical diagnostics are urgently needed.
Aptamers are single-stranded DNA or RNA oligonucleotides that are selected through an in-vitro combinatorial selection process with high affinity and specificity.6 Aptamers have been developed against a wide variety of targets ranging from proteins7 to whole cells,8 and have been integrated in a variety of biosensors.9, 10 As such, aptamers constitute promising alternatives to antibodies in diagnostic assays due to their stability, robustness and in-vitro preparation methods.
We describe here a label-free optical method, i.e. surface-enhanced Raman spectroscopy (SERS), to detect viral nucleoprotein binding to a polyvalent influenza aptamer. SERS is a signal amplification technique based on electromagnetic enhancement where the Raman excitation wavelength is in resonance with a metallic surface plasmon.11 The advantage of SERS-based sensing is that, unlike other label-free methods that rely on a general signal response for the captured analyte,12–14 Raman spectra provide a unique chemically specific molecular fingerprint.15, 16
Raman and SERS have previously been used to directly probe sequence information in DNA/RNA complexes.17–19 Recent reports demonstrated direct detection of DNA/RNA sequences using SERS,20–25 including hybridization of target sequences to oligo-modified substrates,21, 26 with detection of single nucleotide polymorphisms possible.24, 26 SERS spectra were reported sensitive to conformational changes, molecular orientation and packing density in ssDNA and dsDNA oligomers upon ligand binding.21, 22
Most of the Raman aptamer studies published to date have used surface-enhanced resonance Raman (SERRS), a method that employs Raman-active synthetic labels. These studies include adenosine binding to dsDNA,27 as well as detection of cocaine,28 and the protein thrombin.29 Relatively few reports have appeared that focus on non-resonant SERS detection of aptamer-target binding in a direct, label-free fashion.30–32 However, these latter studies have established the proof of principle that aptamer-target binding can be detected from the intrinsic SERS spectra of the complex.
The current work expands on these previous model studies and presents results showing that intrinsic SERS spectra can detect the binding of a real diagnostic aptamer to its influenza target in a complex biological matrix. We studied a commercial polyvalent anti-influenza aptamer that was developed against the nucleoprotein constituents of the viruses contained in a commercially available split-virion inactivated influenza vaccine. The intrinsic SERS spectra provide direct spectral signatures for the aptamer-nucleoprotein complexes. A preliminary investigation on the detection of this polyvalent influenza aptamer using SERS has been reported.33 The present work extends on this investigation and introduces i) two separate negative controls, ii) detailed chemometric spectral analysis, and iii) AFM imaging of the binding events, to demonstrate the specificity of binding for the viral nucleoproteins to the influenza aptamer. These results provide the first detailed evidence for the use of aptamer-modified SERS substrates as diagnostic tools for influenza virus detection in a real, complex biological system.
Experimental Methods
Material and Reagents
(1-Mercaptoundec-11-y)tetra(ethyleneglycol) (95%) and [Arg8]-Vasopressin (>95%) were purchased from Sigma-Aldrich (St. Louis, MO). All other chemicals were of analytical grade and used without any further purification.
Preparation of SERS-Active Ag Nanorod Arrays and Microwell Array Fabrication
Aligned Ag nanorods used as SERS-active substrates were prepared by an oblique-angle vapor deposition (OAD) technique using a custom-designed electron-beam/sputtering evaporation system according to previously published procedures.34 Full details on the nanofabrication, cleaning, and patterning of the aligned Ag nanorod arrays are provided in Supporting Information.
Anti-Influenza Aptamer
The polyvalent 5′-C6-disulfide anti-influenza aptamer was provided by AptaRes AG, Mittenwalde, Germany. The AptaRes polyvalent anti-influenza aptamer used in this study was isolated against the nucleoprotein constituents of a commercially available inactivated influenza vaccine using a cell-free, MonoLex™ combinatorial process.35 The single-stranded DNA aptamer consisted of 63 base pairs with a molecular weight of 25,987 Daltons. The as-received lyophilized aptamer was dissolved in molecular biology grade water to a final concentration of 100 μM.
Influenza and RSV Virus Samples
The AptaRes polyvalent influenza aptamer used in this study was isolated against the nucleoprotein constituents of the viruses contained in the commercially available Fluarix™ Influsplit SSW® 2009/2010 split-virion inactivated influenza vaccine (GlaxoSmithKline, GmbH & Co., KG, Munich, Germany). The three monovalent split viruses used in this vaccine are the i) A/Uruguay/716/2007 NYMC X-175C, ii) B/Brisbane/60/2008, and iii) A/Brisbane/59/2007 IVR-148, influenza strains. The Robert Koch Institut (Berlin, Germany) supplied the influenza nucleoproteins from these viruses in the form of purified, whole virus cell lysates. Respiratory Syncytial Virus (RSV) was used as a negative viral control. Further detailed information on the influenza virus samples and RSV isolation are provided in Supporting Information.
Anti-Vasopressin Aptamer
An anti-vasopressin aptamer HS-C6H12-5′TCA CGT GCA TGA TAG ACG GCG AAG CCG TCG AGT TGC TGT GTG CCG ATG CAC GTA3′ was purchased from Integrated DNA Technologies (IDT, Coralville, IA).
Binding Buffer
The buffer used in the binding experiments was prepared by dissolving 20 mM Tris HCl, 15 mM NaCl, 4 mM KCl, 1 mM MgCl2 and 1 mM CaCl2 in molecular biology grade water at pH 7.3 and stored at 4°C. The buffer and working tools were DNase free.
Raman Spectroscopy
Raman measurements were performed using a confocal Raman microscope (InVia, Renishaw, Inc., Gloucestershire, United Kingdom). A 785 nm near-IR diode laser provided laser excitation. The sample was illuminated through a 20× (N.A. = 0.40) objective resulting in a spot size of approximately 4.8 × 27.8 μm. The laser power used was ~0.36 mW, as measured at the sample. SERS spectra were collected from ten different spots within a given microwell using a 30 second acquisition time with one accumulation. Spectra were collected between 2000 and 500 cm−1.
Data Analysis
Off-line spectral manipulation and analysis were performed using GRAMS 32/AI Version 6.0 (Galactic Industries Corporation, Nashua, NH). Multivariate statistical analysis of the samples, including principal components analysis, hierarchical cluster analysis, and partial least-squares discriminant analysis, was performed with PLS Toolbox version 6.2 (Eigenvector Research Inc., Wenatchee, WA), operating in a MATLAB environment (R2011a, The Mathworks Inc., Natick, MA).
AFM Measurements
Template stripped Ag nanofilms were used as AFM substrates and prepared according to previously published procedures.36 AFM measurements were performed using an Agilent 5500 AFM system equipped with an inverted light microscope (ILM) system (Agilent, Chandler, AZ) combined with an Agilent multi-purpose AFM scanner with a scanning area of 10 μm2. Full details on AFM substrate fabrication and measurements are provided in Supporting Information.
Results and Discussion
Experimental Design
The disulfide-derivatized anti-influenza ss-DNA aptamer was immobilized onto the individual Ag nanorod microwells, resulting in the formation of self-assembled monolayers of DNA oligonucleotides.37 A second immobilization step added the spacer molecule (1-mercaptoundec-11-y)tetra(ethyleneglycol), HS(CH2)11(OCH2CH2)4OH. The functionalized microwells of the Ag nanorod substrate were then incubated with each of the three influenza virus strains or controls. Complete details of these experimental procedures used for immobilization and binding are provided in Supporting Information, as well as a figure (S.1) outlining the various steps in the experimental design.
SERS Spectra of the Aptamer Complex
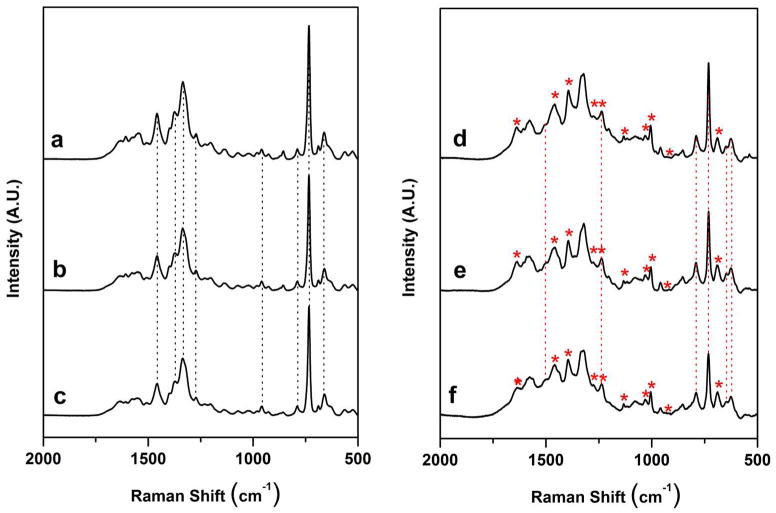
Fig. 1. a (top left) shows the SERS spectrum of the anti-influenza aptamer-spacer complex used in this study, while Fig. 1. b (middle left) is the spectrum of the blank control, i.e. the aptamer complex incubated only with the binding buffer. Fig. 1. b shows that no detectable spectral changes are observed when the aptamer complex is treated with a blank control. The number and position of the bands in Figs. 1. a and 1. b are in agreement with those reported from SERS studies of DNA bound to Ag and Au nanoparticles.17, 38 The dominant features present in Fig. 1. a and 1. b are bands that can be primarily attributed to nucleic acid bases. 17 In addition, a band at 687 cm−1 is indicative of the C-S stretching vibration of the thiol group, and confirms cleavage of the disulfide bond after adsorption and immobilization of the aptamer to the Ag nanorod array.39 A complete table of the observed SERS bands in the spectrum of the anti-influenza aptamer, along with their assignments based on literature precedent, is provided in Table S.1 in Supporting Information.
Figure 1.
(a) SERS spectrum of the anti-influenza aptamer (1000 nM) - PEG spacer (100 nM) complex on a Ag nanorod substrate, (b) anti-influenza aptamer complex and binding buffer blank control, (c) anti-influenza aptamer complex and RSV (105 PFU/mL) negative control, (d) anti-influenza aptamer complex and nucleoproteins from A/Uruguay, (e) anti-influenza aptamer complex and nucleoproteins from A/Brisbane, and (f) anti-influenza aptamer complex and nucleoproteins from B/Brisbane. Virus concentrations in (d) – (f) were adjusted to 1μg/mL (relative to HA content). Each spectrum shown is an average of 10 individual spectra for each particular sample. The dashed vertical lines in (a), (b), and (c) indicate the characteristic oligonucleotide bands for the influenza aptamer and its controls. The dashed vertical lines in (d), (e), and (f) indicate the oligonucleotide bands that changed after binding of the nucleoproteins. Asterisks indicate the presence of new bands in the aptamer complex corresponding to binding of the protein target.
Surface Coverage and Reproducibility
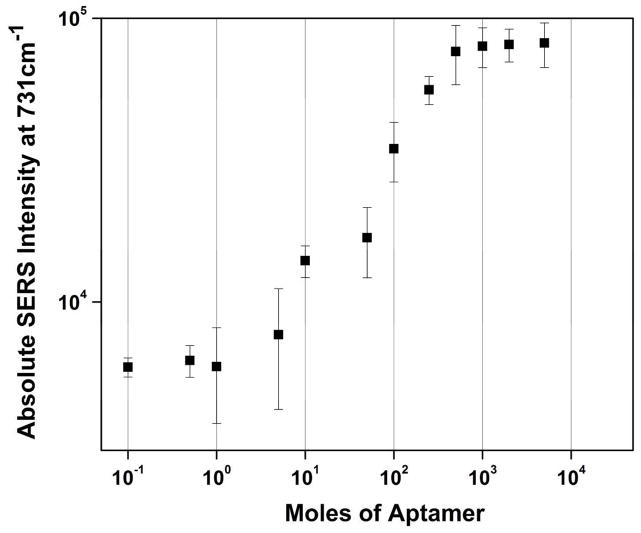
The surface coverage of the disulfide aptamer on the Ag nanorod array was investigated by varying the concentration of the aptamer and monitoring the concentration-dependent A ring-breathing mode at 731 cm−1.22 Fig. 2 shows the log-log plot of the absolute SERS intensity of the 731 cm−1 band as a function of the aptamer concentration. The SERS response displays a sigmoidal increase over three orders of magnitude (10−1 to 102 mol) in the concentration range 0.1 – 500 nM, followed by an intensity plateau. The error bars in Fig. 2 indicate a high degree of spectral reproducibility from the Ag nanorods SERS substrates used in these experiments.
Figure 2.
Log-log plot of the absolute SERS intensity of the 731 cm−1 band of the anti-influenza aptamer as a function of concentration in the range from 0.1 to 5000 nM.
The plateau effect of the band intensity seen in Fig. 2 has been previously observed in Ag nanorod SERS studies and is related to the saturation of the SERS signal with increasing surface coverage.40 The surface density of the aptamer on the surface of the Ag nanorod substrate was calculated by estimating the number of DNA molecules contained in the 20 μL of a 1000 nM DNA solution deposited on the surface of a microwell; we included the estimated surface area of the nanorods in these calculations. The surface density was found to be ~3.7 × 105 molecules/μm2. This estimate is comparable (within a factor of ten) with previous studies of the surface density of DNA adsorbed onto smooth Au films.41 These results are also in agreement with previous surface density estimates of thiolated DNA on metal films.42 Some variation in surface density should be expected in the present case due to the differences in substrate composition (Au vs. Ag) and morphology (nanorods vs. smooth); however, our results agree well with previously published reports on DNA packing density.
Nucleoprotein Binding to the Polyvalent Anti-Influenza Aptamer
Aptamer-target binding leads to addition of proteins to the aptamer-modified surface, which should be reflected as changes in oligonucleotide secondary structure, both of which can be sensed via SERS.30–32
Consistent with this hypothesis, distinct changes are seen in the spectra of the anti-influenza aptamer after incubation with the cell lysates from the three monovalent influenza virus strains. Figure 1. d shows the SERS spectrum of the aptamer and spacer on the Ag nanorod array incubated for 8 hours at 37°C with 1 μg/mL (HA) content of the influenza virus strain A/Uruguay. Major changes in bands attributed to the DNA aptamer are noticed upon nucleoprotein binding. The changes in the oligonucleotide bands due to nucleoprotein binding are highlighted by the dashed vertical lines in Figs. 1. d, 1. e and 1. f.
In addition to nucleic acid spectral features, bands present in the SERS spectra of the aptamer-nucleoprotein complex can be attributed to the nucleoproteins themselves. Prominent bands indicative of protein binding seen in the spectrum in Fig. 1. d include bands attributed to amide vibrations and amino acid side chains.43, 44 Characteristic protein bands present in the spectrum of the influenza nucleoprotein – aptamer complex are marked with asterisks in Fig. 1. d, 1. e, and 1. f. A complete table of the observed SERS bands in the influenza nucleoprotein – aptamer complex along with their literature-based assignments is provided in Table S.2 of Supporting Information.
To test the polyvalent nature of the aptamer, the nucleoproteins from the influenza strains A/Brisbane and B/Brisbane that are also contained in the split-virion influenza vaccine were incubated with the aptamer-spacer complex in the same manner as the A/Uruguay strain. These spectra are shown in Fig. 1. e (A/Brisbane, middle right) and Fig. 1. f (B/Brisbane, bottom right). The identical spectral changes that are seen in the A/Uruguay complex (Fig. 1. d) also occur in the complexes for the A/Brisbane and B/Brisbane strains, indicating a high degree of reproducibility in the nucleoprotein binding of the split virion strains to the polyvalent aptamer.
Negative Control: RSV With Influenza Aptamers
We incorporated respiratory syncytial virus (RSV) as a negative control virus in these experiments to determine the specificity of binding of the influenza nucleoproteins to the anti-influenza aptamer. RSV is an important and ubiquitous respiratory virus, and as such is a good candidate to act as a control virus for influenza.45 Incubation of the RSV control sample was accomplished by adding 20 μL of the lysed RSV virus solution to an aptamer-coated Ag nanorod microwell. The virus sample was adjusted to a concentration of ~105 PFU/mL in the binding buffer, and the binding conditions were kept identical to those used for the incubation of the influenza virus samples.
Figure 1. c shows the spectrum of the RSV virus applied to the aptamer-modified substrate. Only minor spectral intensity changes were observed in Fig. 1. c after incubation of the RSV virus sample, indicating no apparent binding of the RSV lysate to the anti-influenza aptamer. This is readily seen when comparing Fig. 1. c to the aptamer and blank spectra in Figs. 1. a and 1. b. The spectrum of RSV (Fig. 1. c) also contrasts sharply with those of the nucleoprotein - aptamer complex spectra on the right panel (Fig. 1. d, 1. e and 1. f). The SERS spectra in Fig. 1 show a high degree of specificity for the binding of only influenza viral nucleoproteins to the polyvalent anti-influenza aptamer.
Chemometric Analysis of Influenza Aptamer – Nucleoprotein Spectra
Multivariate statistical analysis was used to establish statistically significant differences between the SERS spectra of the samples and controls in Fig. 1 and was used to confirm the binding specificity of the aptamer to its viral nucleoprotein targets.
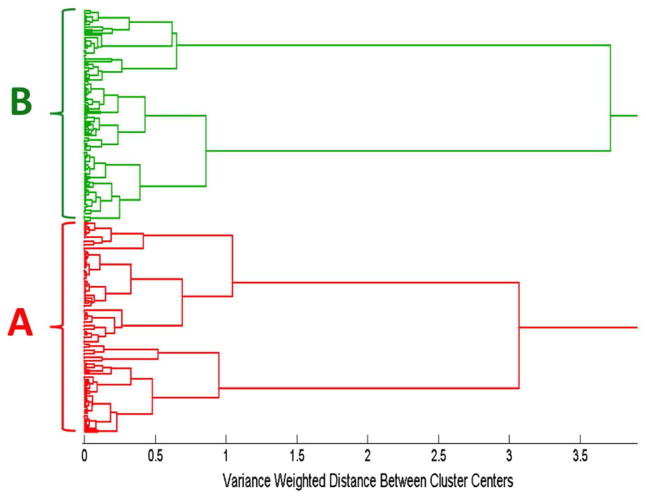
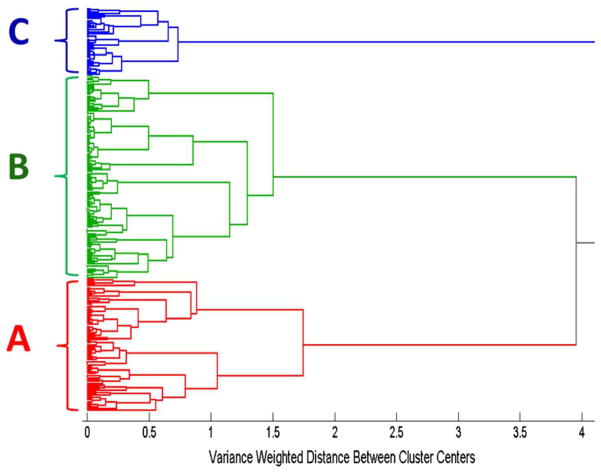
Figure 3 illustrates the hierarchical cluster analysis (HCA) dendrogram based on the spectra of the aptamer, controls, plus influenza nucleoprotein samples, calculated using Ward’s method. Two hundred forty spectra, corresponding to 40 spectra in each sample category, were used to generate this dendrogram. Figure 3 reveals two distinct clusters. The first red cluster (A) corresponds to the spectra in Figs. 1. a, 1. b, and 1. c, which include the aptamer complex (Fig. 1. a), the blank control (Fig. 1. b), and the negative control RSV virus (Fig. 1. c). The second green cluster (B) corresponds to the spectra in Figs. 1. d, 1. e, and 1. f, which refer to the aptamer incubated with all three influenza virus strains, A/Uruguay (Fig. 1. d), A/Brisbane (Fig. 1. e) and B/Brisbane (Fig. 1. f). This classification by HCA provides unambiguous separation between the SERS spectra of the unbound aptamers and its controls with the aptamer-nucleoprotein complexes.
Figure 3.
Dendrogram produced by hierarchical cluster analysis (HCA) of the SERS spectra shown in Fig. 1 using Ward’s method and label-coded as follows: (A) anti-influenza aptamer, blank buffer, and RSV negative control, (B) anti-influenza aptamer incubated with influenza nucleoproteins from A/Uruguay, A/Brisbane, and B/Brisbane. A total of 240 spectra were used to generate this dendrogram, corresponding to 40 spectra in each of the 6 sample categories.
Additional multivariate statistical tests were used to analyze the spectra of the influenza nucleoprotein complexes. These include principal components analysis (PCA) and partial least squares discriminant analysis (PLS-DA). Both methods confirmed the findings of the cluster analysis shown in Fig. 3, i.e. that the SERS spectra of the influenza nucleoprotein complexes can be completely differentiated from those of the blanks and negative control with 100% accuracy. A further discussion of these results is included as Figures S.2 and S.3 in Supporting Information.
Specificity of Binding: Influenza Nucleoproteins With Non-Influenza Aptamer
We further investigated the binding specificity of the influenza viral nucleoproteins to the anti-influenza aptamer using a separate DNA oligonucleotide as a negative control. We utilized a 5′-C6-thiolated anti-vasopressin aptamer for this purpose. This aptamer was developed to bind and inhibit the neuropeptide vasopressin, a peptide hormone regulating the extracellular fluid volume in the body secreted by the posterior lobe of the pituary gland,46 and has no affinity for influenza.
The HS-C6-5′ anti-vasopressin aptamer was immobilized onto the Ag nanorod array substrate via the thiolate group, and incubated with i) binding buffer, ii) vasopressin and iii) all three influenza virus strains, using the procedures described in Supporting Information. Fig. 4 shows representative SERS spectra of the resulting complexes.
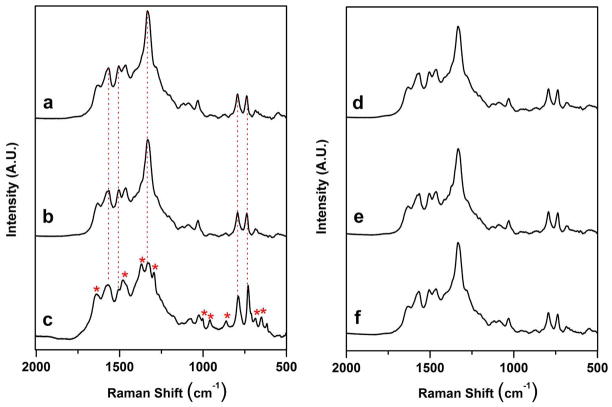
Figure 4.
SERS spectra of the anti-vasopressin aptamer, controls, vasopressin, and influenza nucleoproteins on a Ag nanorod substrate after incubation at 37 °C for 8 hours. (a) anti-vasopressin aptamer (1000 nM) - spacer (100 nM), (b) vasopressin aptamer with blank binding buffer control, (c) vasopressin aptamer with vasopressin (1μg/mL), (d) vasopressin aptamer with nucleoproteins from A/Uruguay, (e) vasopressin aptamer with nucleoproteins from A/Brisbane, and (f) vasopressin aptamer with nucleoproteins from B/Brisbane. Virus concentrations in (d) – (f) are 1μg/mL (relative to HA content). Each spectrum shown is an average of 10 individual spectra for each particular sample. The dashed vertical lines in (a), (b), and (c) indicate the characteristic oligonucleotide bands for the anti-vasporessin aptamer. Asterisks indicate the presence of new bands in the aptamer complex corresponding to binding of the protein target.
The interpretation of the anti-vasopressin aptamer spectrum in Fig. 4. a follows directly from the discussion of the anti-influenza aptamer in Fig. 1. a. The main features in the spectrum in Fig. 4. a are bands attributable to nucleic acid vibrations.17, 38 Fig. 4. b shows the SERS spectrum of the anti-vasopressin aptamer incubated with the binding buffer as a blank control. Fig. 4. b shows that no detectable changes are apparent in the spectra of the anti-vasopressin aptamer incubated with the binding buffer when compared to the aptamer alone, similar to that observed with the spectra of the buffer blank with the anti-influenza aptamer in Figs. 1. a and 1. b.
The anti-vasopressin aptamer was treated with the three monovalent influenza virus strains. Figure 4. d shows the SERS spectrum of the anti-vasopressin aptamer on the Ag nanorod array incubated with the influenza virus strain A/Uruguay. A comparison of the spectrum in Fig. 4. d (top right) with the spectra of the anti-vasopressin aptamer and blank (Figs. 4. a and 4. b) shows that no spectral differences are apparent, indicating a lack of influenza nucleoprotein binding to the anti-vasopressin aptamer. This effect also occurs in the spectra that result from the addition of the influenza strains A/Brisbane (Fig. 4. e, middle right) and B/Brisbane (Fig. 4. f, bottom right). Relative to the aptamer and blank spectra, no discernable spectral changes are observed when any of the three types of influenza nucleoproteins are incubated with the anti-vasopressin aptamer.
These conclusions are confirmed when the anti-vasopressin aptamer is incubated with 1 μg/mL vasopressin. The spectrum of the vasopressin – aptamer complex is shown in Fig. 4. c. This spectrum illustrates that the formation of a vasopressin – anti-vasopressin aptamer complex resulted in changes of the characteristic protein and nucleic acid bands.43, 44 The oligonucleotide bands associated with the changing spectral features due to vasopressin binding are highlighted with dashed vertical lines in Figs. 4. a, 4. b, and 4. c. The characteristic protein bands present in the vasopressin complex are marked with asterisks in Fig. 4. c. A list of the observed SERS bands in the vasopressin aptamer – vasopressin complex with their tentative band assignments is provided in Table S.3 of the Supporting Information.
The presence of significant spectral changes upon the formation of a vasopressin – anti-vasopressin aptamer complex, combined with the absence of any spectral changes upon incubation of the three influenza virus samples with the anti-vasopressin aptamer, strongly suggests that selective binding of the influenza viral nucleoproteins occurs only with the anti-influenza aptamer.
Chemometric Analysis of Negative Control Aptamer – Nucleoprotein Spectra
Figure 5 shows the HCA dendrogram calculated from the anti-vasopressin spectra described above using Ward’s algorithm. Two hundred forty spectra, corresponding to 40 spectra in each sample category, were used to generate this dendrogram. As seen in Fig. 5, the anti-vasopressin control spectra separated into three distinct clusters. The bottom red cluster (A) resulted from the spectra of the anti-vasopressin aptamer incubated with the three influenza virus samples (Fig. 4. d, 4. e and 4. f). The middle green cluster (B) represents the spectra of the anti-vasopressin aptamer alone (Fig. 4. a) along with the spectra of the unbound aptamer treated only with the binding buffer (Fig. 4. b). Finally, the top blue cluster (C) represents the spectra of the anti-vasopressin aptamer (Fig. 4. c) incubated with vasopressin. The results indicate that cluster analysis provides excellent (100%) statistical differentiation of the aptamer controls from the vasopressin target. In particular, the spectra of the influenza nucleoproteins are completely distinguished from that of the vasopressin complex and its controls, confirming the selectivity of binding of the influenza nucleoproteins only to the anti-influenza aptamer (Fig. 1 and Fig. 3).
Figure 5.
Dendrogram produced by hierarchical cluster analysis (HCA) of the SERS spectra shown in Fig. 4 calculated using Ward’s method and label-coded as follows: (A) anti-vasopressin aptamer complex and influenza nucleoproteins from A/Uruguay, A/Brisbane, B/Brisbane, (B) anti-vasopressin aptamer (1000 nM) - spacer (100 nM) complex, along with blank buffer control, and (C) anti-vasopressin aptamer complex and vasopressin (1μg/mL). A total of 240 spectra were used to generate this dendrogram, corresponding to 40 spectra in each of the 6 sample categories.
Additional PCA and PLS-DA statistical tests confirm that the SERS spectra of the influenza nucleoproteins incubated with the anti-vasopressin aptamer show no evidence of binding, and can be completely distinguished from those of the blank spectra and the vasopressin aptamer complex. A further discussion of these results is included as Figures S.4 and S.5 in Supporting Information.
Characterization of the Influenza Aptamer – Nucleoprotein Complex Using AFM
Atomic force microscopy was used to follow the in-situ evolution of viral nucleoprotein binding to the anti-influenza aptamer. A specially designed flow cell was used to image the immobilization steps as the aptamer, spacer, and nucleoproteins were introduced to the cell. This technique allowed imaging of the same surface area throughout the immobilization process between the aptamer probe and the binding to its nucleoprotein target.
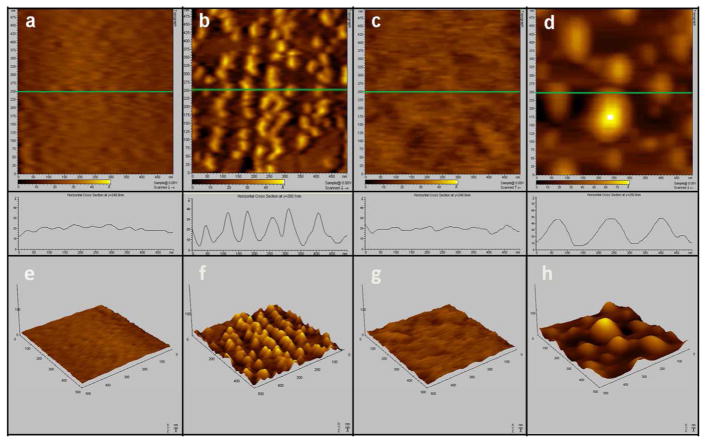
Figure 6 shows i) two-dimensional AFM images, ii) surface roughness line scans, and iii) three-dimensional AFM images, of the four steps in the surface immobilization of the influenza nucleoproteins. The first column of Fig. 6 (images 6. a and 6. e) illustrates the bare Ag substrate prepared by template stripping. The Ag substrate has a homogeneously flat surface with an average height of 1 nm and mean roughness of 0.5 nm. The second column of Fig. 6 (images 6. b and 6. f) represents the same Ag surface after injection of the disulfide anti-influenza aptamer at a concentration of 100 pM for 3 hours, followed by rinsing. The resulting images show a distribution of aggregates with an approximate average height of 6 nm, a mean roughness of 2.7 nm, and an average diameter of 50 nm, consistent with the changes expected upon formation of the self-assembly of flexible thiolated aptamers.47
Figure 6.
(Top) 500 nm × 500 nm 2-D AFM images of (a) bare Ag nanofilm substrate, (b) Ag substrate after injection of the disulfide anti-influenza aptamer at a concentration of 100 pM for 3 hours, (c) Ag substrate after injection of (1-Mercaptoundec-11-y)tetra(ethyleneglycol) at a concentration of 10 pM for 3 hours, (d) Ag substrate after injection of A/Uruguay diluted in the binding buffer to a concentration of 1 pg/mL (HA) content at 37°C for 3 hours.
(Middle) The plots show the surface roughness line scans corresponding to the green line on each of the 2-D AFM images immediately above it in Panels (a), (b), (c) and (d).
(Bottom) Panels (e), (f), (g), and (h) are the representative 3-D AFM images of the 2-D images in (a), (b), (c) and (d), respectively.
The third column in Fig. 6 (images 6. c. and 6. g) are the images obtained of the aptamer-modified surface following treatment with (1-mercaptoundec-11-y)tetra(ethyleneglycol) at a concentration of 10 pM for 3 hours, followed by rinsing of the surface. The addition of the PEG-like spacer molecule induced rearrangement of the aptamers and produced a uniform distribution of the self-assembled monolayers adsorbed on the Ag surface. As a result, the average height and mean roughness of the imaged surface decreased to 0.8 nm and 0.3 nm, respectively.
The last column in Fig. 6 (images 6. d and 6. h) illustrate the aptamer-functionalized surface after incubation with the influenza virus strain A/Uruguay at a concentration of 1 pg/mL (HA) content for 3 hours. Incubation of the aptamer-modified surface with the influenza virus sample resulted in surface topography changes and the appearance of bright spots on the AFM images shown in Figs. 6. d and 6. h. Binding of the aptamer to the nucleoproteins is characterized by the formation of large features on the AFM images and with an average diameter of about 150 nm.
These AFM images do not depict single binding events between the aptamer and the influenza nucleoprotein targets on a molecular level; however, they do support the interpretation that the features in Figs. 6. d and 6. h are due to the aptamer binding with its target. The variations in average height (increase to 2.6 nm) and mean roughness (increase to 1.3 nm) are consistent with the formation of an aptamer-nucleoprotein complex upon incubation with influenza virus lysate. The dimensions of these features are smaller than expected for an intact influenza nucleoprotein complex,48 which exists in a polymerase complex (ribonucleoprotein) composed of multiple (usually 10 or 12) monomeric nucleoproteins.49, 50 However, it is likely that the experimental conditions used here resulted in the dissociation of the polymerase nucleoprotein complex, resulting in the attachment of monomeric nucleoproteins to the surface. This hypothesis is supported by the fact that each individual aggregate shown in Figs. 6. d and 6. h has similar dimensions, consistent with monomeric nucleoprotein binding to the aptamer.
As a control experiment, we repeated the binding experiment by incubating the aptamer-spacer complex with the binding buffer solution (data not shown). After extensive washing, no change in the images of the aptamer-modified surface (Figs. 6. c and 6. g) could be seen. This negative blank experiment further supports the interpretation that the images seen in Figs 6. d and 6. h result from the binding of the influenza viral nucleoprotein to the aptamer-covered surface.
The use of a flat Ag surface to mimic the binding of the nucleoproteins to the aptamer-covered Ag nanorods is imperfect. Nevertheless, the images in Fig. 6 show that aptamer binding to nucleoprotein targets can be characterized using AFM, and that these AFM images support the conclusions obtained from the SERS spectra that influenza nucleoprotein binding occurs on substrates derivatized with the anti-influenza aptamer.
Conclusions
We report here the first evidence for the use of aptamer-modified SERS substrates as diagnostic tools for influenza virus detection in a complex biological matrix. In these studies we used a commercially prepared aptamer isolated against a commercial influenza vaccine. We showed that SERS provides a label-free optical method to detect the specific binding of the nucleoprotein target to the anti-influenza aptamer. This was demonstrated by the distinct spectral differences seen upon aptamer-target binding, while the negative control (RSV) showed a lack of any spectral changes associated with binding (Fig. 1). Statistical analysis of the data confirmed that the intrinsic SERS spectra were able to differentiate binding to the aptamer (Fig. 3). An additional negative control confirmed the lack of binding of the influenza nucleoproteins to a non-influenza aptamer (Figs. 4 and 5). AFM images showed changes consistent with the interpretation that the influenza nucleoproteins bind to the anti-influenza aptamer (Fig. 6).
Supplementary Material
Acknowledgments
The work described here was supported by the U.S. Army Research Laboratory (Cooperative Agreement # W911NF-07-R-001-04), as well as the National Institutes of Health (GM102546).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Supporting Information. 1) A schematic diagram illustrating the overall aptamer immobilization, nucleoprotein binding, and detection strategy (Figure S.1). 2) Additional multivariate analysis, PCA and PLS-DA, of the anti-influenza-nucleoprotein complex (Figures S.2 and S.3). 3) Additional multivariate analysis, PCA and PLS-DA, of the vasopressin aptamer-vasopressin complex (Figures S.4 and S.5). 4) Tables of the observed SERS peaks, including band assignments to particular vibrational modes, in the spectrum of the anti-influenza aptamer (Table S.1), the influenza nucleoprotein-aptamer complex (Table S.2), and the vasopressin aptamer-vasopressin complex (Table S.3).
References
- 1.Basler CF, Aguilar PV. Antivir Res. 2008;79:166–178. doi: 10.1016/j.antiviral.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y, Rambaut A. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 3.Watcharatanyatip K, Boonmoh S, Chaichoun K, Songserm T, Woratanti M, Dharakul T. J Virol Methods. 2010;163:238–243. doi: 10.1016/j.jviromet.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 4.Amano Y, Cheng Q. Anal Bioanal Chem. 2005;381:156–164. doi: 10.1007/s00216-004-2927-0. [DOI] [PubMed] [Google Scholar]
- 5.Uhlendorff J, Matrosovich T, Klenk HD, Matrosovich M. Arch Virol. 2009;154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 7.Gopinath SCB, Sakamaki Y, Kawasaki K, Kumar PKR. J Biochem. 2006;139:837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 8.Tang JJ, Yu T, Guo L, Xie JW, Shao NS, He ZK. Biosens Bioelectron. 2007;22:2456–2463. doi: 10.1016/j.bios.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 9.James W. J Gen Virol. 2007;88:351–364. doi: 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Yigit MV, Mazumdar D, Lu Y. Adv Drug Deliver Rev. 2010;62:592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kneipp K, Moskovits M, Kneipp H, editors. Surface-Enhanced Raman Scattering. Springer-Verlag; Berlin: 2006. [Google Scholar]
- 12.Huang E, Satjapipat M, Han SB, Zhou FM. Langmuir. 2001;17:1215–1224. [Google Scholar]
- 13.Ostatna V, Vaisocherova H, Homola J, Hianik T. Anal Bioanal Chem. 2008;391:1861–1869. doi: 10.1007/s00216-008-2133-6. [DOI] [PubMed] [Google Scholar]
- 14.Rowe AA, Miller EA, Plaxco KW. Anal Chem. 2010;82:7090–7095. doi: 10.1021/ac101491d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maquelin K, Choo-Smith L-P, Kirschner C, Ngo-Thi NA, Naumann D, Puppels GJ. In: Handbook of Vibrational Spectroscopy. Chalmers JM, Griffiths PR, editors. Vol. 5. John Wiley & Sons, Ltd; Chichester: 2002. pp. 3308–3334. [Google Scholar]
- 16.Naumann D. In: Biomedical Vibrational Spectroscopy. Lasch P, Kneipp J, editors. J. Wiley & Sons; Hoboken: 2008. pp. 1–8. [Google Scholar]
- 17.Suh JS, Moskovits M. J Am Chem Soc. 1986;108:4711–4718. [Google Scholar]
- 18.Bonham AJ, Braun G, Pavel I, Moskovits M, Reich NO. J Am Chem Soc. 2007;129:14572. doi: 10.1021/ja0767837. [DOI] [PubMed] [Google Scholar]
- 19.Sun L, Irudayaraj J. Biophys J. 2009;96:4709–4716. doi: 10.1016/j.bpj.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green M, Liu FM, Cohen L, Kollensperger P, Cass T. Faraday Discuss. 2006;132:269–280. doi: 10.1039/b506636k. [DOI] [PubMed] [Google Scholar]
- 21.Barhoumi A, Zhang D, Tam F, Halas NJ. J Am Chem Soc. 2008;130:5523–5529. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 22.Barhoumi A, Zhang D, Halas NJ. J Am Chem Soc. 2008;130:14040–14041. doi: 10.1021/ja804367c. [DOI] [PubMed] [Google Scholar]
- 23.Levin CS, Kundu J, Barhoumi A, Halas NJ. Analyst. 2009;134:1745–1750. doi: 10.1039/b909080k. [DOI] [PubMed] [Google Scholar]
- 24.Moody B, McCarty G. Anal Chem. 2009;81:2013–2016. doi: 10.1021/ac802247y. [DOI] [PubMed] [Google Scholar]
- 25.Neumann O, Zhang D, Tam F, Lal S, Wittung-Stafshede P, Halas NJ. Anal Chem. 2009;81:10002–10006. doi: 10.1021/ac901849k. [DOI] [PubMed] [Google Scholar]
- 26.Green M, Liu FM, Cohen L, Kollensperger P, Cass T. Faraday Discuss. 2006;132:269–280. doi: 10.1039/b506636k. [DOI] [PubMed] [Google Scholar]
- 27.Chen JW, Liu XP, Feng KJ, Liang Y, Jiang JH, Shen GL, Yu RQ. Biosens Bioelectron. 2008;24:66–71. doi: 10.1016/j.bios.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Chen JW, Jiang JH, Gao X, Liu GK, Shen GL, Yu RQ. Chem-Eur J. 2008;14:8374–8382. doi: 10.1002/chem.200701307. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Wei H, Li B, Ren W, Guo S, Dong S, Wang E. Chem Commun. 2007:5220–5222. doi: 10.1039/b709492b. [DOI] [PubMed] [Google Scholar]
- 30.Pagba CV, Lane SM, Cho H, Wachsmann-Hogiu S. J Biomed Opt. 2010;15:047006. doi: 10.1117/1.3465594. [DOI] [PubMed] [Google Scholar]
- 31.Pagba CV, Lane SM, Wachsmann-Hogiu S. J Raman Spectrosc. 2010;41:241–247. [Google Scholar]
- 32.Ochsenkuhn MA, Campbell CJ. Chem Comm. 2010;46:2799–2801. doi: 10.1039/b920941g. [DOI] [PubMed] [Google Scholar]
- 33.Negri P, Kage A, Nitsche A, Naumann D, Dluhy RA. Chem Comm. 2011;47:8635–8637. doi: 10.1039/c0cc05433j. [DOI] [PubMed] [Google Scholar]
- 34.Chaney SB, Shanmukh S, Zhao YP, Dluhy RA. Appl Phys Lett. 2005;87:31908–31910. [Google Scholar]
- 35.Nitsche A, Kurth A, Dunkhorst A, Panke O, Sielaff H, Junge W, Muth D, Scheller F, Stocklein W, Dahmen C, Pauli G, Kage A. BMC Biotechnol. 2007;7 doi: 10.1186/1472-6750-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hegner M, Wagner P, Semenza G. Surface Science. 1993;291:39–46. [Google Scholar]
- 37.Basu S, Jana S, Pande S, Pal T. J Coll Inter Sci. 2008;321:288–293. doi: 10.1016/j.jcis.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Kneipp K, Flemming J. J Mol Struct. 1986;145:173–179. [Google Scholar]
- 39.Rycenga M, McLellan JM, Xia YN. Chem Phys Lett. 2008;463:166–171. doi: 10.1016/j.cplett.2008.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Driskell JD, Shanmukh S, Liu Y, Chaney SB, Tang XJ, Zhao YP, Dluhy RA. J Phys Chem C. 2008;112:895–901. [Google Scholar]
- 41.Braun G, Lee SJ, Dante M, Nguyen TQ, Moskovits M, Reich N. J Am Chem Soc. 2007;129:6378. doi: 10.1021/ja070514z. [DOI] [PubMed] [Google Scholar]
- 42.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R, Viswanadham G. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 43.Stewart S, Fredericks PM. Spectrochim Acta A. 1999;55:1615–1640. [Google Scholar]
- 44.Podstawka E, Ozaki Y, Proniewicz LM. Appl Spectros. 2004;58:570–580. doi: 10.1366/000370204774103408. [DOI] [PubMed] [Google Scholar]
- 45.Simoes EA. Lancet. 1999;354:847. doi: 10.1016/S0140-6736(99)80040-3. [DOI] [PubMed] [Google Scholar]
- 46.Purschke WG, Eulberg D, Buchner K, Vonhoff S, Klussmann S. Proc Natl Acad Sci USA. 2006;103:5173–5178. doi: 10.1073/pnas.0509663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Culha M, Stokes D, Allain LR, Vo-Dinh T. Anal Chem. 2003;75:6196–6201. doi: 10.1021/ac0346003. [DOI] [PubMed] [Google Scholar]
- 48.Klumpp K, Ruigrok RWH, Baudin F. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin-Benito J, Area E, Ortega J, Llorca O, Valpuesta JM, Carrascosa JL, Ortin J. EMBO Reports. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruigrok RWH, Baudin F. J Gen Virol. 1995;76:1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.